Abstract
Results from genome-wide association studies (GWAS) have indicated that strong single-gene effects are the exception, not the rule, for most diseases. We assessed the joint effects of germline genetic variations through a pathway-based approach that considers the tissue-specific contexts of GWAS findings. From GWAS meta-analyses of lung cancer (12 160 cases/16 838 controls), breast cancer (15 748 cases/18 084 controls) and prostate cancer (14 160 cases/12 724 controls) in individuals of European ancestry, we determined the tissue-specific interaction networks of proteins expressed from genes that are likely to be affected by disease-associated variants. Reactome pathways exhibiting enrichment of proteins from each network were compared across the cancers. Our results show that pathways associated with all three cancers tend to be broad cellular processes required for growth and survival. Significant examples include the nerve growth factor (P = 7.86 × 10−33), epidermal growth factor (P = 1.18 × 10−31) and fibroblast growth factor (P = 2.47 × 10−31) signaling pathways. However, within these shared pathways, the genes that influence risk largely differ by cancer. Pathways found to be unique for a single cancer focus on more specific cellular functions, such as interleukin signaling in lung cancer (P = 1.69 × 10−15), apoptosis initiation by Bad in breast cancer (P = 3.14 × 10−9) and cellular responses to hypoxia in prostate cancer (P = 2.14 × 10−9). We present the largest comparative cross-cancer pathway analysis of GWAS to date. Our approach can also be applied to the study of inherited mechanisms underlying risk across multiple diseases in general.
Introduction
There are approximately 14 million new cancer cases and 8 million cancer-related deaths in the world each year (1). In the United States, cancer is the second most common cause of death, accounting for nearly one in every four deaths (2). Genome-wide association studies (GWAS) have significantly improved our understanding of cancers by uncovering novel associations of germline genetic variations known as single-nucleotide polymorphisms (SNPs) with disease risk in a high throughput and agnostic manner (3). GWAS have highlighted the polygenic nature of susceptibility as well as the importance of genetic loci that are not directly related to carcinogenesis (4). However, extending GWAS findings to mechanistic hypotheses about disease development and to clinical predictions has been an ongoing challenge for several reasons. Due to the need for multiple testing correction, the stringent requirements for cancer-associated SNPs to achieve acceptable significance (e.g. P < 5 × 10−8) overlook many potentially genuine signals (5). The significant SNPs discovered, along with the loci in which they are located, also usually offer little insight into disease biology without further investigation. In addition, most SNPs implicated in GWAS have small effect sizes, are low-penetrance, and poorly predict disease occurrence, so cancer-related GWAS findings have not yet been deemed medically actionable (6). Lastly, the products of genes do not function independently, but rather in concert as part of biologic pathways. Therefore it is essential to study the influence of genetic variations on diseases in terms of pathway effects (5).
Substantial progress has been made to overcome the aforementioned issues. Imputation and meta-analysis of multiple GWAS have helped identify SNPs that display weak effects and/or that are not commonly genotyped: rare variants of BRCA2 and CHEK2 affecting susceptibility to lung cancer (7), novel loci associated with hormone receptor status-specific subtypes of breast cancer (8), and a locus conferring modest risk for aggressive prostate cancer (9), were all detected by meta-analyses of many GWAS with imputed genotypes. Incorporation of tissue-specific data has benefitted GWAS analysis as well. For example, re-prioritization of GWAS findings guided by tissue-specific gene co-expression and protein interaction data has been shown to better predict disease genes in the Online Mendelian Inheritance in Man (OMIM) catalog compared with GWAS alone (10). Correlating GWAS results with RNA-seq expression from lymphoblastoid cell lines has demonstrated that protective variants linked to follicular lymphoma exert their effects through cis-regulation of HLA-DBQ1 expression (11). Heart quantitative interaction proteomics in combination with GWAS of long QT syndrome has also elucidated the functions of proteins that contribute to irregular heart rhythm (12).
Pathway analysis both improves the detection of risk variants with modest effect sizes and derives biologically meaningful results from GWAS. Pathway analysis considers whether a group of SNPs, affecting genes that encode functionally related proteins, is jointly associated with a trait (13). For example, groups of genetic loci were shown to collectively influence susceptibility to breast cancer (14), Crohn's disease (5), rheumatoid arthritis (15) and bipolar disorder (16), even when most of the individual loci do not exhibit genome-wide significance. Testing for associations at the pathway level is robust to genetic heterogeneity within study populations, as detection of the cumulative small effects of various germline signals in pathways is more reliable than detection of each individual small effect (17). For gene sets that represent the components of known cellular processes, conducting pathway analysis thus not only helps capture weak signals from GWAS, but also proposes mechanisms through which germline variations may affect disease risk.
Given the potential of pathway analysis to extract mechanistic insights from genomic data, there has been surging interest in developing integrative pathway analysis methods that leverage additional types of data to better understand the bridge between genotypes and phenotypes. Pathway analyses of GWAS coupled with gene expression datasets have identified new pathways associated with type 2 diabetes (18), cardiovascular disease (19,20), Alzheimer's disease (21) and a variety of immune-related disorders (22,23). Similarly, network-guided pathway analyses make use of results from focused and high-throughput experiments. Derived biomolecular interactions between pairs of gene products are portrayed as graphs, where nodes denote gene products and connecting edges denote their relationships. Topologic analyses of these networks, usually taking into account GWAS findings as attributes of the relevant nodes, have also been successful in implicating biologic pathways that mediate genetic risk for a variety of psychiatric (24–27) and immune-related disorders (28–30).
While GWAS of immune-related disorders are frequently studied by these integrative pathway analysis approaches (23,31), the majority of GWAS have yet to garner as much attention, including those involving cancer. A recent review of GWAS-based pathway analysis methods showed that most in fact do not take into account tissue-specific contexts (32). Since protein expression levels and interactions are well known to vary across different types of tissue (33,34), tests of association between biologic pathways and cancer should consider the relevant tissue context of the primary tumor development site. For example, suppose 10 proteins in a pathway are encoded by genes implicated in breast cancer GWAS. And suppose this finding indicates that there is a statistically significant association between the pathway and breast cancer. However, only six of these proteins are found to be appreciably expressed in the breast and to interact with other breast-expressed proteins. If such a finding (6 instead of 10 proteins) is no longer statistically significant, then the original pathway association is invalid, at least in breast tissue. Therefore association results from many previous cancer GWAS pathway analyses that map GWAS hits to pathways without regard for tissue-specific context (14,35–39) are liable to feature false positives. Nevertheless, it is certainly possible for germline variants to indirectly exert their influence on cancer development outside of the primary cancer tissue site. For example, mutation-driven alterations in synthesis, circulating bioavailability, and metabolism of endogenous hormones have been shown to modulate breast cancer risk (40). Here, we focus on the pathway-level effects of genetic variations in the primary cancer tissue site only, as such an approach is less likely to generate false positive results that are difficult to experimentally validate (41).
We applied a novel integrative pathway analysis method to GWAS of common cancers: lung cancer, breast cancer and prostate cancer. Each of the three GWAS meta-analyses used herein is among the largest reported with respect to its corresponding cancer among individuals of European ancestry. For pathways in the Reactome database, we computed statistical enrichment by proteins expressed in the tissue of tumor origin that are linked to disease-associated SNPs, and by the tissue-specific interaction partners of these proteins. Identified susceptibility pathways were also compared across the three cancers to highlight shared and unique pathways, as well as the overlapping and distinct gene members within shared pathways that influence risk for each cancer. This study is the largest comparative cross-cancer GWAS-based pathway analysis of its kind and takes into consideration tissue-specific context as well. In addition, it is the first to account for proteins that may affect disease risk through their interactions (42) with the protein products of GWAS-implicated genes.
Results
The three cancer GWAS meta-analyses used in this study consist of 12 160 lung cancer cases and 16 838 controls, 15 748 breast cancer cases and 18 084 controls, and 14 160 prostate cancer cases and 12 724 controls of European descent (Table 1). Top independently associated SNPs for each cancer were derived from the corresponding meta-analysis summary results through stepwise-selection conditional analysis (43). SNPs were mapped to genes within 10 kb to account for the majority of potential intragenic and transcription regulatory effects driven by variants (32,44). The products of these genes were further filtered for participation in tissue-specific protein interaction networks using the TissueNet database (45). From these networks, we retained for pathway analysis only the members encoded by genes likely to be influenced by independent cancer-associated SNPs (denoted ‘key proteins’ hereafter) and other proteins that interact with at least three key proteins (denoted ‘linking proteins’ hereafter). Key proteins and linking proteins (Supplementary Material, Figs S1‒S3) were then assessed for statistical enrichment of pathways in the Reactome database (46) using the hypergeometric test. The hypergeometric test reports the probability (a P-value) that a random list of proteins would better represent the proteins in a pathway compared with those in each cancer susceptibility interaction network (Table 2 and Fig. 1). We applied false discovery rate (FDR) adjustments (47) to the nominal pathway P-values to correct for evaluation of the network proteins against every Reactome pathway.
Table 1.
Component studies of the cancer GWAS meta-analyses
| Cases | Controls | |
|---|---|---|
| Lung cancer | ||
| MDACCa | 1150 | 1134 |
| UKICRb | 1952 | 5200 |
| IARCc | 2533 | 3791 |
| NCId | 5713 | 5736 |
| Torontoe | 331 | 499 |
| Germanyf | 481 | 478 |
| Totals | 12 160 | 16 838 |
| Breast cancer | ||
| BCACg | 8785 | 10 142 |
| BPC3h | 1998 | 2305 |
| TNBCCi | 1479 | 3180 |
| BCFRj | 3486 | 2457 |
| Totals | 15 748 | 18 084 |
| Prostate cancer | ||
| BPC3 | 2068 | 3011 |
| CRUK1k | 1854 | 1894 |
| CRUK2 | 3706 | 3884 |
| PEGASUSl | 4600 | 2941 |
| CAPS1m | 474 | 482 |
| CAPS2 | 1458 | 512 |
| Totals | 14 160 | 12 724 |
aMD Anderson Cancer Center.
bUnited Kingdom Institute of Cancer Research.
cInternational Agency for Research on Cancer.
dNational Cancer Institute.
eUniversity of Toronto and Lunenfeld-Tanenbaum Research Institute.
fHelmholtz-Gemeinschaft Deutscher Forschungszentren.
gBreast Cancer Association Consortium.
hBreast and Prostate Cancer Cohort Consortium.
iTriple Negative Breast Cancer Consortium.
jBreast Cancer Family Registry.
kCancer Research UK.
lProstate Cancer Genome-wide Association Study of Uncommon Susceptibility Loci.
mCancer of the Prostate in Sweden.
Table 2.
Descriptive statistics of pathway analysis workflow
| Lung cancer | Breast cancer | Prostate cancer | ||
|---|---|---|---|---|
| (A) | Total SNPs in GWAS meta-analysis | 8 945 877 | 7 728 735 | 9 760 429 |
| (B) | From (A), SNPs identified as independently associated with each cancer | 816 | 843 | 1157 |
| (C) | Genes within 10 kb of the SNPs from (B) | 784 | 795 | 1138 |
| (D) | Proteins expressed in the relevant tissue from genes in (C) | 168 | 163 | 231 |
| (E) | Other proteins expressed in the relevant tissue that interact with at least three proteins in (D) | 184 | 174 | 220 |
| (F) | All proteins from (D) + (E) in each tissue-specific cancer susceptibility interaction network | 352 | 337 | 451 |
| (G) | Tissue-specific PPIs among proteins in (F) | 844 | 766 | 1088 |
| (H) | Proteins from (F) that participate in at least one Reactome pathway found to be significant | 167 | 125 | 174 |
Figure 1.
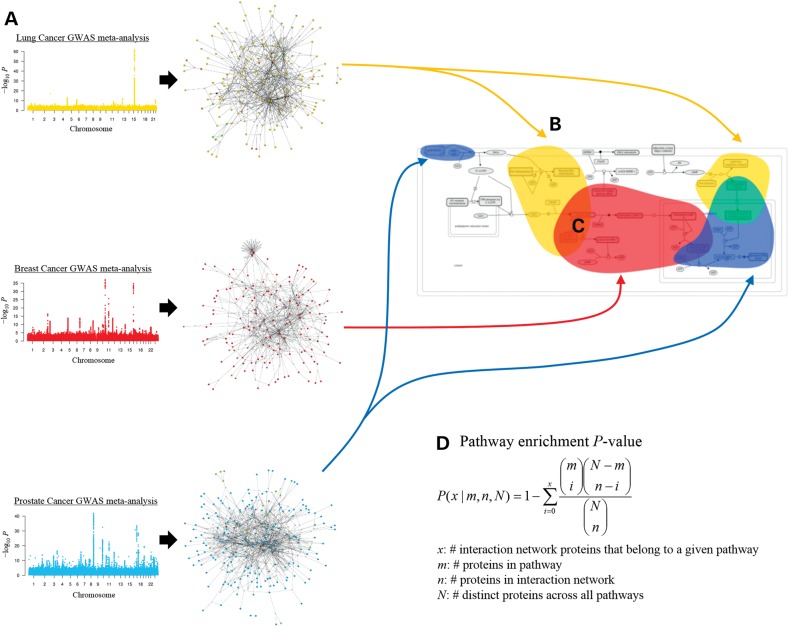
Schematic overview of study design. Lung cancer is used as an illustrative example. (A) The lung cancer susceptibility network was constructed from the lung-expressed products of genes that are located within 10 kb of independent lung cancer-associated SNPs (‘key proteins’) along with mutual tissue-specific interaction partners (‘linking proteins’). (B) This cartoon depicts protein participants in a pathway from the Reactome database. Key proteins for lung cancer are bubbled in yellow. (C) Key proteins for both lung cancer and breast cancer involved in the same pathway are bubbled in orange, the intersection of yellow and red. Shared and unique key proteins between other cancer pairs are portrayed analogously. (D) Formula for the hypergeometric pathway enrichment P-value.
Some key proteins possess far more known interacting partners than average (e.g. epidermal growth factor receptor and p53). Network topology studies have shown that in human protein interaction networks, about 10% of nodes (proteins) have 10+ fold greater degree compared with the average node degree (48). Key proteins with this property are more likely to have linking proteins that also fall into the same pathways. These pathways, as well as larger pathways in general (5,49,50), have a greater tendency to be significantly enriched relative to pathways that are smaller or involve proteins with fewer known interacting partners. We accounted for these two biases by comparing the number of proteins in each pathway that comes from the observed cancer susceptibility networks to the corresponding distribution of counts from networks generated by applying the same pathway analysis workflow to randomly chosen genes (‘randomization rank’, see Methods). For example, if a pathway contains 18 proteins on average from simulated null networks and 20 proteins from an observed cancer susceptibility network (which achieves only a 60th percentile rank in the former distribution), that pathway result would be discarded. Therefore the strength of association between a pathway and cancer has been not only computed in terms of statistical enrichment, but also referenced against a benchmark that is specific to the pathway.
Table 3 presents the significant pathways (P-value <0.05 and randomization rank percentile >95), and organizes them according to shared and unique associations with risk for lung cancer, breast cancer, and prostate cancer (see Supplementary Material, Tables S1‒S4 for details on individual-cancer pathway P-values, network proteins that participate in each pathway, and randomization ranks). Most proteins from the cancer susceptibility networks that participate in significant pathways are linking proteins. In addition, most of the proteins that belong to at least two cancer susceptibility networks and participate in a common pathway are also linking proteins. Pathways that affect risk for multiple cancers were found to be much more strongly associated with each cancer than pathways that are unique to a single cancer. These findings collectively suggest that the potential germline-based mechanisms most important to predisposing risk for cancers of the lung, breast, and prostate tend to be shared across the cancers rather than be unique to any single cancer; yet key proteins for the three cancers tend to be different, even if they contribute to the functions of the same overall pathway.
Table 3.
Results from pathway analysis
| Pathway name | Size | P-value | Enrichment |
Proteins in common |
||||
|---|---|---|---|---|---|---|---|---|
| LC | BC | PC | Key | Linking | ||||
| All three cancers | 1. NGF signaling via TRKA from the plasma membranea,* | 207 | 7.86 × 10−33 | 37 | 34 | 28 | 3 | 11 |
| 2. Signaling by EGFRa,* | 180 | 1.18 × 10−31 | 34 | 30 | 27 | 3 | 12 | |
| 3. Signaling by FGFRa,* | 167 | 2.47 × 10−31 | 29 | 32 | 26 | 3 | 11 | |
| 4. Signaling by PDGFa,* | 185 | 7.47 × 10−26 | 30 | 30 | 23 | 3 | 10 | |
| 5. DAP12 signalingb,* | 165 | 3.60 × 10−25 | 28 | 27 | 23 | 3 | 10 | |
| 6. Signaling by SCF-KITa,* | 144 | 2.46 × 10−24 | 29 | 24 | 18 | 2 | 8 | |
| 7. Platelet activation, signaling, and aggregation* | 221 | 5.12 × 10−24 | 34 | 20 | 35 | 1 | 12 | |
| 8. Fcgamma receptor (FCGR) dependent phagocytosisb,* | 132 | 1.78 × 10−20 | 23 | 17 | 24 | 2 | 10 | |
| 9. GRB2 events in ERBB2 signalinga,* | 30 | 4.79 × 10−16 | 11 | 9 | 9 | 2 | 5 | |
| 10. FCERI mediated MAPK activationa,* | 88 | 5.71 × 10−15 | 17 | 13 | 15 | 0 | 8 | |
| 11. Costimulation by the CD28 familyb,* | 75 | 1.56 × 10−14 | 16 | 12 | 13 | 0 | 6 | |
| 12. Constitutive Signaling by Aberrant PI3K in Cancera,* | 61 | 2.59 × 10−11 | 13 | 12 | 7 | 2 | 4 | |
| 13. Antigen activation of B cell receptor generates 2nd messengersb,* | 58 | 1.29 × 10−9 | 11 | 8 | 11 | 0 | 6 | |
| 14. Circadian Clock* | 63 | 6.46 × 10−9 | 9 | 11 | 10 | 0 | 6 | |
| 15. Repression of WNT target genesa,c,* | 14 | 3.41 × 10−8 | 4 | 5 | 6 | 0 | 3 | |
| 16. Megakaryocyte development and platelet production* | 125 | 7.89 × 10−7 | 14 | 11 | 14 | 0 | 8 | |
| LC and BC | 17. Signaling by ERBB2a,* | 164 | 5.62 × 10−24 | 30 | 29 | 1 | 13 | |
| 18. Toll Like Receptor 2 (TLR2) Cascadeb,* | 96 | 1.45 × 10−16 | 21 | 17 | 0 | 9 | ||
| 19. TRAF6 induction of NFkB and MAP kinases upon TLR activationa,b,* | 86 | 2.24 × 10−16 | 19 | 17 | 0 | 9 | ||
| 20. SHC1 events in ERBB2 signalinga,* | 32 | 2.31 × 10−15 | 13 | 10 | 1 | 6 | ||
| 21. Netrin-1 signalingc,* | 42 | 6.15 × 10−11 | 10 | 11 | 0 | 8 | ||
| 22. Role of LAT2/NTAL/LAB on calcium mobilizationb,* | 152 | 4.00 × 10−10 | 19 | 18 | 1 | 7 | ||
| 23. Translocation of GLUT4 to the plasma membrane | 60 | 6.61 × 10−9 | 11 | 11 | 0 | 9 | ||
| 24. MAPK targets/nuclear events mediated by MAP kinasesa,* | 30 | 1.03 × 10−8 | 7 | 9 | 0 | 3 | ||
| 25. Signaling by Insulin receptor* | 117 | 2.91 × 10−5 | 13 | 11 | 0 | 6 | ||
| LC and PC | 26. VEGFA-VEGFR2 Pathwaya,c,* | 107 | 1.08 × 10−26 | 28 | 28 | 0 | 18 | |
| 27. NOTCH1 Intracellular Domain Regulates Transcriptionc,* | 47 | 8.33 × 10−18 | 16 | 15 | 2 | 10 | ||
| 28. RHO GTPases Activate WASPs and WAVEsc,* | 34 | 2.27 × 10−11 | 10 | 11 | 0 | 8 | ||
| 29. TGF-beta receptor signaling activates SMADsb,* | 32 | 4.87 × 10−10 | 9 | 10 | 0 | 4 | ||
| 30. EPH-Ephrin signalingc,* | 94 | 1.81 × 10−9 | 16 | 14 | 0 | 6 | ||
| 31. Membrane Trafficking* | 197 | 5.10 × 10−9 | 21 | 23 | 0 | 13 | ||
| 32. Regulation of actin dynamics for phagocytic cup formation | 107 | 4.32 × 10−8 | 14 | 16 | 0 | 9 | ||
| 33. Cell surface interactions at the vascular wallc,* | 99 | 2.83 × 10−7 | 15 | 12 | 0 | 10 | ||
| 34. PCP/CE pathwayc,* | 91 | 3.65 × 10−7 | 11 | 15 | 2 | 3 | ||
| 35. RMTs methylate histone arginines* | 45 | 3.43 × 10−6 | 7 | 10 | 0 | 3 | ||
| 36. Interferon gamma signalingb,* | 92 | 3.05 × 10−5 | 13 | 9 | 0 | 4 | ||
| 37. Folding, assembly, and peptide loading of class I MHCb,* | 24 | 3.13 × 10−3 | 4 | 5 | 2 | 1 | ||
| BC and PC | 38. Activation of BH3-only proteinsd,* | 25 | 5.79 × 10−18 | 12 | 12 | 0 | 8 | |
| 39. VEGFR2 mediated cell proliferationa,c,* | 33 | 5.71 × 10−13 | 8 | 14 | 0 | 6 | ||
| 40. Oxidative Stress Induced Senescenced,* | 91 | 1.20 × 10−8 | 12 | 16 | 0 | 6 | ||
| 41. Apoptotic cleavage of cellular proteinsd,* | 38 | 2.22 × 10−7 | 8 | 9 | 0 | 5 | ||
| 42. PLCG1 events in ERBB2 signalinga,* | 35 | 2.98 × 10−6 | 7 | 8 | 1 | 4 | ||
| 43. Opioid signaling* | 81 | 6.31 × 10−6 | 10 | 12 | 0 | 4 | ||
| 44. RHO GTPases Activate Forminsc,* | 114 | 1.01 × 10−4 | 11 | 13 | 1 | 6 | ||
| 45. DAG and IP3 signaling* | 32 | 9.24 × 10−4 | 5 | 6 | 1 | 2 | ||
| LC only | 46. Signaling by Interleukinsb,* | 111 | 1.69 × 10−15 | 28 | ||||
| 47. Interleukin-3, 5 and GM-CSF signalingb,* | 45 | 1.21 × 10−12 | 17 | |||||
| 48. G2/M DNA damage checkpointd,* | 16 | 6.86 × 10−9 | 9 | |||||
| 49. Signaling to ERKsa,* | 44 | 1.26 × 10−8 | 13 | |||||
| 50. Integrin alphaIIb beta3 signaling | 27 | 9.56 × 10−8 | 10 | |||||
| 51. Cellular response to heat stress* | 96 | 7.65 × 10−7 | 16 | |||||
| 52. DCC mediated attractive signalingc,* | 14 | 1.26 × 10−6 | 7 | |||||
| 53. RHO GTPases activate PAKsc,* | 21 | 2.61 × 10−5 | 7 | |||||
| 54. Double-Strand Break Repaird,* | 23 | 4.87 × 10−5 | 7 | |||||
| 55. Activation of Raca,c,* | 14 | 4.04 × 10−4 | 5 | |||||
| 56. L1CAM interactionsc,* | 97 | 4.10 × 10−4 | 12 | |||||
| 57. Adherens junctions interactionsc,* | 30 | 1.23 × 10−2 | 5 | |||||
| 58. Interferon alpha/beta signalingb,* | 67 | 2.46 × 10−2 | 7 | |||||
| 59. Golgi Associated Vesicle Biogenesis* | 54 | 3.16 × 10−2 | 6 | |||||
| BC only | 60. Activation of BAD and translocation to mitochondriad,* | 15 | 3.14 × 10−9 | 9 | ||||
| 61. Misspliced GSK3beta mutants stabilize beta-catenina,c,* | 15 | 6.08 × 10−8 | 8 | |||||
| 62. Deactivation of the beta-catenin transactivating complexa,c,* | 42 | 3.15 × 10−6 | 10 | |||||
| 63. Protein folding* | 54 | 4.16 × 10−6 | 11 | |||||
| 64. Transcriptional activation of mitochondrial biogenesis* | 42 | 2.70 × 10−5 | 9 | |||||
| 65. Signaling by Leptin* | 26 | 7.02 × 10−5 | 7 | |||||
| 66. Mitotic Prophased,* | 106 | 1.65 × 10−3 | 11 | |||||
| 67. Assembly of the primary cilium* | 179 | 3.95 × 10−3 | 14 | |||||
| 68. Acetylcholine regulates insulin secretion* | 10 | 1.37 × 10−2 | 3 | |||||
| PC only | 69. Cellular response to hypoxia* | 25 | 2.14 × 10−9 | 12 | ||||
| 70. RHO GTPases activate PKNsd,* | 60 | 5.49 × 10−6 | 13 | |||||
| 71. Thrombin signaling through proteinase activated receptors* | 32 | 2.79 × 10−5 | 9 | |||||
| 72. CTLA4 inhibitory signalingb,* | 22 | 1.32 × 10−4 | 7 | |||||
| 73. Platelet sensitization by LDL | 17 | 2.77 × 10−4 | 6 | |||||
| 74. G alpha (z) signaling events* | 46 | 4.81 × 10−4 | 9 | |||||
| 75. ADP signaling through P2Y purinoceptor 1* | 25 | 2.28 × 10−3 | 6 | |||||
| 76. CDO in myogenesis | 29 | 4.93 × 10−3 | 6 | |||||
| 77. Sema4D in semaphorin signalingc,* | 27 | 1.86 × 10−2 | 5 | |||||
| 78. Signaling by Hippoa,c,* | 20 | 3.15 × 10−2 | 4 | |||||
| 79. Dectin-2 family | 11 | 3.59 × 10−2 | 3 | |||||
| 80. Glucagon signaling in metabolic regulation* | 33 | 3.67 × 10−2 | 5 | |||||
The hypergeometric test was used to compute the probability that randomly selected proteins would better represent each pathway in the Reactome database compared with proteins in the three cancer susceptibility networks. Size: total number of proteins in a pathway. P-value: the hypergeometric probabilities were adjusted to account for multiple comparisons across all Reactome pathways using the Benjamini–Hochberg FDR method; for a pathway implicated in at least two cancers, a combined P-value was reported following Fisher's method; the pathways presented herein possess adjusted P < 0.05 and randomization rank percentile >95. Enrichment: the number of proteins in each cancer susceptibility network that participates in a pathway. Proteins in Common: the number of key proteins or linking proteins from at least two cancer susceptibility networks that are involved in the same pathway. Pathway categories: acell growth and proliferation, bimmunologic signaling, ccell fate specification and migration, dregulation of cell cycle or cell death. *denotes pathway has been experimentally shown to influence tumor biology of the relevant cancer(s).
LC, lung cancer; BC, breast cancer; PC, prostate cancer.
As labeled in Table 3, the majority of implicated pathways fall under categories of cellular processes that are well-known to promote oncogenesis in a diversity of tissues. These categories describe cell growth and proliferation, immunologic signaling, cell fate specification and migration, and regulation of cell cycle or cell death. Lung cancer risk is mainly associated with immune-related and cell organization pathways (pathway #s 18, 19, 21, 22, 26‒30, 33, 34, 36, 37, 46, 47, 52, 53 and 55‒58 in Table 3), breast cancer risk with growth signaling and cell cycle regulation pathways (pathway #s 17, 19, 20, 24, 38‒42, 60‒62 and 66 in Table 3), and prostate cancer risk with cell organization and platelet-related pathways (pathway #s 26‒28, 30, 33, 34, 39, 44, 71, 73, 75, 77 and 78 in Table 3). In order to avoid misrepresenting cancer risk pathway trends due to the presence of redundant pathways, we removed Reactome pathways that have largely similar protein members (see Methods). These trends are consistent with cancer progression findings in the experimental literature (51–64).
Some key and linking proteins play roles in multiple susceptibility pathways for each cancer. Among pathways associated with lung cancer, breast cancer and prostate cancer, the most frequently observed key proteins transduce extracellular stimuli (genes EGFR, CHUK, ERBB4 and KIT), are involved in calcium-regulated kinase activity (genes PIK3R1, PRKCA, PRKCE and CAMK4) and facilitate signaling by heterotrimeric G proteins (genes ADCY8, GNG2, GNG7 and GNG12), respectively. In other words, many pathways that are associated with a given cancer contain a recurring set of the same key proteins (Table 4 and Supplementary Material, Table S5). In lung cancer for example, conserved helix-loop-helix ubiquitous kinase (CHUK) is a key protein component in a variety of pathways that perform different functions, such as growth factor signaling, inflammation mediation and regulation of leukocyte activity (pathway #s 1‒6, 17‒19, 22 and 46 in Supplementary Material, Table S4). This offers a valuable illustration that alterations in the function or abundance of a few genes have the potential to influence a wide array of biologic processes. In contrast to common key proteins, common linking proteins among implicated pathways exhibit significant overlap across the three cancers (genes MAPK1, PTPN11, SRC, FYN and GRB2). For proper count comparisons, we ensured that gene occurrences in pathways were not driven by SNPs mapping to multiple genes. Indeed, all key proteins from the lung cancer, breast cancer and prostate cancer susceptibility networks are encoded by genes that were mapped one-to-one from independently associated SNPs.
Table 4.
Genes encoding key proteins and linking proteins that participate in the most susceptibility pathways
| Lung cancer |
Breast cancer |
Prostate cancer |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Key | Pathways | Linking | Pathways | Key | Pathways | Linking | Pathways | Key | Pathways | Linking | Pathways |
| EGFR | 12 | GRB2 | 24 | PIK3R1 | 14 | MAPK1 | 21 | ADCY8 | 10 | MAPK1 | 17 |
| CHUK | 11 | PTPN11 | 24 | PRKCA | 14 | PTPN11 | 19 | PRKCE | 10 | SRC | 17 |
| ERBB4 | 11 | MAPK1 | 23 | ERBB4 | 11 | GRB2 | 17 | NRG1 | 8 | FYN | 16 |
| KIT | 9 | MAPK3 | 23 | IRS2 | 11 | YWHAB | 16 | GNG12 | 6 | GRB2 | 16 |
| TNRC6A | 8 | JAK2 | 21 | NRG1 | 11 | AKT1 | 14 | GNG2 | 6 | MAPK3 | 16 |
| PAK2 | 6 | FYN | 20 | FGFR2 | 10 | FYN | 14 | GNG7 | 6 | PTPN11 | 16 |
| BCAR1 | 4 | RAC1 | 19 | GAB1 | 10 | RAF1 | 14 | PLCG2 | 6 | RAC1 | 16 |
| DUSP3 | 4 | PIK3R1 | 18 | PRKCE | 10 | CALM1 | 13 | PPP2R5E | 4 | PRKCD | 15 |
| BLNK | 3 | SRC | 18 | CAMK4 | 9 | PLCG1 | 13 | WASF2 | 4 | PRKCA | 14 |
| BRAF | 3 | MAP2K1 | 17 | GSK3B | 9 | EGFR | 12 | ELMO1 | 3 | YWHAB | 14 |
Discussion
GWAS have been successful in identifying many genetic variants that are significantly associated with human diseases. However, a gap has emerged between the ability to detect these associations and the ability to meaningfully interpret their biologic significance. By incorporating protein interaction and pathway annotations in post-GWAS analysis, we sought to determine the likely mechanisms through which germline genetic variations confer risk for cancers of the lung, breast and prostate in a tissue-specific manner. We identified pathways that are statistically enriched with proteins expressed in the lung, breast and prostate from cancer GWAS-implicated genes, along with mutually interacting partner proteins in the respective tissues. These pathways were compared across the three cancers to highlight shared and unique findings. This study is the largest comparative cross-cancer GWAS-based pathway analysis to date. Furthermore, it is the first to consider the importance of not only the products of genes influenced by disease-associated variants (‘key proteins’), but also their tissue-specific interaction partners (‘linking proteins’).
Our network-guided approach was motivated by the fact that most disease phenotypes are rarely the consequence of a single genetic abnormality. In the complex interconnected network of biomolecules within cells, genetic variations not only impact the gene products whose activity and expression are directly under regulation, but also can spread their effects along links of the network to many other components (65). For example, a study combined GWAS with accurate models of immunologic signaling cascades to identify NF-κb as an important integrator of upstream genetic changes that increase risk for the activated B-cell subtype of diffuse large B-cell lymphoma (66,67). Subunits of NF-κb were not implicated by GWAS, but were shown to interact with and respond to changes in the products of GWAS hits. Similarly, GWAS have indicated that genetic risk loci associated with Alzheimer's disease are related to immune functions, synaptic transmission and lipid processing (68). A subsequent study of gene co-expression then demonstrated that a gene not directly tied to any risk-conferring variants, TYROBP, is a mutual regulator of these mechanisms and affects amyloid-β turnover and neuronal damage in microglia (21). Both discoveries may have been expedited had a preliminary bioinformatics analysis proposed disease pathways based on the involvement of GWAS hits and their interaction partners.
Nevertheless, pathway analysis is only as reliable as the pathways being studied. We chose the Reactome pathways for enrichment analysis because Reactome documents the interrelated biochemical reactions and transformations in which every pathway member participates (46). Another concern in integrative pathway analysis of GWAS data is compromise of the agnostic value of GWAS. While GWAS do not take into consideration any knowledge of diseases or genetic associations, and thus provide unbiased results, coupling GWAS results with databases of tissue-specific protein interactions and pathways may preferentially implicate some genes and their affiliated pathways over others. For example, if a particular gene expressed in a tissue is studied more often than other genes of the tissue, more of that gene product's interactions will be discovered. Pathways encompassing those interactions are then more likely to be detected. Larger pathways also have a greater chance of spuriously being enriched by a random list of proteins. We accounted for these two biases by computing pathway enrichment for tissue-specific protein interaction networks constructed from randomly sampled genes. Of all Reactome pathways found to be statistically enriched using the hypergeometric test, only those containing significantly more proteins from the observed networks compared with the distribution of protein counts from simulated null networks were deemed to exhibit association with cancer risk.
Shared pathways
Shared susceptibility pathways across all three cancers primarily consist of processes that mediate cell proliferation (pathway #s 1‒4, 6, 9, 10, 12, 15 in Table 3). Within these shared pathways, however, key proteins are mostly distinct for each cancer. Therefore in a given individual, germline-based pathway influences on oncogenesis are not the same in every tissue, despite identical genomes in all cells. Tissue-specific contexts, such as patterns of gene expression, protein interactions, and exposures, are likely to make the cancer predisposing effects of dysregulated growth signaling pathways more relevant in certain tissues than in others. Furthermore, even if a shared pathway has the potential to promote tumor growth in multiple tissues, the precise affected components of the pathway tend to differ by cancer tissue type.
Immunologic signaling is the other predominant category of shared pathways. Cancer cells have been shown to acquire enhanced evasion of immune surveillance. Improper secretion of transforming growth factor β (TGFβ), various interleukins and/or interferon γ (IFNγ) is believed to disrupt the antigen recognition, antigen presentation and stimulation events required for lymphocyte activation (69,70) (pathways #s 5, 11, 13, 29, 36, 37, 46 and 72 in Table 3). Aberrant immunologic signaling appears to have a greater relative importance in lung cancer predisposition. We found significantly more immune-related pathways to be associated with lung cancer than with breast cancer and prostate cancer. This observation supports the most common mechanism of lung carcinogenesis: inhaled compounds from tobacco smoking provoke inflammation and DNA damage in the lung. Therefore individuals, especially smokers, with the genetic risk of over-suppressing local immune surveillance in response to inflammation or inadequately halting cell cycle progression in the presence of biochemically altered DNA (pathway #s 18, 19, 48 and 54 in Table 3) are expected to have a higher chance of developing lung cancer compared with individuals without these genetic risk factors (71,72).
Unique pathways
Susceptibility pathways that have been implicated exclusively in lung cancer, breast cancer or prostate cancer usually have fewer member proteins and perform a narrower range of functions compared with the shared pathways. For example, the pathways found to be associated with specifically breast cancer mainly involve β-catenin, cell cycle progression and regulation of satiety (pathway #s 60‒62, 65‒66 and 68 in Table 2). Elevated leptin, which often characterizes a state of obesity and insulin resistance, has been shown to inhibit apoptosis and induce epithelial-mesenchymal transition in breast cancer cells by stabilizing β-catenin in the Wnt signaling pathway (73). The likely presence of such crosstalks between mechanisms of metabolic homeostasis and oncogenic signals is in line with the higher incidence, greater aggressiveness, and poorer prognosis of breast cancer observed in obese females (74).
An abundance of pathways related to platelet functions is associated with prostate cancer risk. We actually identified three platelet receptor pathways associated with prostate cancer risk (pathway #s 8, 71 and 75 in Table 2), against which pharmacologic blockades have demonstrated attenuation of protective aggregation and growth factor secretion by platelets around prostate cancer cells (75–77). More generally, the key proteins driving statistical enrichment of many pathways associated with prostate cancer are known to facilitate signaling by heterotrimeric G proteins (Supplementary Material, Table S3). This pathway-level insight has not been reported in previous bioinformatics analyses of GWAS. Overactive signaling by G protein-coupled receptors (GPCRs) synergistically enhances the tumorigenic effects of PTEN loss (78–80), the most frequently observed tumor-suppressor gene inactivation in prostate cancer. The only GPCR-related highlight from other GWAS is the implication of GPCR family C group 6 member A in three GWAS of prostate cancer among Asian men (81).
Overall, the majority of associated pathways (74 out of 80 total pathways in Table 3) exhibit some involvement in the development of at least one cancer type (51–64). Moreover, we identified associations between metabolism regulation and breast cancer and between platelet signaling and prostate cancer that are in agreement with results from ex vivo studies (75,76,78,80,81), but that have not been detected in previous cancer GWAS analyses. Although encountering matching pathway descriptions in the experimental literature for each cancer's progression hardly suffices as validation of the predicted risk-influencing pathways, these broad corroborations indicate that our findings still provide meaningful direction for new more specific studies. For example, genome editing using CRISPR-Cas9 recently elucidated the mechanism of gene expression alterations that repress white adipocyte browning and thermogenesis in favor of lipid storage caused by an obesity-predisposing variant (82). Guidance for this study design came from the suspected roles of adipocyte differentiation in obesity put forward by many previous investigations, including pathway analyses of obesity GWAS (83). Our network-based pathway analysis is useful for pointing out not only pathways to further pursue, but also the pathway components whose potential alterations may have a genetic basis or may be secondary to interactions with other such components.
Differences from prior studies
It is important to emphasize that just because a pathway was not identified to influence risk for a certain cancer by this analysis does not imply that the pathway plays no role in the cancer's development. For example, leptin signaling was found to be associated with only breast cancer. However, molecular studies have indicated that leptin also affects lung cancer and prostate cancer progression (84–86). These are not necessarily conflicting findings, as we sought to characterize using pathways the effects of germline related risk factors in the initiation of three cancers. To this end, we infer that leptin signaling is important to the earliest stages of breast cancer predisposition, whereas it is likely to become more relevant to lung cancer and prostate cancer later in the neoplastic transformation process. This temporal variation of pathway relevance in cancer cell evolution is described in a recent study of skin cancer (87).
There are also some pathways that were not detected in the present study, but have garnered experimental support and been implicated in other pathway analyses of cancer GWAS (14,35–39). For example, hormone influences were highlighted in previous pathway analyses of breast and prostate cancer GWAS (35,39). Indeed, the interplay among environment, diet and endocrine homeostasis is believed to significantly influence risk for breast cancer and prostate cancer (88,89). In our analysis, the susceptibility pathways found to be shared between breast cancer and prostate cancer are fewer and have a weaker unifying theme (pathway #s 38‒45 in Table 3) compared with those between the other cancer pairs. Hormone-related pathways are not featured altogether. This discrepancy may be attributed to a few reasons. First, the aforementioned GWAS-based pathway analyses mapped SNPs to genes within a much more liberal window (±50–500 kb, versus ±10 kb in the present study). Therefore more genes were used for pathway analysis per SNP implicated. More pathways would then have the opportunity to be statistically enriched compared with our method. In addition, those methods disregarded tissue specificity. Similar to using a wide SNP-gene mapping window, lacking a tissue-specific focus will lead to the detection of more false positives. However, it will also be more proficient in detecting potentially genuine multi-organ factors that affect disease risk. These certainly exemplify endocrine- and diet-driven effects. Lastly, a simple expansion of the list of pathways being assessed may offer insights beyond those presented here due to Reactome's peculiar pathway coverage. Of its 1675 pathways, fewer than 20 involve hormones and almost all of them are unrelated to the mechanisms of sex hormones (46).
Limitations and future directions
Our approach would be improved by implementing SNP-to-gene mapping that more accurately reflect intragenic and transcription regulatory effects. We deemed a gene to be affected by independent cancer-associated SNPs if the SNPs reside either inside the gene or within 10 kb of its boundaries. It has been estimated that 95% of all SNPs that regulate gene expression, also known as expression quantitative trait loci (eQTLs), are located within 20 kb of genes (32,44). Although applying a fixed mapping window for all SNP-gene pairs in all tissues does not reflect true biology, we adhered to this approximate guideline given its common use among existing GWAS-based pathway analysis methods and given the nascent status of reliable tissue-specific SNP-gene effect annotations at this time (90). More complete tissue-specific annotations would be able to quantify the strength of all SNP-gene relationships and capture far-reaching trans effects, such as alterations in transcription factor genes or chromatin organization, that are missed by regional window mapping altogether. Such eQTL datasets have long existed for leukocytes, owing to their ease in sampling from human subjects, and have facilitated many integrative, comparative GWAS-based pathway analyses of immune-related disorders (23,31). Due to difficulty obtaining tissues, eQTL results for normal tissues other than peripheral blood are just beginning to be publically released through the Genotype-Tissue Expression project (91). The insights from the present study, and potential future studies that utilize a similar pathway analysis workflow, will become more refined with the incorporation of more comprehensive tissue-specific eQTL findings.
Even so, leveraging tissue-specific eQTLs, gene expression patterns, and protein interactions in pathway analysis boosts the ability to detect overarching pathways that are relevant to a heterogeneous mix of cells within the tissue, but dilutes the ability to discern cell-specific associations. The microenvironment surrounding and supporting tumors is an ecosystem composed of many cell types, including fibroblasts, endothelial cells, adipocytes, and cells of the immune system (92). For example, reciprocal signaling between fibroblasts and mammary cells is critical for breast cancer progression and invasion (93), while the processing of inflammatory biomolecules by tumor cells and leukocytes is more important to lung carcinogenesis (94). However, GWAS-based pathway analyses alone cannot yet attribute susceptibility pathways to specific cells with high confidence. This pertains to cells of both the microenvironment and the main cancer lineage. Therefore collapsing complex cancers with multiple subtypes, such as cancers of the lung and breast, into single diseases also facilitates implicating overarching pathways across cancer subtypes, but not subtype-specific mechanisms. Pathway analyses of GWAS that integrate findings from tissue-specific profiles offer promising new insights into disease risk mechanisms. Nevertheless, the pursuit of progressively more cell-specific studies also prompts the need for more specific complementary datasets that require additional time and new technologies to be generated accurately.
Materials and Methods
Description of GWAS meta-analyses and component studies
Lung cancer
Fixed effects meta-analysis with inverse variance weighting was conducted for summary results from previously reported GWAS of 12 160 lung cancer cases and 16 838 controls of European ancestry. Subjects were from the MD Anderson Cancer Center lung cancer study (95); the UK lung cancer GWAS by the Institute of Cancer Research (96) which includes cases from the Genetic Lung Cancer Predisposition Study (GELCAPS) (97) and controls from the 1958 Birth Cohort (98); the IARC lung cancer GWAS (99) which includes the Central Europe GWAS (100); the National Cancer Institute (NCI) lung cancer GWAS which includes the Environment and Genetics in Lung Cancer Etiology (EAGLE) study (101) and the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial (102); the Toronto lung cancer GWAS by the University of Toronto and the Lunenfeld-Tanenbaum Research Institute (99); and the Helmholtz-Gemeinschaft Deutscher Forschungszentren (HGF) lung cancer GWAS (103). These data represent part of the Transdisciplinary Research in Cancer of the Lung (TRICL) project and are being uploaded to the Database of Genotypes and Phenotypes (dbGaP) for public availability to interested scientists.
Breast cancer
Summary statistics from a fixed-effects inverse variance weighted GWAS meta-analysis of 15 748 breast cancer cases and 18 084 controls of European ancestry were downloaded from the Genetic Associations and Mechanisms in Oncology (GAME-ON) Sharepoint website on 14 November 2013. Subjects were from studies by the Breast Cancer Association Consortium (BCAC) which consist of the Australian Breast Cancer Family Study (ABCFS) (104), the Amsterdam Breast Cancer Study (ABCS) (105), the Helsinki Breast Cancer Study (HBCS) (106), the British Breast Cancer Study (BBCS) (107), the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) study (108), the UK2 study (109), the Singapore and Sweden Breast Cancer (SASBAC) study (110), and the Mammary Carcinoma Risk Factor Investigation (MARIE) study (111); studies by the Breast and Prostate Cancer Cohort Consortium (BPC3) which consist of the American Cancer Society Cancer Prevention Study II (CPS-II) (112), the European Prospective Investigation Into Cancer and Nutrition (EPIC) study (113), the Multi-Ethnic Cohort (MEC) study (114), the Nurses’ Health Study (NHS) and NHSII (115), the Polish Breast Cancer Study (PBCS) (116), and the PLCO Cancer Screening Trial (117); studies by the Triple Negative Breast Cancer Consortium (TNBCC) (118–120); and the Breast Cancer Family Registry (BCFR) study (121). These data represent part of the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) project and are publically available.
Prostate cancer
Summary statistics from a fixed-effects inverse variance weighted GWAS meta-analysis of 14 160 prostate cancer cases and 12 724 controls of European ancestry were downloaded from the GAME-ON Sharepoint website on 29 July 2013. Subjects were from studies by BPC3 (122), Cancer Research UK (CRUK) (123,124), the Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility loci (PEGASUS) arm of the PLCO Cancer Screening Trial (125), and Cancer of the Prostate in Sweden (CAPS) (126). These data represent part of the Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) project and are publically available.
Description of GWAS with individual-level genotype data used for linkage disequilibrium estimation
All GWAS datasets have been filtered to exclude individuals with more than 10% missing genotypes, SNPs with minor allele frequency less than 5%, SNPs with genotyping rate less than 90%, and SNPs that fail the Hardy–Weinberg test at the 0.0001 significance level.
NCI EAGLE GWAS
EAGLE is a case-control study that was conducted to investigate the genetic and environmental determinants of lung cancer and smoking persistence in Italians (101). After data quality control, 501 658 SNPs (Illumina HumanHap550v3.0) in 3900 unrelated individuals consisting of 1923 lung cancer cases and 1977 controls were retained for further analysis.
Cancer Genetic Markers of Susceptibility breast cancer GWAS
The Cancer Genetic Markers of Susceptibility (CGEMS) breast cancer GWAS was conducted on postmenopausal women of European ancestry with invasive breast cancer and controls from NHS (127). After data quality control, 493 677 SNPs (Illumina HumanHap550v1.1) in 2287 unrelated females consisting of 1145 breast cancer cases and 1142 controls were retained for further analysis.
CGEMS prostate cancer GWAS
The CGEMS prostate cancer GWAS was conducted on prostate cancer patients and controls of European ancestry from the PLCO Cancer Screening Trial (128). After data quality control, 531 892 SNPs (Illumina HumanHap300v1.1 and HumanHap240Sv1.0) in 2293 unrelated males consisting of 1148 prostate cancer cases and 1145 controls were retained for further analysis.
Tissue-specific data
We obtained tissue-specific protein–protein interactions (PPIs) from the TissueNet database (45). TissueNet has assembled PPIs from the Biological General Repository for Interaction Datasets (BioGRID) (129), the Database of Interacting Proteins (DIP) (130), IntAct (131), and the Molecular INTeraction database (MINT) (132) as pairs of interacting proteins. These PPIs were experimentally detected by high throughput yeast two-hybrid tests and/or more focused studies, including co-immunoprecipitation, affinity chromatography, and affinity immuno-electrophoresis. The database then assigned PPIs to 16 major human tissues only if each partner of a PPI had passed at least one of the following tissue-specific thresholds: mRNA intensity value greater than 100 in BioGPS (133), positive immunohistochemistry expression value with a medium or high antibody reliability score in the Human Protein Atlas (HPA) (134), or RNA-seq measurement of at least 1 RPKM in Illumina Body Map 2.0 (135).
All tissue samples that contributed to TissueNet come from healthy individuals, which is ideal for this type of study. We are interested in the interplay among genes in originally non-tumor tissues that collectively contribute to an eventual cancer phenotype.
Bioinformatics analyses
To the GWAS meta-analysis summary results of each cancer, we applied the software Genome-wide Complex Trait Analysis (GCTA) (136) to perform approximate conditional analysis and determine independently associated SNPs through stepwise selection. This was done to limit the inclusion of false positive associations due to SNP correlations within individuals. In the absence of individual-level genotype data for the three large meta-analyses, GCTA estimated linkage disequilibrium (LD) structure (43) from the corresponding GWAS described above. Specifically, SNP LD of the lung cancer meta-analysis was represented by the NCI EAGLE GWAS, SNP LD of the breast cancer meta-analysis by the CGEMS breast cancer GWAS, and SNP LD of the prostate cancer meta-analysis by the CGEMS prostate cancer GWAS. The total individual-level genotype data evaluated by GCTA consist of the separate GWAS in addition to SNPs imputed from the 1000 Genomes Project (Phase 3 integrated release, October 2014) (137) with haplotype phasing by SHAPEIT (138) using IMPUTE2 v2.3.1 (139). The best-guess genotypes of imputed SNPs with information measure greater than 0.9 were converted to PLINK format (140), which is the required input format for GCTA, by the software fcGENE (141). SNPs with meta-analysis P-value less than 0.05 and conditional P-value less than 0.001 were mapped to genes in the National Center for Biotechnology Information (NCBI) database Build 38 using the R package NCBI2R (https://cran.r-project.org/web/packages/NCBI2R/index.html). A gene was considered to be affected by a SNP if the SNP is located within 10 kb upstream and downstream of the gene. This window was chosen to reflect potential SNP influences on both the structure (when SNP resides in the gene) and abundance (when SNP resides in a regulatory region near the gene) of transcription products.
For each set of genes to which independent association signals were mapped, we retained for further analysis only the genes encoding proteins predicted to be expressed in lung, breast, or prostate tissue and to interact with at least one other such protein. The TissueNet database (45) provided the reference for this filtering. Protein interaction networks were also constructed (Supplementary Material, Figs S1–S3) from TissueNet's tissue-specific datasets of PPI pairs and plotted using the R package qgraph (https://cran.r-project.org/web/packages/qgraph/index.html). These networks contain both ‘key proteins’ (products of genes that are likely to be affected by cancer-associated SNPs) and ‘linking proteins’ (proteins that interact with at least three key proteins). The inclusion of linking proteins is important because their interactions with key proteins, although not necessarily as part of any established pathway, may indirectly perturb the functions of a given pathway (42). Statistical enrichment of network proteins in pathways from the Reactome database (46) was assessed using the hypergeometric test; Reactome contained 7854 human proteins in 1675 pathways at the time of this analysis. The obtained nominal P-values were adjusted for FDR using the Benjamini–Hochberg method (47). Pathways that do not contain any key proteins of a cancer were omitted from consideration for that cancer, regardless of pathway enrichment significance due to linking proteins alone.
Pathways that are larger or involve key proteins with more interacting partners have a greater tendency to be enriched due to chance. We accounted for these two biases by randomly sampling 50 000 gene sets from NCBI Build 38 of size equal to the number of genes mapped from independently associated SNPs for each of the three cancers. Tissue-specific interaction networks were then created from the products of these genes following the same procedure above. For every Reactome pathway exhibiting an FDR-adjusted hypergeometric P-value (denoted simply ‘P-value’ hereafter) less than 0.05 with respect to a cancer, we compared the number of proteins from the observed network in the pathway against the null distribution of corresponding counts from networks generated by random gene selection. If the observed value ranks higher than the 95th percentile (‘Randomization Rank’ metric in Supplementary Material, Tables S1–S4), that pathway was deemed significantly associated with the cancer at hand. For each pathway associated with at least two cancers, we combined their separate P-values using Fisher's method (142) to produce overall P-values that facilitate sorting. We then highlighted shared and unique susceptibility pathways across the studied cancers (Table 3 and Supplementary Material, Table S4). Within shared pathways, distinct and overlapping key proteins and linking proteins from the cancer susceptibility interaction networks were also noted.
Reactome features many pathways with similar protein constituents. Some pathways are even entirely subsets of others. For pathways A and B with P-values less than 0.05, we discarded the less significant of the two pathways if their intersection represents greater than 80% of either A or B. Examples include ‘Constitutive signaling by aberrant PI3K in cancer’, ‘PI3K/AKT activation’, ‘PI-3K cascade:FGFR1’, ‘PI-3K cascade:FGFR2’, and ‘PI3K events in ERBB4 signaling’; only the first pathway has been retained in our results. We also removed uninformatively broad pathways. We identified such pathways as those for which Reactome does not illustrate pathway diagrams at the protein level in its Pathway Browser tool (46). Examples include ‘Apoptosis’ and ‘Cell–Cell communication’.
Supplementary Material
Funding
This work was supported by the National Cancer Institute (U19 CA148127, U19 CA148537, U19 CA148065), the National Science Foundation Graduate-K12 Fellowship in collaboration with Kimball Union Academy (DGE-0947790 to D.C.Q.), and an anonymous donor to the Geisel School of Medicine at Dartmouth.
Supplementary Material
Acknowledgements
The authors would like to thank all members of the Transdisciplinary Research in Cancer of the Lung (TRICL), the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE), and the Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) projects of the Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative. Their clinical data collection and bioinformatics contributions have made this study possible.
Conflict of Interest statement. None declared.
References
- 1.International Agency for Research on Cancer (2012) GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012.
- 2.American Cancer Society (2014) Cancer Facts & Figures 2014.
- 3.Figueiredo J.C., Stram D.O., Haiman C.A. (2014) The impact of GWAS findings on cancer etiology and prevention. Curr. Epidemiol. Rep., 1, 130–137. [Google Scholar]
- 4.Chung C.C., Magalhaes W.C., Gonzalez-Bosquet J., Chanock S.J. (2010) Genome-wide association studies in cancer—current and future directions. Carcinogenesis, 31, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K., Li M., Hakonarson H. (2010) Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet., 11, 843–854. [DOI] [PubMed] [Google Scholar]
- 6.Stadler Z.K., Thom P., Robson M.E., Weitzel J.N., Kauff N.D., Hurley K.E., Devlin V., Gold B., Klein R.J., Offit K. (2010) Genome-wide association studies of cancer. J. Clin. Oncol., 28, 4255–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., McKay J.D., Rafnar T., Wang Z., Timofeeva M.N., Broderick P., Zong X., Laplana M., Wei Y., Han Y. et al. (2014) Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet., 46, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddiq A., Couch F.J., Chen G.K., Lindstrom S., Eccles D., Millikan R.C., Michailidou K., Stram D.O., Beckmann L., Rhie S.K. et al. (2012) A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet., 21, 5373–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin Al Olama A., Kote-Jarai Z., Schumacher F.R., Wiklund F., Berndt S.I., Benlloch S., Giles G.G., Severi G., Neal D.E., Hamdy F.C. et al. (2013) A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet., 22, 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C. et al. (2015) Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet., 47, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conde L., Bracci P.M., Richardson R., Montgomery S.B., Skibola C.F. (2013) Integrating GWAS and expression data for functional characterization of disease-associated SNPs: an application to follicular lymphoma. Am. J. Hum. Genet., 92, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundby A., Rossin E.J., Steffensen A.B., Acha M.R., Newton-Cheh C., Pfeufer A., Lynch S.N., Olesen S.P., Brunak S. et al. , QT Interval International GWAS Consortium, (2014) Annotation of loci from genome-wide association studies using tissue-specific quantitative interaction proteomics. Nat. Methods, 11, 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia P., Zhao Z. (2014) Network-assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum. Genet., 133, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menashe I., Maeder D., Garcia-Closas M., Figueroa J.D., Bhattacharjee S., Rotunno M., Kraft P., Hunter D.J., Chanock S.J., Rosenberg P.S. et al. (2010) Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res., 70, 4453–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eleftherohorinou H., Hoggart C.J., Wright V.J., Levin M., Coin L.J. (2011) Pathway-driven gene stability selection of two rheumatoid arthritis GWAS identifies and validates new susceptibility genes in receptor mediated signalling pathways. Hum. Mol. Genet., 20, 3494–3506. [DOI] [PubMed] [Google Scholar]
- 16.Askland K., Read C., Moore J. (2009) Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet., 125, 63–79. [DOI] [PubMed] [Google Scholar]
- 17.Mooney M.A., Wilmot B. (2015) Gene set analysis: a step-by-step guide. Am. J. Med. Genet., 168, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong H., Yang X., Kaplan L.M., Molony C., Schadt E.E. (2010) Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet., 86, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C. et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol., 6, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makinen V.P., Civelek M., Meng Q., Zhang B., Zhu J., Levian C., Huan T., Segre A.V., Ghosh S., Vivar J. et al. (2014) Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet., 10, e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R. et al. (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell, 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunyavanich S., Schadt E.E., Himes B.E., Lasky-Su J., Qiu W., Lazarus R., Ziniti J.P., Cohain A., Linderman M., Torgerson D.G. et al. (2014) Integrated genome-wide association, coexpression network, and expression single nucleotide polymorphism analysis identifies novel pathway in allergic rhinitis. BMC Med. Genomics, 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkes M., Cortes A., van Heel D.A., Brown M.A. (2013) Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet., 14, 661–673. [DOI] [PubMed] [Google Scholar]
- 24.Pedroso I., Lourdusamy A., Rietschel M., Nothen M.M., Cichon S., McGuffin P., Al-Chalabi A., Barnes M.R., Breen G. (2012) Common genetic variants and gene-expression changes associated with bipolar disorder are over-represented in brain signaling pathway genes. Biol. Psychiatry, 72, 311–317. [DOI] [PubMed] [Google Scholar]
- 25.Jia P., Wang L., Fanous A.H., Pato C.N., Edwards T.L., Zhao Z., International Schizophrenia Consortium and (2012) Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS Comput. Biol., 8, e1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj T., Shulman J.M., Keenan B.T., Chibnik L.B., Evans D.A., Bennett D.A., Stranger B.E., De Jager P.L. (2012) Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am. J. Hum. Genet., 90, 720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Detera-Wadleigh S.D., Akula N. (2011) A systems approach to the biology of mood disorders through network analysis of candidate genes. Pharmacopsychiatry, 44, S35–S42. [DOI] [PubMed] [Google Scholar]
- 28.Baranzini S.E., Srinivasan R., Khankhanian P., Okuda D.T., Nelson S.J., Matthews P.M., Hauser S.L., Oksenberg J.R., Pelletier D. (2010) Genetic variation influences glutamate concentrations in brains of patients with multiple sclerosis. Brain, 133, 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossin E.J., Lage K., Raychaudhuri S., Xavier R.J., Tatar D., Benita Y., Cotsapas C., Daly M.J., International Inflammatory Bowel Disease Genetics Consortium, (2011) Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet., 7, e1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotsapas C., Voight B.F., Rossin E., Lage K., Neale B.M., Wallace C., Abecasis G.R., Barrett J.C., Behrens T., Cho J. et al. (2011) Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet., 7, e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menon R., Farina C. (2011) Shared molecular and functional frameworks among five complex human disorders: a comparative study on interactomes linked to susceptibility genes. PLoS One, 6, e18660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mooney M.A., Nigg J.T., McWeeney S.K., Wilmot B. (2014) Functional and genomic context in pathway analysis of GWAS data. Trends Genet., 30, 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Las Heras J.I., Meinke P., Batrakou D.G., Srsen V., Zuleger N., Kerr A.R., Schirmer E.C. (2013) Tissue specificity in the nuclear envelope supports its functional complexity. Nucleus, 4, 460–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M., King V., Lim W.K. (2013) Assembling cell context-specific gene sets: a case in cardiomyopathy. J. Integr. Bioinform., 10, 234. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y.H., Kim J.H., Song G.G. (2014) Genome-wide pathway analysis of breast cancer. Tumour Biol., 35, 7699–7705. [DOI] [PubMed] [Google Scholar]
- 36.Fehringer G., Liu G., Briollais L., Brennan P., Amos C.I., Spitz M.R., Bickeboller H., Wichmann H.E., Risch A., Hung R.J. (2012) Comparison of pathway analysis approaches using lung cancer GWAS data sets. PLoS One, 7, e31816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee D., Lee G.K., Yoon K.A., Lee J.S. (2013) Pathway-based analysis using genome-wide association data from a Korean non-small cell lung cancer study. PLoS One, 8, e65396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia P., Liu Y., Zhao Z. (2012) Integrative pathway analysis of genome-wide association studies and gene expression data in prostate cancer. BMC Syst. Biol., 6, S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Y.Z., Xu Z., Xu L.W., Yu P., Zhao Y., Xin H., Wu R., Tan S.J., Song Q., Wu J.P. et al. (2014) Pathway analysis of genome-wide association study on serum prostate-specific antigen levels. Gene, 551, 86–91. [DOI] [PubMed] [Google Scholar]
- 40.Martin A.M., Weber B.L. (2000) Genetic and hormonal risk factors in breast cancer. J. Natl. Cancer Inst., 92, 1126–1135. [DOI] [PubMed] [Google Scholar]
- 41.Edwards S.L., Beesley J., French J.D., Dunning A.M. (2013) Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet., 93, 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Agarwal P., Rajagopalan D. (2008) A global pathway crosstalk network. Bioinformatics, 24, 1442–1447. [DOI] [PubMed] [Google Scholar]
- 43.Yang J., Ferreira T., Morris A.P., Medland S.E. Genetic Investigation of ANthropometric Traits (GIANT) Consortium, DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W. et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veyrieras J.B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. (2008) High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet., 4, e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barshir R., Basha O., Eluk A., Smoly I.Y., Lan A., Yeger-Lotem E. (2013) The TissueNet database of human tissue protein-protein interactions. Nucleic Acids Res., 41, D841–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R. et al. (2014) The Reactome pathway knowledgebase. Nucleic Acids Res., 42, D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y., Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann. Stat., 29, 1165–1188. [Google Scholar]
- 48.Fadhal E., Gamieldien J., Mwambene E.C. (2014) Protein interaction networks as metric spaces: a novel perspective on distribution of hubs. BMC Syst. Biol., 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmans P., Green E.K., Pahwa J.S., Ferreira M.A., Purcell S.M., Sklar P., Owen M.J., O'Donovan M.C., Craddock N., Wellcome Trust Case-Control Consortium, (2009) Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet., 85, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramanan V.K., Shen L., Moore J.H., Saykin A.J. (2012) Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet., 28, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng L., Alexander R.E., Maclennan G.T., Cummings O.W., Montironi R., Lopez-Beltran A., Cramer H.M., Davidson D.D., Zhang S. (2012) Molecular pathology of lung cancer: key to personalized medicine. Mod. Pathol., 25, 347–369. [DOI] [PubMed] [Google Scholar]
- 52.Pao W., Girard N. (2011) New driver mutations in non-small-cell lung cancer. Lancet Oncol., 12, 175–180. [DOI] [PubMed] [Google Scholar]
- 53.Guedj M., Marisa L., de Reynies A., Orsetti B., Schiappa R., Bibeau F., MacGrogan G., Lerebours F., Finetti P., Longy M. et al. (2012) A refined molecular taxonomy of breast cancer. Oncogene, 31, 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eroles P., Bosch A., Perez-Fidalgo J.A., Lluch A. (2012) Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat. Rev., 38, 698–707. [DOI] [PubMed] [Google Scholar]
- 55.da Silva H.B., Amaral E.P., Nolasco E.L., de Victo N.C., Atique R., Jank C.C., Anschau V., Zerbini L.F., Correa R.G. (2013) Dissecting major signaling pathways throughout the development of prostate cancer. Prostate Cancer, 2013, 920612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molloy N.H., Read D.E., Gorman A.M. (2011) Nerve growth factor in cancer cell death and survival. Cancers, 3, 510–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polivka J. Jr., Janku F. (2014) Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol. Ther., 142, 164–175. [DOI] [PubMed] [Google Scholar]
- 58.Gordon M.D., Nusse R. (2006) Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem., 281, 22429–22433. [DOI] [PubMed] [Google Scholar]
- 59.Nasarre P., Potiron V., Drabkin H., Roche J. (2014) Guidance molecules in lung cancer. Cell Adh. Migr., 4, 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lachej N., Didžiapetrienė J., Kazbariene B., Kanopiene D., Jonusiene V. (2012) Association between Notch signaling pathway and cancer. Acta Medica Lithuanica, 19, 427–437. [Google Scholar]
- 61.Ouyang L., Shi Z., Zhao S., Wang F.T., Zhou T.T., Liu B., Bao J.K. (2012) Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif., 45, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciocca D.R., Calderwood S.K. (2005) Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones, 10, 86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lennon F.E., Moss J., Singleton P.A. (2012) The mu-opioid receptor in cancer progression: is there a direct effect? Anesthesiology, 116, 940–945. [DOI] [PubMed] [Google Scholar]
- 64.Wang M., Kaufman R.J. (2014) The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer, 14, 581–597. [DOI] [PubMed] [Google Scholar]
- 65.Barabasi A.L., Gulbahce N., Loscalzo J. (2011) Network medicine: a network-based approach to human disease. Nat. Rev. Genet., 12, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Compagno M., Lim W.K., Grunn A., Nandula S.V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A. et al. (2009) Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature, 459, 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenz G., Wright G., Dave S.S., Xiao W., Powell J., Zhao H., Xu W., Tan B., Goldschmidt N., Iqbal J. et al. (2008) Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med., 359, 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bettens K., Sleegers K., Van Broeckhoven C. (2013) Genetic insights in Alzheimer's disease. Lancet Neurol., 12, 92–104. [DOI] [PubMed] [Google Scholar]
- 69.Lippitz B.E. (2013) Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol., 14, e218–e228. [DOI] [PubMed] [Google Scholar]
- 70.Mocellin S., Marincola F.M., Young H.A. (2005) Interleukin-10 and the immune response against cancer: a counterpoint. J. Leukoc. Biol., 78, 1043–1051. [DOI] [PubMed] [Google Scholar]
- 71.Walser T., Cui X., Yanagawa J., Lee J.M., Heinrich E., Lee G., Sharma S., Dubinett S.M. (2008) Smoking and lung cancer: the role of inflammation. Proc. Am. Thorac. Soc., 5, 811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caramori G., Adcock I.M., Casolari P., Ito K., Jazrawi E., Tsaprouni L., Villetti G., Civelli M., Carnini C., Chung K.F. et al. (2011) Unbalanced oxidant-induced DNA damage and repair in COPD: a link towards lung cancer. Thorax, 66, 521–527. [DOI] [PubMed] [Google Scholar]
- 73.Yan D., Avtanski D., Saxena N.K., Sharma D. (2012) Leptin-induced epithelial-mesenchymal transition in breast cancer cells requires beta-catenin activation via Akt/GSK3- and MTA1/Wnt1 protein-dependent pathways. J. Biol. Chem., 287, 8598–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Battle M., Gillespie C., Quarshie A., Lanier V., Harmon T., Wilson K., Torroella-Kouri M., Gonzalez-Perez R.R. (2014) Obesity induced a leptin-Notch signaling axis in breast cancer. Int. J. Cancer, 134, 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitrugno A., Williams D., Kerrigan S.W., Moran N. (2014) A novel and essential role for FcgammaRIIa in cancer cell-induced platelet activation. Blood, 123, 249–260. [DOI] [PubMed] [Google Scholar]
- 76.Gao L., Smith R.S., Chen L.M., Chai K.X., Chao L., Chao J. (2010) Tissue kallikrein promotes prostate cancer cell migration and invasion via a protease-activated receptor-1-dependent signaling pathway. Biol. Chem., 391, 803–812. [DOI] [PubMed] [Google Scholar]
- 77.Choe K.S., Cowan J.E., Chan J.M., Carroll P.R., D'Amico A.V., Liauw S.L. (2012) Aspirin use and the risk of prostate cancer mortality in men treated with prostatectomy or radiotherapy. J. Clin. Oncol., 30, 3540–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu L.L., Sun C., Petrovics G., Makarem M., Furusato B., Zhang W., Sesterhenn I.A., McLeod D.G., Sun L., Moul J.W. et al. (2006) Quantitative expression profile of PSGR in prostate cancer. Prostate Cancer Prostatic Dis., 9, 56–61. [DOI] [PubMed] [Google Scholar]
- 79.Weng J., Wang J., Cai Y., Stafford L.J., Mitchell D., Ittmann M., Liu M. (2005) Increased expression of prostate-specific G-protein-coupled receptor in human prostate intraepithelial neoplasia and prostate cancers. Int. J. Cancer, 113, 811–818. [DOI] [PubMed] [Google Scholar]
- 80.Rodriguez M., Siwko S., Zeng L., Li J., Yi Z., Liu M. (2015) Prostate-specific G-protein-coupled receptor collaborates with loss of PTEN to promote prostate cancer progression. Oncogene (epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang M., Liu F., Hsing A.W., Wang X., Shao Q., Qi J., Ye Y., Wang Z., Chen H., Gao X. et al. (2012) Replication and cumulative effects of GWAS-identified genetic variations for prostate cancer in Asians: a case-control study in the ChinaPCa consortium. Carcinogenesis, 33, 356–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. et al. (2015) FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med., 373, 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Magi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E. et al. (2015) New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ribeiro R., Lopes C., Medeiros R. (2006) The link between obesity and prostate cancer: the leptin pathway and therapeutic perspectives. Prostate Cancer Prostatic Dis., 9, 19–24. [DOI] [PubMed] [Google Scholar]
- 85.Malli F., Papaioannou A.I., Gourgoulianis K.I., Daniil Z. (2010) The role of leptin in the respiratory system: an overview. Respir. Res., 11, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vansaun M.N. (2013) Molecular pathways: adiponectin and leptin signaling in cancer. Clin. Cancer Res., 19, 1926–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martincorena I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D.C., Fullam A., Alexandrov L.B., Tubio J.M. et al. (2015) High burden and pervasive positive selection of somatic mutations in normal human skin. Science, 348, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Risbridger G.P., Davis I.D., Birrell S.N., Tilley W.D. (2010) Breast and prostate cancer: more similar than different. Nat. Rev. Cancer, 10, 205–212. [DOI] [PubMed] [Google Scholar]
- 89.Grover P.L., Martin F.L. (2002) The initiation of breast and prostate cancer. Carcinogenesis, 23, 1095–1102. [DOI] [PubMed] [Google Scholar]
- 90.The GTEx Consortium (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.The GTEx Consortium (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gajewski T.F., Schreiber H., Fu Y.X. (2013) Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol., 14, 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mao Y., Keller E.T., Garfield D.H., Shen K., Wang J. (2013) Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev., 32, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinrich E.L., Walser T.C., Krysan K., Liclican E.L., Grant J.L., Rodriguez N.L., Dubinett S.M. (2012) The inflammatory tumor microenvironment, epithelial mesenchymal transition and lung carcinogenesis. Cancer Microenviron., 5, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amos C.I., Wu X., Broderick P., Gorlov I.P., Gu J., Eisen T., Dong Q., Zhang Q., Gu X., Vijayakrishnan J. et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y., Broderick P., Webb E., Wu X., Vijayakrishnan J., Matakidou A., Qureshi M., Dong Q., Gu X., Chen W.V. et al. (2008) Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eisen T., Matakidou A., Houlston R. and Gelcaps Consortium (2008) Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS). BMC Cancer, 8, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Power C., Elliott J. (2006) Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol., 35, 34–41. [DOI] [PubMed] [Google Scholar]
- 99.Hung R.J., McKay J.D., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P. et al. (2008) A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637. [DOI] [PubMed] [Google Scholar]
- 100.Scelo G., Constantinescu V., Csiki I., Zaridze D., Szeszenia-Dabrowska N., Rudnai P., Lissowska J., Fabianova E., Cassidy A., Slamova A. et al. (2004) Occupational exposure to vinyl chloride, acrylonitrile and styrene and lung cancer risk (Europe). Cancer Causes Control, 15, 445–452. [DOI] [PubMed] [Google Scholar]
- 101.Landi M.T., Consonni D., Rotunno M., Bergen A.W., Goldstein A.M., Lubin J.H., Goldin L., Alavanja M., Morgan G., Subar A.F. et al. (2008) Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health, 8, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hayes R.B., Sigurdson A., Moore L., Peters U., Huang W.Y., Pinsky P., Reding D., Gelmann E.P., Rothman N., Pfeiffer R.M. et al. (2005) Methods for etiologic and early marker investigations in the PLCO trial. Mutat. Res., 592, 147–154. [DOI] [PubMed] [Google Scholar]
- 103.Sauter W., Rosenberger A., Beckmann L., Kropp S., Mittelstrass K., Timofeeva M., Wolke G., Steinwachs A., Scheiner D., Meese E. et al. (2008) Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol. Biomarkers Prev., 17, 1127–1135. [DOI] [PubMed] [Google Scholar]
- 104.Dite G.S., Jenkins M.A., Southey M.C., Hocking J.S., Giles G.G., McCredie M.R., Venter D.J., Hopper J.L. (2003) Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J. Natl. Cancer Inst., 95, 448–457. [DOI] [PubMed] [Google Scholar]
- 105.Schmidt M.K., Tollenaar R.A., de Kemp S.R., Broeks A., Cornelisse C.J., Smit V.T., Peterse J.L., van Leeuwen F.E., Van't Veer L.J. (2007) Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J. Clin. Oncol., 25, 64–69. [DOI] [PubMed] [Google Scholar]
- 106.Fagerholm R., Hofstetter B., Tommiska J., Aaltonen K., Vrtel R., Syrjakoski K., Kallioniemi A., Kilpivaara O., Mannermaa A., Kosma V.M. et al. (2008) NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat. Genet., 40, 844–853. [DOI] [PubMed] [Google Scholar]
- 107.Fletcher O., Johnson N., Palles C., dos Santos Silva I., McCormack V., Whittaker J., Ashworth A., Peto J. (2006) Inconsistent association between the STK15 F31I genetic polymorphism and breast cancer risk. J. Natl. Cancer Inst., 98, 1014–1018. [DOI] [PubMed] [Google Scholar]
- 108.Frank B., Hemminki K., Wappenschmidt B., Meindl A., Klaes R., Schmutzler R.K., Bugert P., Untch M., Bartram C.R., Burwinkel B. (2006) Association of the CASP10 V410I variant with reduced familial breast cancer risk and interaction with the CASP8 D302H variant. Carcinogenesis, 27, 606–609. [DOI] [PubMed] [Google Scholar]
- 109.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S. et al. (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet., 42, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J., Humphreys K., Heikkinen T., Aittomaki K., Blomqvist C., Pharoah P.D., Dunning A.M., Ahmed S., Hooning M.J., Martens J.W. et al. (2011) A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res. Treat., 126, 717–727. [DOI] [PubMed] [Google Scholar]
- 111.Flesch-Janys D., Slanger T., Mutschelknauss E., Kropp S., Obi N., Vettorazzi E., Braendle W., Bastert G., Hentschel S., Berger J. et al. (2008) Risk of different histological types of postmenopausal breast cancer by type and regimen of menopausal hormone therapy. Int. J. Cancer, 123, 933–941. [DOI] [PubMed] [Google Scholar]
- 112.Calle E.E., Rodriguez C., Jacobs E.J., Almon M.L., Chao A., McCullough M.L., Feigelson H.S., Thun M.J. (2002) The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer, 94, 500–511. [DOI] [PubMed] [Google Scholar]
- 113.Riboli E., Hunt K.J., Slimani N., Ferrari P., Norat T., Fahey M., Charrondiere U.R., Hemon B., Casagrande C., Vignat J. et al. (2002) European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr., 5, 1113–1124. [DOI] [PubMed] [Google Scholar]
- 114.Kolonel L.N., Henderson B.E., Hankin J.H., Nomura A.M., Wilkens L.R., Pike M.C., Stram D.O., Monroe K.R., Earle M.E., Nagamine F.S. (2000) A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol., 151, 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Colditz G.A., Hankinson S.E. (2005) The Nurses’ Health Study: lifestyle and health among women. Nat. Rev. Cancer, 5, 388–396. [DOI] [PubMed] [Google Scholar]
- 116.Garcia-Closas M., Brinton L.A., Lissowska J., Chatterjee N., Peplonska B., Anderson W.F., Szeszenia-Dabrowska N., Bardin-Mikolajczak A., Zatonski W., Blair A. et al. (2006) Established breast cancer risk factors by clinically important tumour characteristics. Br. J. Cancer, 95, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hayes R.B., Reding D., Kopp W., Subar A.F., Bhat N., Rothman N., Caporaso N., Ziegler R.G., Johnson C.C., Weissfeld J.L. et al. (2000) Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control. Clin. Trials, 21, 349S–355S. [DOI] [PubMed] [Google Scholar]
- 118.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T. et al. (2010) A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet., 42, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C., Wang X., Ademuyiwa F., Ahmed S., Ambrosone C.B. et al. (2011) A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat. Genet., 43, 1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stevens K.N., Vachon C.M., Lee A.M., Slager S., Lesnick T., Olswold C., Fasching P.A., Miron P., Eccles D., Carpenter J.E. et al. (2011) Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer Res., 71, 6240–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.John E.M., Hopper J.L., Beck J.C., Knight J.A., Neuhausen S.L., Senie R.T., Ziogas A., Andrulis I.L., Anton-Culver H., Boyd N. et al. (2004) The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res., 6, R375–R389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schumacher F.R., Berndt S.I., Siddiq A., Jacobs K.B., Wang Z., Lindstrom S., Stevens V.L., Chen C., Mondul A.M., Travis R.C. et al. (2011) Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet., 20, 3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J. et al. (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet., 40, 316–321. [DOI] [PubMed] [Google Scholar]
- 124.Eeles R.A., Kote-Jarai Z., Al Olama A.A., Giles G.G., Guy M., Severi G., Muir K., Hopper J.L., Henderson B.E., Haiman C.A. et al. (2009) Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet., 41, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Andriole G.L., Crawford E.D., Grubb R.L. III, Buys S.S., Chia D., Church T.R., Fouad M.N., Gelmann E.P., Kvale P.A., Reding D.J. et al. (2009) Mortality results from a randomized prostate-cancer screening trial. N. Engl. J. Med., 360, 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Duggan D., Zheng S.L., Knowlton M., Benitez D., Dimitrov L., Wiklund F., Robbins C., Isaacs S.D., Cheng Y., Li G. et al. (2007) Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl. Cancer Inst., 99, 1836–1844. [DOI] [PubMed] [Google Scholar]
- 127.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A. et al. (2007) A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet., 39, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N. et al. (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet., 39, 645–649. [DOI] [PubMed] [Google Scholar]
- 129.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L. et al. (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res., 43, D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. (2004) The Database of Interacting Proteins: 2004 update. Nucleic Acids Res., 32, D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U. et al. (2012) The IntAct molecular interaction database in 2012. Nucleic Acids Res., 40, D841–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A.P., Santonico E. et al. (2012) MINT, the molecular interaction database: 2012 update. Nucleic Acids Res., 40, D857–D861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C.L., Haase J., Janes J., Huss J.W. III et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol., 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Berglund L., Bjorling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C.A., Persson A., Ottosson J., Wernerus H., Nilsson P. et al. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteomics, 7, 2019–2027. [DOI] [PubMed] [Google Scholar]
- 135.Petryszak R., Burdett T., Fiorelli B., Fonseca N.A., Gonzalez-Porta M., Hastings E., Huber W., Jupp S., Keays M., Kryvych N. et al. (2014) Expression Atlas update—a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res., 42, D926–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Delaneau O., Marchini J., and The 1000 Genomes Project Consortium (2014) Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun., 5, 3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Delaneau O., Marchini J., Zagury J.F. (2012) A linear complexity phasing method for thousands of genomes. Nat. Methods, 9, 179–181. [DOI] [PubMed] [Google Scholar]
- 139.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roshyara N.R., Scholz M. (2014) fcGENE: a versatile tool for processing and transforming SNP datasets. PloS One, 9, e97589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fisher R.A. (1925) Statistical Methods for Research Workers. Oliver and Boyd, Edinburgh, UK. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.