Abstract
Acetylcholinesterase (AChE) that has been covalently inhibited by organophosphate compounds (OPCs), such as nerve agents and pesticides, has traditionally been reactivated by using nucleophilic oximes. There is, however, a clearly recognized need for new classes of compounds with the ability to reactivate inhibited AChE with improved in vivo efficacy. Here we describe our discovery of new functional groups—Mannich phenols and general bases—that are capable of reactivating OPC-inhibited AChE more efficiently than standard oximes and we describe the cooperative mechanism by which these functionalities are delivered to the active site. These discoveries, supported by preliminary in vivo results and crystallographic data, significantly broaden the available approaches for reactivation of AChE.
Keywords: drug discovery, high-throughput screening, medicinal chemistry, neurological agents, structure–activity relationships
Introduction
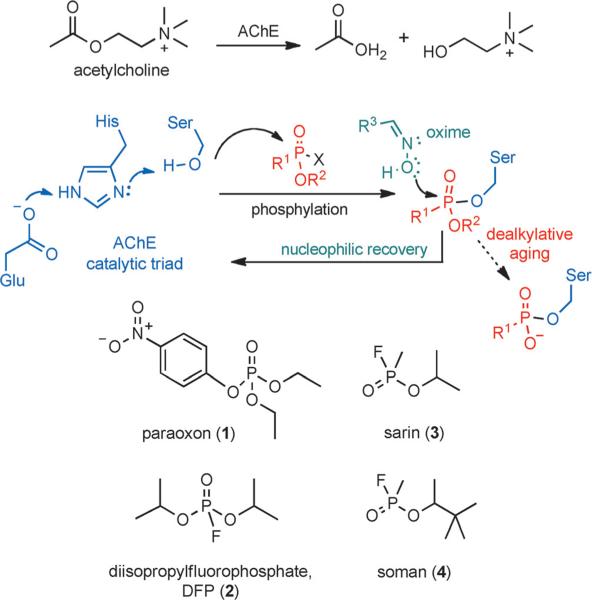
Acetylcholinesterase (AChE), a key enzyme in regulation of neurotransmission, catalyzes the hydrolysis of the neurotransmitter acetylcholine (Scheme 1A). The enzyme is vulnerable to organophosphate compounds (OPCs), which readily phosphylate (“phosphylate” denotes both “phosphorylate” and “phosphonylate”) the highly nucleophilic hydroxy group of the serine residue of the catalytic triad of AChE (Scheme 1B).[1] OPCs are commonly used as pesticides or military-grade nerve agents. Shown in Scheme 1C are the common pesticides paraoxon (1, the toxic metabolite of the pesticide parathion) and diisopropylfluorophosphate (DFP, 2), and the nerve agents sarin (3) and soman (4). The overwhelming formation of OPC· AChE adducts in vivo triggers a cholinergic crisis, resulting from the accumulation of acetylcholine and overstimulation of receptors in synapses.
Scheme 1.
AChE activity, inhibition, and reactivation. A) AChE functions to hydrolyze the neurotransmitter acetylcholine. B) The catalytic triad of the active site of AChE is vulnerable to inhibition by organophosphates. C) Examples of common organophosphates used as active principles in pesticides and nerve agents that covalently modify the serine residue in the AChE active site.
Whereas the threat of a nerve agent attack on the civilian population has remained largely theoretical, the broad use of the pesticides in agriculture leads to a large number of cases of pesticide poisoning[2] in the developing world. A lack of accurate reporting makes the data on the actual number of fatalities due to OPC toxicity difficult to determine,[3] but the World Health Organization (WHO) recognizes this as a significant problem that needs to be addressed.[4] Thus, there is a combination of a well-recognized need to expand treatment options for pesticide poisoning[3] and an interest in introducing therapies that would be effective in the case of mass civilian exposure.
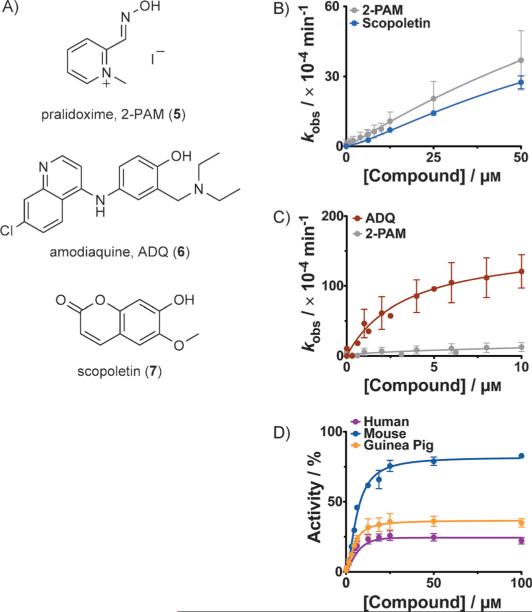
Currently, the only available treatment is administration of atropine (to block cholinergic receptors) in combination with an oxime to reverse the inhibition by nucleophilic reactivation (Scheme 1B).[5] Nucleophilic reactivation was first proposed over sixty years ago by Wilson[6] and is based on the transfer of the phosphylated group from OPC·AChE adducts to oxygen in the oxime. The archetypical oxime—pralidoxime (5, 2-PAM, Figure 1A)[6b]—is currently used for treatment in the United States and is actually the minimal structure that can act both as a nucleophile and as an anchoring group to position the oxime within the active site. The positively charged pyridinium component of 5 interacts with the anionic binding site (Trp86, amino acid numbering given for human enzyme)[7] and positions the oxime for nucleophilic attack, with maximum activity achieved at concentrations that are in the high micromolar to millimolar range.
Figure 1.
Newly identified reactivators of AChE. A) Structures of the traditional oxime reactivator pralidoxime (5) and of the recently identified hits from our screening assay: ADQ (6), and scopoletin (7). B), C) Concentration dependence of reactivation of paraoxon-inhibited (1-inhibited) human AChE: B) by scopoletin (7), or C) by ADQ (6). D) Concentration dependence of reactivation of 1-inhibited human (purple), mouse (blue), or guinea pig (orange) forms of AChE at increasing ADQ (6) concentrations. Reactivation was allowed for 3 h at room temperature. All reactions were repeated in triplicate or in duplicate on multiple days. Errors bars show the standard deviations.
The affinity of 5 can be improved by anchoring it to the peripheral anionic site (PAS);[8] this has led to the design of advanced oximes that are efficient at low micromolar concentrations with more sterically hindered nerve agents.[9] However, these are associated with more severe side effects, and are also stronger inhibitors of native AChE. Recent studies on ways to improve delivery to the central nervous system (CNS) have focused on the development of alternative uncharged species.[10]
Although many oximes worked well in animal models,[11] clear benefits of their use could not be established in controlled clinical trials of pesticide poisoning.[12] Further limitations of oxime-based therapy, such as toxicity of intermediates, potentially dangerous inhibition of the remaining native AChE, inability to be used against all types of nerve agents, various pharmacokinetic and distribution issues, and the inability to reactivate “aged” adducts (Scheme 1B), have been widely recognized.[5a,13] In order to expand the panel of functionalities available for reactivation beyond this single class, we undertook a combination of extensive screening and medicinal chemistry to identify new functional groups.
Results and Discussion
Screening result—Amodiaquine reactivates inhibited AChE
Initially, we screened a library containing 2000 bio-active compounds and approved drugs (for details of the screening platform see Figure S1 in the Supporting Information) and identified two compounds: the antimalarial drug amodiaquine (ADQ, 6, Figure 1A) and scopoletin (7). Scopoletin had only slightly lower activity than pralidoxime (5) in recovering paraoxon-inhibited AChE (Figure 1B); however, it did not reactivate DFP-inhibited AChE (Figure S2), and therefore we focused on 6, which was significantly more active. Reactivation rate constants for 6 were consistently faster than with 5 (Figure 1C and Figure S3) and it facilitated the recovery of human, mouse, and guinea pig isoforms of 1-inhibited AChE (Figure 1D). See the Supporting Information for details on experimental determination of kinetic rate constants: kinetic parameters are listed in Table S1, and controls, showing negligible background hydrolysis of substrate due to just the compound under identical reaction conditions, can be found in Figure S4.
Mannich phenol is a minimal reactivating functionality in ADQ (6)
Although preliminary in vivo studies with ADQ (6) were encouraging (Figure S5), we did not pursue its further development because it was active at doses that would be considered unsafe for broad prophylaxis, due to reported cases of hepatotoxicity and agranulocytosis,[14] as a result of p-aminoquinone metabolite formation in liver and white blood cells.[15]
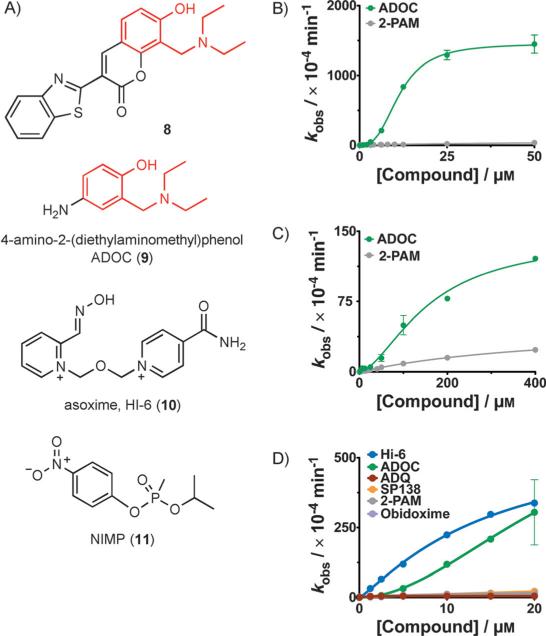
Instead, we decided to investigate further which part of the molecule was responsible for reactivation. We noticed during our continued screening that other hits had a common functionality with ADQ (6): 2-(diethylaminomethyl)phenol (Mannich phenol), which was attached to different hydrophobic groups (see compound 8 in Figure 2 and Figure S6 for the related hits 8a and 8b). Thus, we explored the role of Mannich phenols in reactivation, by testing various analogues (Figure S7, compounds 8c–q), discovering in the process that 4-amino-2-(diethylaminomethyl)phenol (ADOC, 9), which is ADQ without the hydrophobic anchor, reactivated OPC-inhibited AChE (Figures 2 and S8), thus indicating that this group is responsible for reactivation. ADOC (9) was also of particular interest because it lacked the hydrophobic anchor that is responsible for the toxic effects (agranulocytosis) on white blood cells, and thus it was likely to be less toxic in vivo (similarly to paracetamol).
Figure 2.
Additional investigation into reactivation by these compounds. A) Structures are shown for: compound 8, an example of a compound containing a Mannich phenol that was identified to have significant reactivation activity, ADOC (9), the minimal functional group that is highly active, HI-6 (10), the oxime currently used as a therapeutic in Europe, and NIMP (11), a safer nerve agent analogue that leaves the same adduct as sarin (3) in the AChE active site. B), C) Concentration dependence of reactivation by 9 of B) 1-inhibited, or C) 2-inhibited human AChE. Reactivation by 9 is much faster than reactivation by pralidoxime (5, gray) and is best fit by a sigmoidal curve, indicative of cooperativity. D) Reactivation of the sterically hindered adducts left by 11 on huAChE. ADOC (9, green) is much more efficient than standard oximes pralidoxime (5, gray) and obidoxime (purple) and reaches the activity of HI-6 (10, blue).
Upon further characterization, we determined that only ADOC (9), but neither of its positional isomers with the amino group in the 5- or 6-positions (compounds 9a and 9b in Figure S9), was capable of reactivating 1-inhibited enzyme. In addition, we tested an analogue with the hydroxy functionality blocked by a methyl group (compound 9c, Figure S9) and found that it was inactive.
The following immediately stood out upon full kinetic characterization of reactivation by ADOC (9):
-
1)
The apparent rate constants (kobs) were exceptionally high in comparison with pralidoxime (5, Figure 2B) at concentrations above ≈10 μm until they reached saturation. At saturation, the maximum rate constant for reactivation (k2) of 1-inhibited human AChE by ADOC (9, ≈0.14 min−1) was similar to the reported k2 value for 5 (≈0.27 min−1).[16]
-
2)
A much lower concentration was needed in order to reach k2 than for 5. The KR value of 1-inhibited human AChE was ≈ (11±9.1) μm for 9 whereas the reported KR for 5 was 1.8 mm.[16] Because much lower concentrations of these compounds are needed for maximum activity, this might translate into a lower therapeutic dose.[17]
-
3)
The apparent rate constant versus concentration curve indicated a high degree of cooperativity; that is, there was a sharp increase in apparent rate constant with only a small increase in concentration of compound, thus indicating that more than one molecule is needed to interact with the enzyme in order to achieve maximum activity. The concentration dependence curve was best fit to a sigmoidal curve with a Hill coefficient indicative of cooperativity (for human AChE inhibited by 1, h=2.8±0.23, or by 2, h=1.6 0.1, Figure 2B, C). See Figure S10 for reactivation of mouse AChE.
This raises the possibility that more than one molecule of ADOC (9) could be positioned within the active site by hydrophobic groups in a way that is analogous to the anchoring of the minimal nucleophilic group of pralidoxime (5) in advanced reactivators that are dimeric or pseudodimeric, such as asoxime (HI-6, 10, see Figure 2, currently recommended instead of 5 in Europe).
We observed that the Ki value for inhibition of free AChE [≈(30±12) μm, Figure S11] was similar to the half-maximal concentration of reactivation (KR) with 1 [≈(11±9.1) μm, Figure 2B], which can be explained by a model in which binding by reactivator is not blocked by the paraoxon adduct on the serine residue. We note that we have observed significant, irreversible inhibition of AChE at concentrations above 500 μm, possibly due to either the induction of conformational changes in AChE or more drastic denaturation.
In addition, we characterized the ability of ADOC (9) to facilitate the recovery of AChE from adducts with highly sterically hindered organophosphates, such as 2 (Figure 2C) and 11[18] (Figure 2D). NIMP (11) is safer to handle than sarin (3),[19] yet it creates the same adduct with AChE. ADOC (9) was able to reactivate the adducts at lower concentrations than pralidoxime (5) and with rate constants that were much faster than those determined for most of the best available oximes, with the maximum rate constant for reactivation of 11-inhibited AChE approaching that of 10. We also synthesized and tested the reactivation of AChE inhibited by SIMP (11a), an analogue of soman (4),[20] and found similar results (Figure S12).
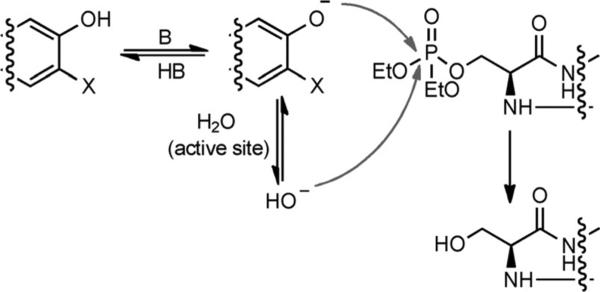
The next question would be, “By what mechanism do these phenols reactivate AChE?”. The suggestion that the mechanism is nucleophilic, similar to that of pralidoxime (5), is tempting, and there is precedence for a proposal that such a mechanism would be possible.[21] The abilities of various phenols to displace fluoride from nerve agents in solution were tested in early attempts to expand reactivation beyond oximes to other nucleophiles, but without subsequent reports of their practical use in reactivating inhibited AChE.[21] Also, nitrophenols[22] and coumarins[23] have been used as either colorimeteric or fluorogenic leaving groups in safer phosphylating agents to mimic nerve agents.[23–24] Thus, it is reasonable to suggest that certain phenols, when present in excess, could push the reaction in the reverse direction: that is, towards reactivation (Scheme 2). Phenols that are used as good leaving groups in phosphylation have significantly lower pKa values (≈7) than the phenol functionalities of 6 and 7 (pKa=8–9,[25] which are more similar to oximes).[26]
Scheme 2.
Two possible mechanisms for reactivation: either nucleophilic regeneration or through the generation of a hydroxy anion by deprotonation of water.
Yet, none of the experiments we performed, including the pH-profile of reactivation (Figure S13), precludes the possibility that the mechanism is the delivery of an external base to the active site of inhibited AChE, instead of a nucleophilic attack. In fact, the additional studies we describe next further supported this possibility.
Basic functionalities reactivate inhibited AChE
Knowing that ADOC was sufficient for reactivation, we expected choloroquine (CQ, 12), an analogue of 6 without a Mannich phenol moiety (Figure 3), to be inactive and attempted to use it as a negative control. To our surprise, our “negative control” also reproducibly reactivated human AChE inhibited by either 1 or 2 (Figure 3A, B and Figure S14). Although 12 is much less toxic than 6, it is still used prophylactically for malaria with minimal side effects;[27] it is reported to distribute to brain tissues,[28] achieving concentrations sufficient for reactivation, on the basis of our in vitro results. However, it was not efficient in reactivating inhibited enzyme from other species (Figure S14). Thus, it would be difficult to establish a reliable animal model for in vivo studies.
Figure 3.
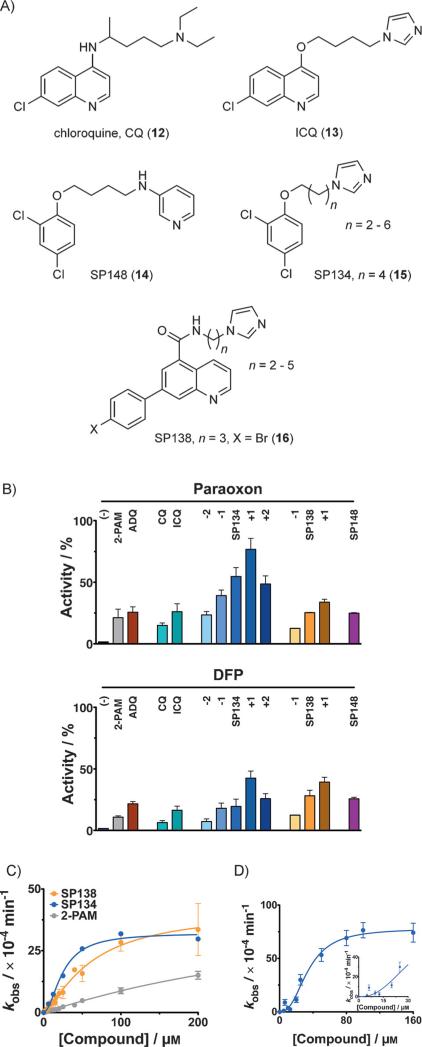
Reactivation of AChE with analogues of 6. A) ADQ (6) was the starting point for optimization, leading to chloroquine (CQ, 12) and ICQ (13), and then the SP series [e.g., SP148 (14), SP134 (15), and SP138 (16)]. B) Reactivation by different analogues of 6 (12.5 μm each) of 1-inhibited (left) and 2-inhibited (right) human AChE. Analogues of 15 and 16 with different linker lengths were synthesized; the difference in the number of carbon atoms (n) in the linker is indicated as +1, +2, etc. Activity is expressed relative to uninhibited enzyme. C) Concentration dependence curve for the reactivation of human AChE inhibited by 2—variously with 15 (blue), 16 (orange), or pralidoxime (5, 2-PAM, gray), with 15 and 16 being more efficient. D) Kinetics of reactivation by 15 of 2-inhibited mouse AChE were best fit to a sigmoidal curve with a Hill coefficient of ≈(2.5±0.31), agreeing with the crystal structure in Figure 4. Inset shows narrow range of the titration curve. All reactivation reactions were done in triplicate or in duplicate on multiple days (minimum of quadruplicate). Error bars show standard deviations. Full kinetic parameters can be found in Table S1.
The activity of CQ (12) indicated that the 9-chloroquinoline ring might have a general role in the delivery of reactivation functionalities into the AChE active site, and that the low activity of 12 might be addressed by substituting the diethylamino group with other bases. Indeed, a chloroquine analogue with an N-alkylimidazole group—ICQ (13), readily synthesized in a two-step procedure from commercial starting materials (Supporting Information)—was a significantly more efficient reactivator than 12 (Figure 3B). This result inspired us to continue our screening and structure–activity relationship (SAR) studies, during which we confirmed reactivation activity with additional compounds containing similar pharmacophoric features: that is, a hydrophobic group connected to a base through linkers of variable lengths. Compounds containing a pyridine system, such as SP148 (14, Figure 3A, B), showed less reactivation than compounds containing imidazole, such as SP134 (15), SP138 (16), and their analogues with different spacers (Figure 3A, B), so 15 and 16 were chosen for full characterization (Figure 3C, D). (Figure S15 shows that background hydrolysis of substrate by reactivator on its own was negligible under conditions identical to those of the reactivation assay.) We found that whereas ADOC (9) had a higher k2 value for reactivation of paraoxon (1), the k2 values for reactivation of DFP (2) inhibition by 15 and 16 were similar to that of 9 (Table S1). We also found that 16 reactivates AChE inhibited by NIMP (11) or SIMP (11a), analogues of the nerve agents sarin and soman, respectively (Figure S16).
It is important to note that compounds that facilitate reactivation, whether through the delivery of a base or a nucleophile to the active site of AChE, might not necessarily completely recover the enzyme. In fact, most oximes do not fully restore the activity of AChE,[29] largely because of the parallel “aging” process (cf. Figure 1B; β-elimination has also been reported).[19] We list the maximum extents of reactivation for these compounds in Table S1.
Crystal structure reveals unique mechanism of reactivation
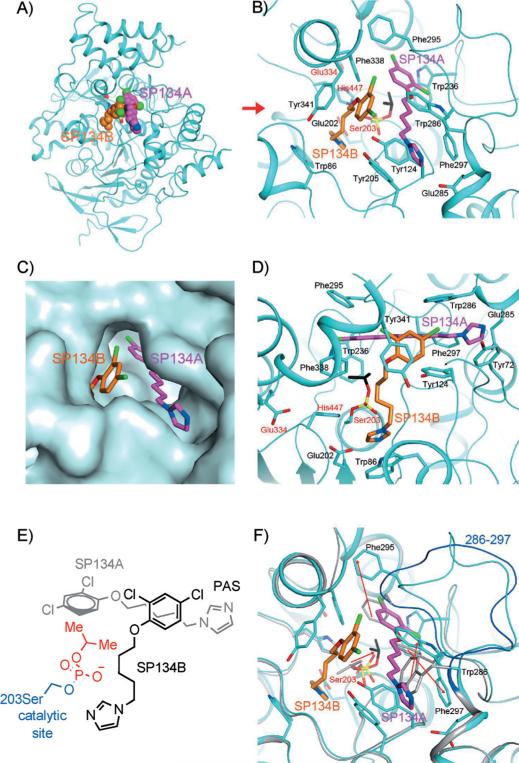
We attempted to co-crystallize all of these compounds with various forms of AChE in order to gain additional insight into the mechanism. We were successful in obtaining a crystal structure at 2.7 Å resolution of the adduct of DFP (2) and murine AChE in a ternary complex with two molecules of SP134 (15; we refer to these as SP134A and SP134B) in the active site region (Figure 4A and Table S2). This agreed with our kinetics data: the concentration dependence of the observed rate constants for reactivation (kobs) of 2-inhibited human AChE by 15 and by 16 were best fit to sigmoidal curves with Hill coefficients greater than 1, indicating cooperativity (15, h=1.8±0.20, and 16, h=1.4±0.13; Figure 3C, D; see Figure S17 for the concentration dependence of the rate of reactivation for 1-inhibited enzyme and for mouse AChE, and full kinetic parameters are listed in Table S1), confirming the functional relevance of this crystal structure. Although previous crystal structures of AChE have shown the ability of the enzyme to accept more than one ligand (for example, tacrine and acetylcholine),[30] this is a unique observation of two molecules within AChE in intimate contact with each other based on hydrophobic interactions.
Figure 4.
Interactions between SP134 (15) and AChE. A) Schematic drawing of the overall structure of murine AChE in complex with 15. The two molecules of 15 in each active site are shown as sphere models in magenta (SP134A) and orange (SP134B) for carbon atoms. B) Detailed interactions between 15 and murine AChE. Residues in the catalytic triad are labeled in red. C) Molecular surface showing the binding of the two molecules of 15 in a deep pocket of AChE. D) Detailed interactions between 15 and murine AChE, viewed along the red arrow of panel B). E) Chemdraw schematic representation of interacting 15 molecules from the same perspective as in C). F) Overlay of the structures of the 15 complex (in cyan) and the “aged” DFP (2) adduct (in gray). A large conformational change for residues 286–297 is visible, and the backbone of these residues in the 2 adduct is shown in blue. The red arrows indicate changes in the positions of the side chains that are important for SP134A binding, as well as a rotation of the remaining isopropyl group of 2 to interact with SP134A.
The binding modes of SP134A and SP134B in the enzyme are best described by the positions of each of their imidazole groups. SP134B is oriented to deliver its imidazole group to the active site (AS), whereas SP134A has its imidazole group at the entryway of the peripheral anion site (PAS, Figure 4B, C). The binding site of AChE is lined by aromatic residues, and the interactions between the compounds and AChE are primarily hydrophobic in nature. For SP134A, the dichlorophenoxy group is surrounded by Trp236, Phe295, and Phe338. The pentamethylene linker is in the fully extended conformation and has good electron density (Figure S18); it is sandwiched between Tyr124 and Trp286 and is close to Phe297. The imidazole is π-stacked with Tyr72 and located near Glu285. For SP134B, the dichlorophenoxy group is sandwiched between the pentamethylene linker of SP134A and Tyr341. The pentamethylene linker of SP134B is not in the fully extended conformation, with weaker electron density (Figure S18), and has some interaction with Phe338. SP134B is positioned almost perpendicular to SP134A. The imidazole group is p-stacked against Trp86, and is close to the His447 of the catalytic triad. It is also close to the side chain of Glu202, just prior to the catalytic Ser203, another highly conserved residue among the AChEs.
Electron density for only one isopropyl group of DFP (2) was observed (Figure S18), suggesting that the adduct of 2 with AChE had “aged” during the course of crystallization, thereby losing one isopropyl group (although we cannot rule out the possibility that the second isopropyl group is disordered in the crystal). The isopropyl group interacts directly with the phenyl group of SP134A, which might explain why compounds of this class have lower affinity for the uninhibited enzyme. In other words, the additional interaction between the isopropyl group left by 2 and SP134A in the inhibited enzyme might increase the binding affinity for reactivator, thus resulting in its KR value [≈(34±35) μm] being much lower than its inhibition constant, or Ki, value [≈ (240±17) μm; see Figure S19]. Note that refinement in the calculation of values for KR is complicated by the high degree of cooperativity, which contributes to the variability in measurements within the narrow range of concentrations that elicits the most significant effect on the reactivation rate.
Binding of SP134B introduces a new basic residue into the active site. Its imidazole group is ≈4.5 Å away from the phosphorus atom of 2-modified Ser203 (Figure 4D), and this imidazole group can activate a water molecule for a nucleophilic attack to initiate reactivation. The proximity of Glu202 to this imidazole group suggests that it might play a role in reactivation as well. The non-extended conformation of the SP134B pentamethylene linker, with multiple gauche interactions, indicates that there is significant structural flexibility to adjust the position of the imidazole group of SP134B in the active site, consistently with our experimental observation that even analogues with shorter linkers are active. Because our structure probably represents an “aged” 2 adduct, it prevents us from hypothesizing about the exact events in the vicinity of the catalytic triad that lead to reactivation of non-“aged” adduct.
For comparison, we determined the structures of “aged”, 2-modified mouse AChE at 2.4 Å resolution and of free AChE at 2.0 Å resolution (Table S2). These two structures have very similar conformations overall, with an rms distance of 0.32 Å for their equivalent Cα atoms. However, a comparison with the structure of the tertiary complex with two SP134 units reveals a large conformational change for residues crucial to the binding of SP134A (residues 286–297, Figure 4F, Figure S18). In fact, residues Phe295 and Phe297 in the free enzyme clash with the bound position of the dichlorophenoxy group of SP134A. The side chain of Phe295 has moved by up to 9 Å, and the side chain of Phe297 has twisted to accommodate a different rotamer. Trp286 has also undergone a conformational change to avoid a steric clash with the compound. In addition, the remaining isopropyl group of 2, with weak electron density, is rotated by ≈° relative to that in the 15 complex.
To the best of our knowledge, such a large conformational change for this segment of AChE has not been observed in other structures of this enzyme. This conformational change creates a hydrophobic pocket in the active site region for the binding of two molecules of 15 (Figure 4E and F). A smaller conformational change for this segment was observed in the structure of “aged”, 2-inhibited Torpedo californica AChE (Figure S18).[31] However, the three aromatic residues in the segment (equivalent to Trp286, Phe295, and Phe297 of murine AChE) still assume conformations that are similar to those in murine AChE alone and would actually clash with the SP134A compound. In addition, the Arg residue that is equivalent to Arg296 of murine AChE assumes a different conformation in the main chain and the side chain.
The rotation of the isopropyl group of 2 to interact with the phenyl group of SP134A suggests that this hydrophobic interaction, unique to the inhibited enzyme, can be taken advantage of to minimize interactions with the active enzyme (Figure 4B). For example, we observed experimentally that SP134 (15) shows very little inhibition of AChE at concentrations that show high reactivation (Figure 3D and inhibition curves in Figure S19). This is in stark contrast to active oximes with anchoring groups, which do not introduce new hydrophobic groups into the active site and are strongly inhibitory.[32]
New reactivators are effective in vivo
Next we wanted to test the effectiveness of these compounds for reactivation of inhibited cholinesterase in vivo. Our initial considerations were the animal species and the OPC to use as a challenge agent. DFP (2) was chosen due to reported difficulty in reversing its effects with other reactivators,[33] and a mouse model was preferred in an academic setting. As a drawback to using mice, however, we had to increase the dose of 2, in order to overwhelm circulating carboxylesterases.[34] We found that a dose of 3 mgkg−1 of 2, when given subcutaneously (s.c.), cumulatively resulted in a 95% mortality rate in untreated mice (33/35 mice expired in less than ≈100 min after exposure; two mice survived, showing signs of severe neurological distress).
Before testing for in vivo reactivation, we established whether the compounds themselves caused acute toxicity. Initial testing showed that whereas ADOC (9) and SP138 (16) were tolerated quite well by the mice, SP134 (15) caused significant neuromuscular relaxant side effects, which eliminated it from further testing. Compounds 9 and 16 were also stable with both mouse and human liver microsomes and did not affect hepatocyte viability (Supporting Information). Of particular concern was the potential for toxicity by 9, because a protein-reactive quinone-imine species could be formed through oxidation of the 4-aminophenol group (cf., paracetamol[35] and amodiaquine).[23] To address this concern, we treated animals with a single dose of 9 at 120 mgkg−1 and examined the histology of liver slices two weeks after treatment (to allow for the development of signs of necrosis), and found that there were no differences between treated animals and untreated controls, (Figure S20).
We then considered dosing strategies and determined that 9 and 16 were best suited for different routes of administration. Whereas 9 was soluble at the high concentrations needed for intraperitoneal (i.p.) injection, making it suitable for either pre- or post-exposure treatment, 16 could only achieve high concentrations as its corresponding salt and so it was used only in a model of prophylaxis through oral administration (mixed in daily chow).
We then demonstrated that two injections of 9 (at 60 mgkg−1 per dose), one given 20 min prior to challenge with a lethal dose of 2 (1.6×LD50, 3 mgkg−1) and then one within 5 min after challenge, offered full protection to the treated mice (nine out of nine mice survived to 24 h; cumulative results can be found in Table S3). However, when a highly inhibitory reactivator is administered before challenge with OPC, it is not possible to distinguish whether the protection afforded is from reactivation or from blocking of the AChE active site to prevent inhibition by the OPC.[36] To minimize this kind of competitive inhibition, we eliminated the pre-challenge dose of 9 and demonstrated that injection of 9 (120 mgkg−1) within five minutes post-exposure was sufficient to offer substantial protection to the mice. Collectively, six out of seven mice survived to 24 h, at which point they were sacrificed for tissue analysis. Furthermore, all mice treated with 9 showed immediate relief of neurological symptoms. This observation is very important as a control as well, because it shows that the activity is on inhibited enzyme, rather than against circulating nerve agent (which is not a likely proposition, but cannot be fully eliminated).
For the previous sets of experiments, we had used an “LD95” dose of DFP (2) (≈1.6×LD50; dose: 3 mgkg−1; in our hands 33/35 animals expired within 100 min at this dose). To explore further the notion that ADOC (9) might be reaching AChE prior to the 2 or reacting with the OPC prior to inhibition of the enzyme, we used a lower (0.8×LD50) dose of 2 that was not lethal, but resulted in severe signs of OPC toxicity, and we delayed treatment with 9 until outward manifestation of toxicity was evident (15 min post-challenge). In all cases, we found that treatment of mice with 9 resulted not only in survival, but also in complete lack of outward physiological signs of OPC intoxication, with administration of 9 actually correlating with cessation of developing tremors and return to nearly normal behavior. Unfortunately, a single dose of 9 was unable to prevent toxicity when the amount of 2 was further increased (e.g., 2×LD50), thus indicating a poor protective ratio of 9; this is likely an indication that pharmacokinetics might need to be optimized through creation of analogues. This has been addressed in oxime research in the past by continuous infusion[37] or by further anchoring.
Next, because SP138 (16) could only be given prophylactically, we addressed the possibility that it could prevent the effects of the OPC by binding at the active site and blocking the agent from gaining access to it. In order to determine the potential for in vivo reactivation by 16, we tested its ability to reactivate cholinesterase ex vivo in brain tissue harvested from untreated mice that had been exposed to DFP (2), and found significant reactivation (Figure S21), indicating that actual reactivation can happen in vivo, in the presence of the compound. We also note that 16 is only mildly inhibitory to AChE (Figure S19), and that the estimated concentration of 16 for the entire treatment dose in the average blood volume of a mouse[38] would be well below the measured Ki value for 16. We fed mice with 16 prior to challenge with an otherwise lethal dose of 2 and observed that it offered them significant protection (cumulatively, with different dosing strategies, 15 out of 17 animals survived to 24 h; see Table S4).
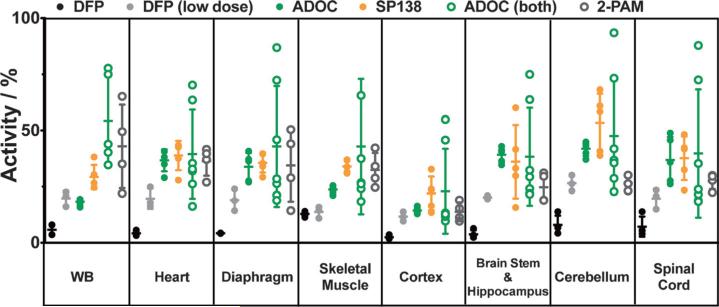
In order to correlate survival with in vivo reactivation, we measured cholinesterase activity across a panel of tissues from both treated and control mice. We show representative experiments (Figure 5), in which all the mice treated with ADOC (9) and SP138 (16) were alive at 24 h and showed no visible signs of neurotoxicity. The animals were sacrificed at 24 h, their tissues were harvested, and cholinesterase activity was measured by a standard Ellman's assay,[39] except for the analysis of whole blood, in which DTP (2,2′-dithiodipyridine)[40] was used in place of DTNB [5,5′-dithiobis(2-nitrobenzoic acid); activity was normalized for the amount of total protein in each sample]. Cholinesterase activity was measured across a panel of tissues and set relative to the activity in tissues from untreated, unchallenged mice. We note that we are unable to distinguish the contribution of activity that was specifically due to AChE from this; some activity might have been due to the presence of other esterases (such as butyrylcholinesterase); however, our control animals still give us an estimate of the activity had they been left untreated as a point of reference. Untreated animals expired within 100 min, and their tissues were harvested immediately afterwards. As an additional control, we challenged mice with a lower dose of 2 (2.5 mgkg−1), after which all three mice survived for 24 h with outward signs of neurological distress. The animals were then euthanized, and their tissues were used to obtain an estimate for the minimal cholinesterase activity needed for survival and as a control for any newly synthesized AChE in the period post-exposure. This control was important because any cholinesterase activity above that seen in mice that survived exposure to a low dose of 2 could be attributed to actual reactivation, rather than de novo synthesis, and the relative absence of neurological sequelae could then be attributed to protective effects of treatment with reactivator.
Figure 5.
Cholinesterase activity in mouse tissues after challenge with DFP (2). Animals were treated with 120 mgkg−1 total of ADOC (9) either 5 min after challenge (solid green circles) or 20 min before and 5 min (60 mgkg−1 per dose) after exposure (open green circles) to an otherwise lethal dose of 2 (3 mgkg−1). Another group of animals were fed 1 mg a day of SP138 (16, orange) for three days prior to exposure. All treated animals survived 24 h, after which tissues were collected and cholinesterase activity was measured. Activity from the treated animals was significantly higher than that from untreated control animals (i.e., 2 only), which expired within 90 min and are shown in black (two animals from each experiment). For comparison, three animals were treated with a lower dose of 2 (solid gray circles); they all survived for 24 h, albeit under visible distress, unlike the treated animals. Activity in tissues from across the BBB was also significantly higher than activity from a group of mice treated with pralidoxime (5) both 20 min prior to and 5 min post exposure (open gray circles). Activity is reported as the % activity in the samples relative to cumulative activity from tissue samples of control animals that were untreated and unchallenged. Activity measurements were repeated in triplicate and averaged; samples were normalized for protein content. Error bars show the standard deviation within each cohort of animals.
In animals treated with either ADOC (9) or SP138 (16), we found that cholinesterase activity was restored in all tissues examined, including CNS tissues across the blood–brain barrier (BBB); activity in tissues from the untreated control groups given DFP (2) was lower in each tissue group examined. In parallel, we treated mice with an equivalent amount of pralidoxime (5), using the dual dosing strategy pre- and post-challenge (120 mgkg−1 total), and found that the average activity in tissues from those animals was consistently lower than that from animals with the dual dose of 9. Importantly, in brain and CNS tissues, the animals treated with either dosing strategy, with 9 or 16, had an average cholinesterase activity that was notably higher than that in mice treated with 5, possibly indicating the potential for penetration across the BBB. We note that that the measurement of AChE activity in recipients of treatment both before and after exposure to 2 had more scatter in activity, possibly due to individual differences in metabolism during the 20 min prior to challenge.
Cumulative experiments (Tables S3 and S4) from multiple experiments support these results within expected variation. Different cohorts of the same strain of mice showed differences in susceptibility to DFP (2);[41] although these variations make it difficult to come to firm conclusions regarding translation to effective dosing and protection strategies for humans, they do not prevent clear conclusions relating to the ability of these compounds to reactivate cholinesterase inhibited by a sterically hindered agent in vivo and ultimately to protect animals against its toxicity.
Conclusion
For the past 60 years, the improvement of oximes has been the sole focus of research into reactivation of AChE. Yet, as a result of a single screen, we have discovered that Mannich phenols and base catalysts are as efficient at reactivating AChE and provide significant protection to animals exposed to otherwise lethal doses of OPCs. This opens up new avenues to address organophosphate toxicity through further medicinal chemistry optimization.[17] Additional knowledge about subtleties of the mechanism and ternary complex formation should serve as the foundation for new optimization efforts, but even this generation of synergistic base-delivering reactivators shows solid activity both in vitro and in vivo.[17]
In retrospect, both classes of compounds could have been predicted to reactivate inhibited AChE. For Mannich phenols, it could have been argued that because phenols are commonly used as leaving groups for phosphylation of AChE,[21] it would be expected that the reaction could be reversed, if properly selected phenols were present in excess. As for basic functionalities, it had been shown previously that site-directed mutagenesis could introduce new basic residues (histidines containing imidazoles), which resulted in modest phosphodiesterase activity,[42] so it is not surprising that similar activity can be achieved by their extraneous delivery.
In terms of function, these are deceptively simple molecules, but because the actual delivery of the functional groups to the active site of AChE is complex, their ability to reach their target might be the reason why their earlier discovery was hindered: notably, the most efficient reactivators in both classes required a cooperative mechanism in order to achieve maximum activity. From our crystal structure, we can see a further level of complexity in that the cooperativity is not just the result of a compound causing a rearrangement of the enzyme, but that the molecules self-assemble within the active site to form a dimer. In addition, as demonstrated in the case of SP138 (16), they can interact in this manner with added efficiency by avoiding inhibition of the native enzyme.
AChE is historically one of the best-studied enzymes, yet self-assembly within its active site has never been reported before, let alone self-assembly that accomplishes a new function. Dimerization within the active site not only delivers the functional group, but it also expands the active site, and rearranges and positions alkyl groups near the phosphylated serine (the absence of which results in decreased affinity, and thus has the advantage of having low inhibition of the native enzyme), while leaving sufficient flexibility for formation of productive intermediates. To the best of our knowledge, similar self-assembly such as this within an active site has not been reported with any other enzymes.
Along with broadly increasing the understanding of conformational flexibilities in enzymes and other proteins,[43] these findings might provide a more general inspiration for a way to increase complexity of functions and biological activities in structurally simple molecules.
Experimental Section
Materials
Purified, recombinant AChE was obtained from Chesapeake PERL, Inc. (C-PERL, Savage, MD, USA). Compounds libraries screened were from MicroSource Discovery Systems (Gaylordsville, CT, USA) and ChemBridge (San Diego, CA, USA). AChE was immobilized on NUNC MicroWell plates with Nunclon Delta surface (for optimization of the screen see the Supporting Information). Acetylthiocholine (ATCh), DTNB, and DTP were from Sigma–Aldrich. All other commercially available compounds were purchased from ChemBridge or Sigma–Aldrich; compounds that were not available were synthesized in-house.
Chemistry
All solvents and reagents were obtained from Sigma–Aldrich and used without further purification. Analytical thin-layer chromatography (TLC) was performed on aluminium plates precoated with silica gel, also obtained from Sigma–Aldrich. Column chromatography was carried out on Merck 938S silica gel. Proton NMR spectra were recorded with a Varian 400 MHz NMR spectrometer. Spectra were referenced to the residual solvent peak, and chemical shifts are expressed in ppm from the internal reference peak. All compounds described were of >95% purity. Purity was confirmed by analytical LC/MS recorded with a Shimadzu system. Elution started with water (95%, +0.1% formic acid) and acetonitrile (5%, +0.1% formic acid) and ended with acetonitrile (95%, 0.1% formic acid) and water (5%, 0.1% formic acid) and used a linear gradient at a flow rate of 0.2 mLmin−1. The molecular ions [M]+, with intensities in parentheses, are given, followed by peaks corresponding to major fragment losses. Melting points were measured with a MEL-TEMP II melting point apparatus and are reported uncorrected. For synthesis details and results see the Supporting Information.
Solid-phase high-throughput screening assay
Enough AChE enzyme was deposited overnight on NUNC MicroWell plates in P-buffer [sodium phosphate (100 mm), NaCl (100 mm), pH 7.4] to achieve a rate of hydrolysis of ATCh that resulted in an OD412 of ≈1 (from breakdown of DTNB)[38] within 7–10 min. The plates were washed three times with P-Buffer to remove excess AChE before addition of the reaction components. Initial screening was performed with paraoxon (1) as the inhibiting OPC; any potential hits underwent additional characterization with DFP (2). Greater than 99% inhibition of the immobilized AChE was achieved after incubation with excess 1 (200 nm) for 10 min, and any remaining free 1 was washed away with 1× PBS. Either P-buffer, final DMSO (0.1%, for control reactions to match DMSO concentration in compound library), or library compounds at a final concentration of 10 μm each were added and allowed to incubate for 3 h. Any products from the reactivation reaction were washed away as previously described. Recovery of AChE activity was measured by use of a standard Ellman's assay[39] to determine the rate of ATCh hydrolysis upon addition of ATCh and DTNB (2 mm each). Controls in which no reactivator compound was added were performed, allowing us to determine if there was any spontaneous reactivation. Those compounds that resulted in an increase in AChE activity greater than three standard deviations above the controls in which no compound was added were considered hits.
Solution-phase reactivation assays
To confirm that our hits would have similar activity on the free enzyme as on the immobilized AChE, we developed a solution-phase assay for reactivation. Firstly, AChE (320 μgmL−1, 4.7 μm active site concentration) was incubated with enough OPC to give >95% inhibition. This varies, based on the type of enzyme (mouse, human, or guinea pig) and the OPC used, and ranged from four- to 350-fold excess. Any excess OPC was removed by applying the mixture to a GE Health-care PD minitrap G-25 column (catalogue number 28–9180–07). After the enzyme had been run over the PD10 column, the protein concentration was too low for direct measurement, so a parallel sample that had not been treated with OPC was also run in order to have a control from which we could calculate maximum AChE activity. The enzyme that was recovered from the column was then diluted 1:25 in P-buffer (see above) and BSA (0.25%) to stabilize the enzyme and reduce residual OPC concentration. All chromatography steps and subsequent handling of the enzyme were done on ice immediately prior to that day's experiment, to reduce the formation of “aged” adducts.
The recovered enzyme was treated with varying concentrations of compound and allowed to incubate at room temperature for 3 h [in some cases, huAChE inhibited by DFP (2) required more time for complete reactivation and was left to incubate overnight]. Many of our stock compounds have DMSO in them; when this was the case, we matched the DMSO concentration in each subsequent dilution, in order to eliminate any background effects of dilution of DMSO (i.e., the concentration of compound was changed, but the concentration of DMSO remained the same in each reaction). Also, equivalent amounts of DMSO were added to control reactions without compound in order to have a direct comparison of the effect of DMSO itself on the enzyme. For this reason, slight differences in enzyme activity in the absence of compound are seen, due to the fact that we adjusted for what else we were adding with the compound. We added a 1:100 dilution step after reactivation in order to eliminate possible negative/inhibitory effects of the compound on AChE (a 100-fold dilution brings even the maximum concentration of compound tested well below the Ki value). After dilution, ATCh and DTNB were added (2 mm each final), and activity was measured at 412 nm. Reactions were done in triplicate or in duplicate on multiple days (minimum of quadruplicate, in those cases). Error bars show the standard deviation.
Activity is expressed in terms of % reactivation, a measure of how much activity is restored relative to the uninhibited control that is run in parallel. In order to calculate % reactivation values, we used the following formula:
where vreact is the rate of the reactivated enzyme as defined by the following:
where vt is the rate of hydrolysis by the OPC-inhibited enzyme at a given time t (after incubation with reactivator), and v0 is the rate of hydrolysis by the control OPC-inhibited enzyme in the absence of reactivator. This would account for any spontaneous reactivation. The bottom denominator, vfull, is the full rate of hydrolysis by the enzyme and takes into account any effect of the reactivator on the enzyme itself, which is obtained by use of the following equation:
where vfree is the activity of the enzyme in the absence of OPC or reactivator and vfree+R is the rate of hydrolysis by the free enzyme incubated with reactivator. We note that with our dilution step, the effect of reactivator on enzyme itself was negligible. In parallel, under identical conditions, we also ran controls with just reactivator, to see background hydrolysis of ATCh due to the reactivator itself (Figures S4 and S15) and found it was also negligible. We note that extended incubation with ADQ (6) or ADOC (9) seemed to have increasing irreversible inhibitory effects on specific forms of AChE. Although this is noteworthy and interesting, we did not pursue this finding further and chose only to use conditions (shorter incubation times or lower concentrations) under which this inhibition was eliminated.
By stopping the reactivation reaction at various time points, we were then able to measure the rate of reactivation over time. Full discussion of the determination of reactivation rate constants can be found in the Supporting Information.
AChE inhibition
The direct effects of these compounds on AChE were tested by firstly adding the compound in question to AChE and then adding ATCh and DTNB to follow enzyme activity. Full substrate concentration-dependence curves were generated in order to calculate Vmax and Km values at increasing concentrations of reactivation compound, and these were then used to determine Ki and mechanism of inhibition.
X-ray crystallography
The recombinant, truncated, untagged mouse acetylcholinesterase (AChE) was obtained from Chesapeake Perl. The apo-AChE protein was crystallized at 4°C in PEG 600 (30%, v/v) and sodium citrate (pH 6.5, 0.1 m), by the sitting drop vapor diffusion method. The concentration of the protein was 6 mgmL−1, and the crystals took approximately three weeks to grow to full size. For the DFP (2) adduct, AChE was incubated with 2 (10 mm) for one hour before crystallization under the same conditions as used for the apo enzyme. To prepare crystals of the DFP·SP134·AChE ternary complex, crystals of the DFP·AChE binary complex were soaked with SP134 (10 mm) for 16 h. The crystals were flash-frozen in liquid nitrogen for data collection at 100 K. Diffraction data for AChE were collected at the X29A beamline of the National Synchrotron Light Source (NSLS), with use of a Quantum 315 CCD X-ray detector (Area Detector System Corp.; (Poway, CA, USA). All data sets were collected at a wavelength of 1.075 Å with 1° oscillation range. The crystals belong to space group P212121 and contain two monomers in the crystallographic asymmetric unit. All the data sets were indexed, integrated, and scaled with the aid of the HKL2000 program suite.[44] The data processing statistics are summarized in Table S2. The crystals are isomorphous to those of mouse AChE reported earlier.[30b,45] The structure refinement was carried out by use of the programs Refmac[46] and Phenix.[47] The atomic model was manually fit into the electron density with the aid of the program Coot.[48] The refinement statistics are shown in Table S2.
Animals
Balb/C mice of approximately 6–8 weeks in age were obtained from Jackson Laboratories. The mice were housed and cared for at the University animal facilities, and all experiments were carried out under approval of the Columbia University Medical Center Institutional Animal Care and Use Committee (protocol #AC-AAAG1818). Animals in distress were euthanized immediately. Mice were challenged with DFP (2), which was given subcutaneously. ADOC (9) or pralidoxime (5) were formulated in saline before administration intraperitoneally, either before and/or after challenge agent, as described. In either dosing strategy, animals were given a total of 120 mgkg−1 of 9 or 5. ADQ (6) and SP138 (16) were mixed with daily chow and fed to mice in the various dosing strategies described and summarized in the Supporting Information (Tables S3 and S4). Animals that survived to 24 h were sacrificed, and their tissues were harvested. Tissues from animals that expired within 100 min due to acute DFP (2) poisoning were harvested immediately. Tissue lysates were prepared by homogenizing in 1× PBS and Triton X-100 (1%), and total cholinesterase activity was measured by a standard Ellman's assay,[39] except in the analysis of whole blood, in which DTP[40] was used in place of DTNB. Total protein in each tissue sample was measured by BCA assay, and activity was normalized for amount of protein added to each reaction. Activity is expressed relative to control animals that were not treated with reactivation compound and not challenged with OPC.
Supplementary Material
Acknowledgements
This work was supported by HDTRA1–11-C-0006 to D.W.L. and by NIH CounterACT grant number 5R21NS080790–02 to D.W.L. Equipment used to characterize synthetic compounds was supplied through ONR grant N00014–11–1–0900 to D.W.L. and NIH grant 1S10OD018121–01 to M.N.S. We are grateful to Timothy Sauder for assistance in developing robotic protocols to facilitate tissue analysis.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201500348.
References
- 1.a Chambers J, Oppenheimer SF. Toxicol. Sci. 2004;77:185–187. doi: 10.1093/toxsci/kfh060. [DOI] [PubMed] [Google Scholar]; b Hardman JG, Limbird LE, Gilman AG, editors. Anticholinesterase Agents. P. Taylor in Goodman and Gilman's The Pharmacological Basis of Therapeutics. 10th ed. McGraw–Hill; New York: 2001. p. 175. [Google Scholar]; c Jennings LL, Malecki M, Komives EA, Taylor P. Biochemistry. 2003;42:11083–11091. doi: 10.1021/bi034756x. [DOI] [PubMed] [Google Scholar]
- 2.Yuantari MG, Van Gestel CA, Van Straalen NM, Widianarko B, Sunoko HR, Shobib MN. Environ. Monit. Assess. 2015;187:142. doi: 10.1007/s10661-015-4371-3. [DOI] [PubMed] [Google Scholar]
- 3.Eyer P. Toxicol. Rev. 2003;22:165–190. doi: 10.2165/00139709-200322030-00004. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. 2015 http://www.who.int/ipcs/assessment/public_health/pesticides/en/
- 5.a Eddleston M, Szinicz L, Eyer P, Buckley N. QJM. 2002;95:275–283. doi: 10.1093/qjmed/95.5.275. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Peter JV, Moran JL, Graham PL. Expert Opin. Pharmacother. 2007;8:1451–1464. doi: 10.1517/14656566.8.10.1451. [DOI] [PubMed] [Google Scholar]
- 6.a Wilson IB. Federation Proceedings. 1959;18:752–758. [PubMed] [Google Scholar]; b Wilson IB, Ginsburg B. Biochim. Biophys. Acta. 1955;18:168–170. doi: 10.1016/0006-3002(55)90040-8. [DOI] [PubMed] [Google Scholar]
- 7.Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Proc. Natl. Acad. Sci. USA. 1993;90:9031–9035. doi: 10.1073/pnas.90.19.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekstrçm F, Pang YP, Boman M, Artursson E, Akfur C, Borjegren S. Biochem. Pharmacol. 2006;72:597–607. doi: 10.1016/j.bcp.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Worek F, Diepold C, Eyer P. Arch. Toxicol. 1999;73:7–14. doi: 10.1007/s002040050580. [DOI] [PubMed] [Google Scholar]
- 10.Okolotowicz KJ, Dwyer M, Smith E, Cashman JR. J. Biochem. Mol. Toxicol. 2014;28:23–31. doi: 10.1002/jbt.21519. [DOI] [PubMed] [Google Scholar]
- 11.a Shih TM, Skovira JW, O'Donnell JC, McDonough JH. Chem.-Biol. Interact. 2010;187:207–214. doi: 10.1016/j.cbi.2010.03.006. [DOI] [PubMed] [Google Scholar]; b Chen R, Raveh L, Zomber G, Rabinovitz I, Cohen G, Adani R, Amitai G. J. Appl. Toxicol. 2001;21:285–291. doi: 10.1002/jat.756. [DOI] [PubMed] [Google Scholar]; c Eyer P, Worek F, Kiderlen D, Sinko G, Stuglin A, Simeon-Rudolf V, Reiner E. Anal. Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]; d Bures J, Kvetina J, Pavlik M, Kunes M, Kopacova M, Rejchrt S, Jun D, Hrabinova M, Kuca K, Tacheci I. Neuro Endocrinol. Lett. 2013;34:79–83. [PubMed] [Google Scholar]
- 12.a Kassa J, Krejcova G, Vachek J. Acta Medica (Hradec Kralove) 2002;45:149–153. [PubMed] [Google Scholar]; b Worek F, Thiermann H. Defence against the Effects of Chemical Hazards: Toxicology, Diagnosis and Medical Countermeasures. 2007:30-31–30-36. [Google Scholar]; c Hatton CS, Peto TE, Bunch C, Pasvol G, Russell SJ, Singer CR, Edwards G, Winstanley P. Lancet. 1986;1:411–414. doi: 10.1016/s0140-6736(86)92371-8. [DOI] [PubMed] [Google Scholar]
- 13.a Worek F, Elsinghorst P, Koller M, Thiermann H. Toxicol. Lett. 2014 doi: 10.1016/j.toxlet.2014.09.012. [DOI] [PubMed] [Google Scholar]; b Georgiev B, Khristoskova E. Veterinarno-Meditsinski Nauki. 1978;15:108–115. [PubMed] [Google Scholar]; c Becker C, Worek F, John H. Drug Testing Anal. 2010;2:460–468. doi: 10.1002/dta.167. [DOI] [PubMed] [Google Scholar]; d Antonijevic B, Stojiljkovic MP. Clin. Med. Res. 2007;5:71–82. doi: 10.3121/cmr.2007.701. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Worek F, Szinicz L, Eyer P, Thiermann H. Toxicol. Appl. Pharmacol. 2005;209:193–202. doi: 10.1016/j.taap.2005.04.006. [DOI] [PubMed] [Google Scholar]; f Petroianu GA, Lorke DE, Athauda G, Darvas F, Kalasz H. J. Environ. Immunol. Toxicol. 2013;1:35–40. [Google Scholar]
- 14.a Neftel KA, Woodtly W, Schmid M, Frick PG, Fehr J. Br. Med. J. 1986;292:721–723. doi: 10.1136/bmj.292.6522.721. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Schulthess HK, von Felten A, Gmur J, Neftel K. Schweiz. Med. Wochenschr. 1983;113:1912–1913. [PubMed] [Google Scholar]
- 15.Tingle MD, Jewell H, Maggs JL, O'Neill PM, Park BK. Biochem. Pharmacol. 1995;50:1113–1119. doi: 10.1016/0006-2952(95)00236-s. [DOI] [PubMed] [Google Scholar]
- 16.Radic Z, Sit RK, Kovarik Z, Berend S, Garcia E, Zhang L, Amitai G, Green C, Radic B, Fokin VV, Sharpless KB, Taylor P. J. Biol. Chem. 2012;287:11798–11809. doi: 10.1074/jbc.M111.333732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stojanovic M, Katz F, Landry DW. WO. 2014;2014210168 [Google Scholar]; Stojanovic M, Katz F, Landry DW. WO. 2014;2014113495 [Google Scholar]
- 18.Baynes JW, Dominiczak MH. Medical Biochemistry. 3rd ed. Mosby, Edinburgh: 2009. [Google Scholar]
- 19.Gilley C, MacDonald M, Nachon F, Schopfer LM, Zhang J, Cash-man JR, Lockridge O. Chem. Res. Toxicol. 2009;22:1680–1688. doi: 10.1021/tx900090m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a Meek EC, Chambers HW, Coban A, Funck KE, Pringle RB, Ross MK, Chambers JE. Toxicol. Sci. 2012;126:525–533. doi: 10.1093/toxsci/kfs013. [DOI] [PubMed] [Google Scholar]; b Li WS, Lum KT, Chen-Goodspeed M, Sogorb MA, Raushel FM. Bioorg. med. Chem. 2001;9:2083–2091. doi: 10.1016/s0968-0896(01)00113-4. [DOI] [PubMed] [Google Scholar]
- 21.Epstein J, Michel HO, Rosenblatt DH, Plapinger RE, Stephani RA, Cook E. J. Am. Chem. Soc. 1964;86:4959–4963. [Google Scholar]
- 22.Muthukrishnan S, Shete VS, Sanan TT, Vyas S, Oottikkal S, Porter LM, Magliery TJ, Hadad CM. J. Phys. Org. Chem. 2012;25:1247–1260. doi: 10.1002/poc.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briseno-Roa L, Hill J, Notman S, Sellers D, Smith AP, Timperley CM, Wetherell J, Williams NH, Williams GR, Fersht AR, Griffiths AD. J. Med. Chem. 2006;49:246–255. doi: 10.1021/jm050518j. [DOI] [PubMed] [Google Scholar]
- 24.Aldridge WN, Reiner E. Biochem. J. 1969;115:147–162. doi: 10.1042/bj1150147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a Hawley SR, Bray PG, O'Neill PM, Park BK, Ward SA. Biochem. Pharmacol. 1996;52:723–733. doi: 10.1016/0006-2952(96)00354-1. [DOI] [PubMed] [Google Scholar]; b Warhurst DC, Steele JC, Adagu IS, Craig JC, Cullander C. J. Antimicrob. Chemother. 2003;52:188–193. doi: 10.1093/jac/dkg319. [DOI] [PubMed] [Google Scholar]
- 26.Kurtz AP, D'Silva TD. J. Pharm. Sci. 1987;76:599–610. doi: 10.1002/jps.2600760805. [DOI] [PubMed] [Google Scholar]
- 27.Michaelides M, Stover NB, Francis PJ, Weleber RG. Arch. Ophthalmol. 2011;129:30–39. doi: 10.1001/archophthalmol.2010.321. [DOI] [PubMed] [Google Scholar]
- 28.Koreeda A, Yonemitsu K, Kohmatsu H, Mimasaka S, Ohtsu Y, Oshima T, Fujiwara K, Tsunenari S. Arch. Toxicol. 2007;81:471–478. doi: 10.1007/s00204-007-0185-6. [DOI] [PubMed] [Google Scholar]
- 29.a Luo C, Tong M, Chilukuri N, Brecht K, Maxwell DM, Saxena A. Biochemistry. 2007;46:11771–11779. doi: 10.1021/bi701002f. [DOI] [PubMed] [Google Scholar]; b Kovarik Z, Radic Z, Berman HA, Simeon-Rudolf V, Reiner E, Taylor P. Biochemistry. 2004;43:3222–3229. doi: 10.1021/bi036191a. [DOI] [PubMed] [Google Scholar]; c Worek F, Eyer P, Szinicz L. Arch. Toxicol. 1998;72:580–587. doi: 10.1007/s002040050546. [DOI] [PubMed] [Google Scholar]; d Maxwell DM, Lieske CN, Brecht KM. Chem. Res. Toxicol. 1994;7:428–433. doi: 10.1021/tx00039a022. [DOI] [PubMed] [Google Scholar]
- 30.a Rydberg EH, Brumshtein B, Greenblatt HM, Wong DM, Shaya D, Williams LD, Carlier PR, Pang YP, Silman I, Sussman JL. J. Med. Chem. 2006;49:5491–5500. doi: 10.1021/jm060164b. [DOI] [PubMed] [Google Scholar]; b Bourne Y, Radic Z, Sulzenbacher G, Kim E, Taylor P, Marchot P. J. Biol. Chem. 2006;281:29256–29267. doi: 10.1074/jbc.M603018200. [DOI] [PubMed] [Google Scholar]
- 31.a Dvir H, Jiang HL, Wong DM, Harel M, Chetrit M, He XC, Jin GY, Yu GL, Tang XC, Silman I, Bai DL, Sussman JL. Biochemistry. 2002;41:10810–10818. doi: 10.1021/bi020151+. [DOI] [PubMed] [Google Scholar]; b Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Biochemistry. 1999;38:7032–7039. doi: 10.1021/bi982678l. [DOI] [PubMed] [Google Scholar]
- 32.a Jokanovic M, Stojiljkovic MP. Eur. J. Pharmacol. 2006;553:10–17. doi: 10.1016/j.ejphar.2006.09.054. [DOI] [PubMed] [Google Scholar]; b Sepsova V, Karasova JZ, Korabecny J, Dolezal R, Zemek F, Bennion BJ, Kuca K. Int. J. Mol. Sci. 2013;14:16882–16900. doi: 10.3390/ijms140816882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a Dube SN, Kumar D, Sikder AK, Sikder N, Jaiswal DK, Das Gupta S. Die Pharmazie. 1992;47:68–69. [PubMed] [Google Scholar]; b Petroianu GA, Lorke DE. Mini Reviews Med. Chem. 2008;8:1328–1342. doi: 10.2174/138955708786369555. [DOI] [PubMed] [Google Scholar]
- 34.a Duysen EG, Koentgen F, Williams GR, Timperley CM, Schopfer LM, Cerasoli DM, Lockridge O. Chem. Res. Toxicol. 2011;24:1891–1898. doi: 10.1021/tx200237a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Maxwell DM, Koplovitz I. J. Pharmacol. Exp. Ther. 1990;254:440–444. [PubMed] [Google Scholar]
- 35.Dahlin DC, Miwa GT, Lu AY, Nelson SD. Proc. Natl. Acad. Sci. USA. 1984;81:1327–1331. doi: 10.1073/pnas.81.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colovic MB, Krstic DZ, Lazarevic-Pasti TD, Bondzic AM, Vasic VM. Curr Neuropharmacol. 2013;11:315–335. doi: 10.2174/1570159X11311030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tush GM, Anstead MI. Annals Pharmacother. 1997;31:441–444. doi: 10.1177/106002809703100411. [DOI] [PubMed] [Google Scholar]
- 38.a Mitruka BM, Rawnsley HM. Clinical Biochemical and Hematological Reference Values in Normal Experimental Animals and Normal Humans. 2nd ed. Masson Pub; USA, New York: 1981. [Google Scholar]; b Novotny J. Vet. Med. Sm Anim. Clin. 1983;78:1130–1130. [Google Scholar]
- 39.Ellman GL, Courtney KD, Andres V, Jr., Feather-Stone RM. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 40.Ceron JJ, Fernandex del Palacio MJ, Bernal LJ, Gutierrez C, AOAC Int J. 1996;79:757–763. [PubMed] [Google Scholar]
- 41.a Chen K, Teo S, Seng KY. Toxicol. Mechanisms Methods. 2009;19:486–497. doi: 10.1080/15376510903300335. [DOI] [PubMed] [Google Scholar]; b Gearhart JM, Jepson GW, Clewell HJ, 3rd, Andersen ME, Conolly RB. Toxicol. Appl. Pharmacol. 1990;106:295–310. doi: 10.1016/0041-008x(90)90249-t. [DOI] [PubMed] [Google Scholar]
- 42.Masson P, Nachon F, Broomfield CA, Lenz DE, Verdier L, Schopfer LM, Lockridge O. Chem.-Biol. Interact. 2008;175:273–280. doi: 10.1016/j.cbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Kokkinidis M, Glykos NM, Fadouloglou VE. Adv. Protein Chem. 2012;87:181–218. doi: 10.1016/B978-0-12-398312-1.00007-X. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.a Hçrnberg A, Tunemalm AK, Ekstrçm F. Biochemistry. 2007;46:4815–4825. doi: 10.1021/bi0621361. [DOI] [PubMed] [Google Scholar]; b Ekstrçm F, Hçrnberg A, Artursson E, Hammarstrçm L-G, Schneider G, Pang YP. PloS One. 2009;4:e5957. doi: 10.1371/journal.pone.0005957. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Berg L, Niemiec MS, Qian W, Andersson CD, Wittung-Stafshede P, Ekstrçm F, Linusson A. Angew. Chem. Int. Ed. 2012;51:12716–12720. doi: 10.1002/anie.201205113. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012;124:12888–12892. [Google Scholar]; d Andersson CD, Forsgren N, Akfur C, Allgardsson A, Berg L, Engdahl C, Qian W, Ekstrçm F, Linusson A. J. Med. Chem. 2013;56:7615–7624. doi: 10.1021/jm400990p. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.