Abstract
MicroRNAs are increasingly seen as targets of drug discovery because they influence gene function acting both to silence and subtly modulate protein translation. Little is known about effects of dynamic physiological states on microRNA regulation in humans. We hypothesized that microRNA expression in peripheral blood mononuclear cells (PBMCs) would be affected by brief exercise. Twelve young men performed brief bouts of heavy exercise. PBMC microRNA was analyzed before and immediately after exercise using the Agilent Human microRNA V2 Microarray. Exercise altered expression level of 34 microRNAs (FDR < 0.05). Many of them play roles in inflammatory processes (e.g., miR‐125b[↓], down‐regulated by proinflammatory factor LPS; and miR‐132[↑], 125b[↓] and let‐7e[↓] involved inTLR4 signaling). Using previous exercise data in PBMCs, we linked the microRNA changes to specific gene pathways. This analysis identified 12 pathways including the TGF‐β and MAPK signaling. We also compared exercise‐associated microRNA changes in PBMCs with the exercise‐associated microRNAs previously identified in neutrophils. Nine microRNAs were affected in both PBMCs and neutrophils, but only six changed in the same direction. A commonly occurring physiologic perturbation, brief heavy exercise, changes microRNA profiles in PBMCs, many of which are related to inflammatory processes. The pattern of change suggests that exercise differentially influences microRNAs in leukocyte subtypes. Clin Trans Sci 2012; Volume #: 1–7
Keywords: leukocytes, epigenetic, immune system, miRNA
Introduction
The study of microRNAs (small noncoding RNA molecules 20–22 nt in length) is rapidly increasing because they are involved in many human diseases. However, very little is known about dynamic changes in microRNAs in response to common physiological perturbations such as physical exercise. Understanding how exercise alters gene regulation at the level of microRNAs will likely prove important in optimizing therapeutic uses of exercise to benefit human health and as an adjunct to specific drug therapies, in particular, for diseases like asthma or coronary artery disease in which both exercise and microRNAs play a key role in pathogenesis. 12 , 13 , 14 , 16
The goal of this study was to test the hypothesis that brief exercise alters microRNA expression level in circulating peripheral blood mononuclear cells (PBMCs) in humans. In contrast to granulocytes (neutrophils, basophils, and eosinophils) that have granules in their cytoplasm and varying shapes of the nucleus, PBMCs are leukocytes that have a round nucleus such as lymphocytes (T cells, B cells), natural killer (NK) cells, and monocytes. PBMCs constitute a critical component of the immune system and function to fight infection and prepare the organism for “danger” 21 that may accompany exercise. A single brief bout of exercise causes a profound perturbation of cellular homeostasis and leads to an increase in the number of circulating PBMCs and a transient, muted activation of immune cells and inflammatory mediators even in healthy humans. 45
Exercise is increasingly used as a naturally occurring and quantifiable tool to assess immune function. 30 In healthy individuals, pro‐inflammatory genes and gene pathways are up‐regulated by exercise, but the inflammatory response is balanced by antiinflammatory genes. This precise regulation, suggested earlier by Ostrowski et al., 28 ensures that exercise‐associated inflammatory processes gone awry, such as exercise‐associated anaphylaxis 37 or exercise‐induced bronchoconstriction, 8 do not occur every time a child or adult engages in vigorous play or exercise. In earlier studies, we reported that brief exercise alters gene expression in PBMCs 32 and in neutrophils, 34 and microRNA in neutrophils 35 in a balanced manner. To our knowledge, the impact of exercise on microRNA in PBMCs has never been studied.
In general, the microRNAs function to mitigate or silence protein translation, often acting as subtle regulators. 5 MicroRNAs also play a role in DNA methylation 10 and chromatin remodeling. 2 A single microRNA may regulate from just a few target genes to literally thousands. A growing number of animal‐model and human studies point toward key regulatory roles for microRNA in the immune system, 3 , 19 and suggest that microRNAs provide an added layer in orchestrating (“fine tuning”) the immune response. 4
The primary purpose of this study was to investigate for the first time whether PBMC microRNAs are altered by brief exercise. A secondary goal was to perform a combined analysis that included the microRNA results of this study with: (1) a previously published data base of PBMC gene expression following the same type of exercise; 33 and (2) a previously published data base of microRNA expression in neutrophils following the same type of exercise. 35
Materials and Methods
Participants
Twelve healthy young men (mean age 22.3 ± 1.0) participated in this study ( Table 1 ). The decision to include only men in this investigation was made primarily because we wanted in these initial studies to minimize possible confounding effects related to gender (e.g., menstrual cycle hormones, often difficult to time precisely, that are known to influence stress and inflammatory responses). Elite athletes, individuals who participated vigorously in competitive sports and anyone with a history of any chronic medical conditions or medication use were excluded from participation. The Institutional Review Board at the University of California Irvine approved the study, and written informed consent was obtained from all participants upon enrollment. Note that 11 of these individuals also participated in our earlier study on neutrophil responses to exercise. 35
Table 1.
Anthropometric characteristics and exercise responses of the 12 subjects.
| Age (years) | 22.3 ± 1.0 |
| Height (cm) | 173.1 ± 2.8 |
| Body Mass (kg) | 74.5 ± 3.3 |
| BMI (kg/m2) | 24.8 ± 0.9 |
| Peak VO2 (mL/kg/min) | 42.7 ± 1.9 |
| Average VO2 during the constant work rate as percent of peak VO2 | 76.4±2.1 |
| Lymphocytes (% change from before to after exercise, p = 0.001) | 39.5 ± 9.2 |
| Monocytes (% change from before to after exercise, p = 0.001) | 30.7 ± 12.8 |
Values are means ± SE. BMI = body mass index; Peak VO2 = peak oxygen uptake.
Anthropometric measurements
Standard calibrated scales and stadiometers were used to determine height and body mass.
Measurement of fitness
Each subject performed a ramp‐type progressive cycle ergometer exercise test using the SensorMedics metabolic system (Ergoline 800S, Yorba Linda, CA, USA). After sitting comfortably without pedaling (“resting”) on the cycle ergometer for 3‐minutes and 1‐minutes of unloaded pedaling, the work rate (WR) was incremented at 20–30 watts/min to the limit of the subject's tolerance. Subjects were vigorously encouraged during the high‐intensity phases of the exercise protocol. Gas exchange was measured breath‐by‐breath and the anaerobic (lactate) threshold and peak ˙VO2 were calculated using standard methods.
Exercise protocol
At least 48 hours, but not exceeding 10 days following the completion of the ramp test, each subject performed 20 minutes of exercise consisting of ten 2‐minute bouts of constant WR cycle ergometry, with 1‐minute rest interval between each bout of exercise (i.e., a 30‐minute interval). The participants were instructed not to perform any type of vigorous physical activity 48 hours prior to the exercise challenge. The WR was individualized for each subject and was calculated to be equivalent to the WR corresponding roughly to 50% of the WR between the anaerobic threshold and the peak oxygen uptake (as determined noninvasively from the ramp‐type test). On average, this WR was equivalent to 76% of the participants’ peak ˙VO2.
Blood sampling and analysis
Participants arrived at the laboratory fasting between 7:30 and 8:30 am. An indwelling catheter was inserted into the antecubital vein. A baseline sample was taken 30 minutes after the placement of the catheter and before the onset of exercise. We waited 30 minutes to ensure that measurable physiological parameters of stress (e.g., heart rate and blood pressure) were at baseline levels. Subjects then completed the 30 minutes of intermittent exercise and additional blood samples were obtained immediately after exercise. Complete blood counts for white blood cell analysis were obtained by standard methods from the clinical hematology laboratory.
PBMC isolation
PBMCs were isolated from EDTA‐treated peripheral blood using OptiPrep® Density Gradient Medium (Sigma‐Aldrich, St. Louis, MO, USA). The duration from blood draw to stabilization of RNA never exceeded 60 minutes.
RNA extraction
Total RNA was extracted using TRIzol® (Gibco BRL Life Technologies, Rockville, MD, USA). RNA pellets were resuspended in diethyl pyrocarbonate‐treated water. RNA integrity was assessed (prior to beginning target processing) using Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Palo Alto, CA, USA). We used for analysis only samples with RNA Integrity Number [RNI] > 9.2).
MicroRNA microarrays
One hundred nanograms of total RNA was labeled with the fluorescent dye Cyanine 3‐pCp (Cy3) using the microRNA Labeling Reagent and Hybridization Kit (Agilent Technologies) following the manufacturer's protocol. Cy3‐labeled RNA from each sample was hybridized to Agilent Human microRNA Version 2 Microarray. The hybridized array was then washed and scanned according to Agilent specifications and data was extracted from the scanned image using Feature Extraction version 10.2 (Agilent Technologies).
The results were analyzed using GeneSpring GX 10.0.2 Software (Agilent Technologies, Inc). All raw signal values lower than 1 were adjusted to 1 and normalized using percentile shift (90th percentile). Only entities that had a present or marginal flag and passed the 20‐percentile filtration in at least 100% of values in any one out of the two conditions were selected for further analysis. Overall, 236 out of 821 entities represented on the array met these criteria. The microRNA data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE28745 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28745). Traditional Student's paired t‐test was first applied to each probe set and fold change (FC, increase or decrease) >1.2 and false discovery rate (FDR < 0.05; Benjamini–Hochberg) procedure was carried out.
The final list of significantly changed microRNAs was then additionally analyzed using TargetScan database, release 5.1 (provided by GeneSpring) http://www.targetscan.org/. We looked for the predicted target genes for each microRNA (context percentile = 50, conserved database). Briefly, TargetScan predicts gene targets of microRNAs by searching for the presence of conserved 8mer and 7mer sites that match the seed region (positions 2–7 of a mature microRNA) of each microRNA.
An “intersecting” analysis of microRNA and gene expression level
To identify genes and pathways that were potentially targeted specifically by microRNAs that had been altered by exercise, we simultaneously analyzed the microRNA target genes and the 1,246 genes whose expression was shown previously to be affected by the same exercise protocol in late pubertal boys (mean age 17.4 ± 0.4; Figure 1 , Table 3 ). 33 We then performed pathway analysis in this overlapping gene set that included microRNAs and their gene targets. We present only pathways with EASE score # 0.05. (n.b., EASE score is a modified Fisher Exact P‐value in the DAVID system used for gene‐enrichment analysis. An EASE score P‐value = 0 represents perfect enrichment; P‐value # 0.05 was considered as significant gene‐enrichment in a specific annotation category.) (http://david.abcc.ncifcrf.gov/helps/functional_annotation.htmlsummary).
Figure 1.
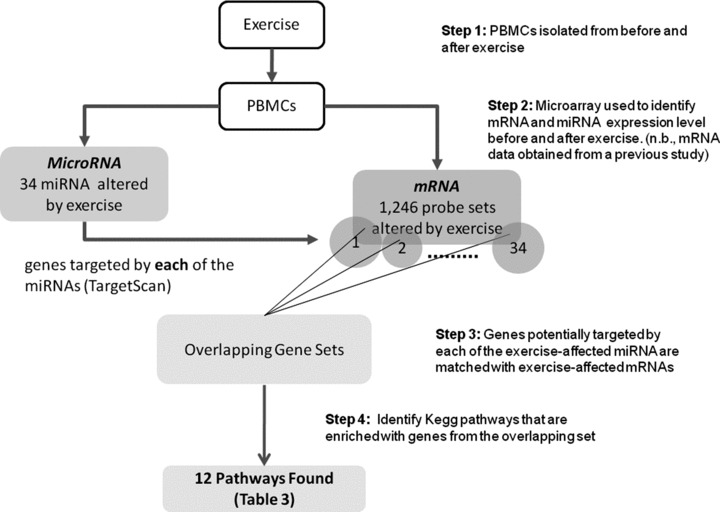
An intersecting analysis of the specific PBMC microRNAs and genes whose expression was significantly altered by exercise and genes whose expression was also significantly altered by exercise. This approach identified 12 significant pathways, and provided direction for further investigation of how microRNAs altered by exercise regulate immune function.
Table 3.
Summary of key findings: association between microRNA and gene expression in response to brief exercise in human PBMCs and gene pathways that were significantly affected.
| microRNA | Fold change | Regulation | Predicted target genes (number) | Genes affected by exercise: (1) known to be targeted by the specific miRNA; and (2) whose expression level was altered by exercise in our previous study | Common genes enrich in Kegg pathways EASE < 0.05 |
|---|---|---|---|---|---|
| hsa‐miR‐132 | 1.3 | Up | 219 Target genes | 26 Gene Symbols: for example, ADCY3, MAPK1, PPP2R5E, PPP2R5C, MAPK1, E2F5, SMAD5, MAPK1 | Oocyte meiosis, TGF‐beta signaling pathway |
| hsa‐miR‐140‐5p | 1.3 | Up | 193 Target genes | 18 Gene Symbols: for example, MAPK1, TGFBR1, YES1 | Adherens junction |
| hsa‐miR‐181a | 1.4 | Up | 661 Target genes | 76 Gene Symbols: for example, E2F5, SMAD7, MAPK1, TGFBR1, DUSP10, DUSP5, PPM1B, TGFBR1, CBLB, TGFBR1 | TGF‐beta signaling pathway, MAPK signaling pathway, chronic myeloid leukemia, pathways in cancer, colorectal cancer |
| hsa‐miR‐181b | 1.7 | Up | 652 Target genes | 76 Gene Symbols: for example, E2F5, SMAD7, MAPK1, TGFBR1, DUSP10, DUSP5, PPM1B, TGFBR1, CBLB, TGFBR1 | TGF‐beta signaling pathway, MAPK signaling pathway, chronic myeloid leukemia, pathways in cancer, colorectal cancer |
| hsa‐miR‐181c | 1.2 | Up | 662 Target genes | 76 Gene Symbols: for example, E2F5, SMAD7, MAPK1, TGFBR1, BCL2, CBLB, RASSF1, ITGA6, DUSP10, DUSP5, PPM1B, CBLB | TGF‐beta signaling pathway, MAPK signaling pathway, chronic myeloid leukemia, pathways in cancer, colorectal cancer |
| hsa‐miR‐7 | 1.4 | Up | 266 Target genes | 22 Gene Symbols: for example, ADCY9, ATP2B4, CAMK2D, SKP1, TCF4 | Oocyte meiosis, calcium signaling pathway |
| hsa‐miR‐130a | 1.2 | Down | 605 Target genes | 62 Gene Symbols: for example, EREG, ITPR1, MAPK1, NR3C2, PDK1, PRKACB, SKP1, SMAD5, TGFA | Oocyte meiosis, long‐term potentiation, TGF‐beta signaling pathway, ErbB signaling pathway, gap junction, GnRH signaling pathway |
Physiological data
The physiological data are presented as mean and standard error (SE). The two‐sided paired t‐test was applied for testing changes from before to after the exercise and the significance level was set at 0.05.
Quantitative real‐time polymerase chain reaction (RT‐PCR)
For confirmation of microRNA microarray expression findings, TaqMan assays were carried out on a subset of microRNAs. All of them are relevant to the immune system; miR‐15a, 181a, and 363 were up‐regulated and miR‐451 was down‐regulated.
Reverse transcriptase reactions were carried out using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Inc., Foster City, CA, USA) according to manufacturer's instructions. Briefly, each RT reaction contained 0.25 mM each dNTPs, 3.33 U/μL MultiScribe Reverse Transcriptase, 1X Reverse Transcription Buffer, 0.25 U RNase Inhibitor, water, 10 ng total RNA and microRNA‐specific RT primer. The 15 μL reactions were incubated in a 96 well plate for 30 minutes at 16°C, 30 minutes at 42°C, 5 minutes at 85°C, and then held at 4°C in a Techne TC‐512 thermal cycler (Burlington, NJ, USA).
RT‐PCR was performed using a standard TaqMan PCR protocol on an Applied Biosystems 7900HT Sequence Detection System. Each TaqMan reaction contained 1X TaqMan microRNA assay (primers/probe) specific to the microRNA of interest, a 1:15 dilution of the RT product for that microRNA of interest, 1X Universal MasterMix (no AmpErase UNG), and water to 20 μL. The reactions were carried out in a 96 well plate at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. All reactions were run in duplicate. The cycle threshold for each sample was determined using SDS software version 2.3 (Applied Biosystems) and was normalized to endogenous control microRNA RNU44.
Results
Anthropometric and physiological characteristics
The anthropometric and physiological characteristics of the 12 subjects appear in Table 1 .
Plasma Lactate
The exercise bout caused a mean increase of 6.3 ± 0.5 mmol/L in lactate levels in the plasma (before [1.7 ± 0.3 mmol/L] versus after [7.9.0 ± 0.3 mmol/L], p< 0.004).
Leukocyte response to exercise
As shown in Table 1 , the number of lymphocytes and monocytes was significantly elevated at peak exercise (p < 0.001).
The Effects of exercise on PBMC microRNA expression
Thirty minutes of intermittent exercise (i.e., the ten 2‐minute bouts) altered the expression level of 34 microRNAs (15 had higher expression, Table 2 ). In silco analysis showed that 31 microRNAs had potentially target genes (3 microRNAs didn't have any known target genes; Table S1).
Table 2.
Thirty‐four microRNAs were altered by exercise (FDR < 0.05).
| microRNA | Fold change | Regulation |
|---|---|---|
| hsa‐miR‐181a‐2* | 2.0 | Up |
| hsa‐miR‐181b | 1.7 | Up |
| hsa‐miR‐181a | 1.4 | Up |
| hsa‐miR‐363 | 1.5 | Up |
| hsa‐miR‐181c | 1.2 | Up |
| hsa‐miR‐338‐3p | 1.4 | Up |
| hsa‐miR‐1225‐5p | 1.5 | Up |
| hsa‐miR‐26b | 1.2 | Up |
| hsa‐miR‐132 | 1.3 | Up |
| hsa‐miR‐15a | 1.3 | Up |
| hsa‐miR‐939 | 1.3 | Up |
| hsa‐miR‐7 | 1.4 | Up |
| hsa‐miR‐140‐5p | 1.3 | Up |
| hsa‐miR‐21* | 1.5 | Up |
| hsa‐miR‐940 | 1.3 | Up |
| hsa‐miR‐125b | 1.4 | Down |
| hsa‐let‐7e | 1.4 | Down |
| hsa‐miR‐451 | 3.8 | Down |
| hsa‐miR‐320 | 1.2 | Down |
| hsa‐miR‐151‐5p | 1.3 | Down |
| hsa‐miR‐486‐5p | 1.7 | Down |
| hsa‐miR‐31 | 1.3 | Down |
| hsa‐miR‐125a‐5p | 1.3 | Down |
| hsa‐miR‐99b | 1.4 | Down |
| hsa‐miR‐652 | 1.2 | Down |
| hsa‐miR‐151‐3p | 1.3 | Down |
| hsa‐miR‐130a | 1.2 | Down |
| hsa‐miR‐126 | 1.3 | Down |
| hsa‐miR‐199b‐3p | 1.3 | Down |
| hsa‐miR‐23b | 1.2 | Down |
| hsa‐miR‐221 | 1.2 | Down |
| hsa‐miR‐199a‐5p | 1.3 | Down |
| hsa‐miR‐584 | 1.3 | Down |
| hsa‐miR‐145 | 1.3 | Down |
Intersecting analysis of microRNA and gene expression level
In Table 3 we present 7 microRNAs, and we show: (1) microRNAs that were affected by exercise; (2) the number of potentially targeted genes for each affected microRNA (in silico analysis); and (3) from the genes identified in #2, we show those genes that we found previously to have actually been affected by exercise 33 (The complete list of all 34 microRNAs can be found in the Table S1). Pathway analysis of these sets of genes showed 12 significant pathways: for example, TGF‐beta signaling pathway, MAPK signaling pathway, Adherens junction, Gap junction, ErbB signaling pathway, and Calcium signaling pathway. Intriguingly, the TGF‐beta signaling pathway was found to be significant in 5 of the 31 microRNAs that had target genes ( Figure 1 ).
RT‐PCR verification of specific microRNAs
We used TaqMan assays to verify the expression level changes from before to after exercise of 4 microRNAs. miR‐15a (FC = 1.5, p= 0.02), 181a (FC = 1.7, p= 0.008), and 363 (FC = 2, p= 0.005) were up‐regulated and miR‐451 (FC = 1.3, p= 0.01) was down‐regulated.
Discussion
We have demonstrated that a relatively brief heavy interval‐type exercise session ( Table 1 ), one that mimics patterns of physical activity naturally found in many sport activities, is sufficient to change the expression pattern of 34 PBMC microRNAs out of the possible 826 entities currently represented on the chip. We used conservative statistical approaches (FDR < 0.05) to identify those altered microRNAs, and further corroborated the results using a different platform, RT‐PCR. The changes in PBMC microRNAs in this study, along with our previous finding in circulating neutrophils, 35 show that brief intense exercise significantly alters key epigenetic regulatory profiles of circulating leukocytes. Our analysis of individual microRNAs uncovered a number of intriguing potential exercise‐related immune mechanisms. In addition, our subsequent in silico analysis involving the microRNA data collected from this cohort of healthy young subjects and mRNA data from our previous investigations, where we used the same exercise protocol 33 further enhanced the utility of the data by specifically identifying 12 known gene pathways in which microRNA and mRNA mechanisms likely interacted. Some of these identified gene pathways are involved in processes that are currently known to link exercise with health and disease.
New insights gained from changes in individual microRNAs
Recent reports regarding a number of the microRNAs that were altered by exercise highlight potentially novel mechanisms by which physical activity influences immune cell function. For example, we found that exercise led to an increase in the expression level of miR‐181a in the PBMCs ( Table 2 ). In an elegant series of in vitro studies using T cells derived from transgenic mice, Li et al. 18 demonstrated that increasing miR‐181a expression in mature T cells augmented the T cell sensitivity to peptide antigens by a posttranscriptional regulation of multiple phosphatases, and, ultimately, T‐cell receptor signaling threshold. Perhaps the greater T‐cell responsiveness and simultaneous reduced susceptibility to infection observed in physically active compared with sedentary elderly adults 40 may ultimately prove to be related to an exercise‐associated up‐regulation of T cell miR‐181a.
We also found that the expression of miR‐132 was up‐regulated while the expression of miR‐125b and let‐7e were down‐regulated following exercise ( Table 2 ). These three microRNAs are known to regulate toll‐like receptors (TLRs) in monocytes. 24 , 41 Moreover, miR‐125b and let‐7a were recently shown to play a key role in mediating the suppressive effect of estradiol on TNF‐α production in human primary macrophages. Activation of macrophages with LPS leads to changes in the expression of miRNAs that regulate molecules involved in TLR signaling. 22 Oliveira and Gleeson 27 recently showed that prolonged cycling at 75% VO2 peak temporarily reduces monocyte TLR4 expression. They speculated that this downregulation might explain the phenomenon of acute postexercise immunodepression. The role of microRNAs in exercise associated TLR regulation, particularly with reference to factors such as exercise dose, gender, and maturation, remain unexplored.
Additional examples in which microRNAs that we found to have been altered by exercise might influence immune cell function include the recent discoveries on the roles of mir‐31 in T‐regulatory lymphocytes 38 and mir‐21 in down‐regulating DNA methylation in CD4 + T cells. 29 miR‐132 and miR‐125b (noted earlier) are overproduced in monocytes exposed to bacterial endotoxin, lipopolysaccharide (LPS), or TNF‐α. 43 mir‐125a and mir‐132 both appear to play a role in diseases whose natural history and pathogenesis are altered by exercise, namely, systemic lupus erythematosis, and rheumatoid arthritis. 9 It seems safe to speculate that this discovery that brief exercise alters a number of individual microRNA expression profiles in circulating PBMCs will lead to new hypothesis on the molecular mechanisms that link physical activity with health.
New insights gained from intersecting gene pathway analysis
Our in silico intersecting analysis of gene expression (mRNA) PBMC responses to exercise (as noted previously using the same exercise protocol), combined with the discoveries of microRNA PBMC responses to exercise in this study, also lead to novel insights into mechanisms that link physical activity with health. As shown in Table 3 , we identified 12 pathways. A number of these gene regulatory pathways can be potentially linked to exercise in both immune and nonimmune tissues. For example, Hoene and Weigert 15 recently demonstrated activation of the MAPK pathway in the liver following vigorous exercise in rodents. TGF‐β proteins (TGF Signaling Pathway) are known to play a role in both muscle and tendon adaptations in response to exercise. 17 , 20 In addition, TGF‐β, secreted by monocytes/macrophages in the lung, contributes to the mechanisms of lung remodeling 7 that accompanies repeated asthma attacks. It is known that exercise stimulates circulating levels of both pro‐ and antiinflammatory cytokines. 23 , 31 The extent to which microRNAs in PBMCs are involved in these mechanisms remains unknown.
Other PBMC exercise‐affected pathways are known to play a central role in immune cell communication. For example, Bopp et al. 6 discuss in their review the mechanism by which T‐reg cells and conventional CD4 + T cells communicate via gap junctions. These investigators support the finding of Ring et al. 36 showing that T‐reg cells not only modulate ongoing CD4(+)T cell‐mediated immune reactions at tissue sites, but also abrogate the de novo induction of CD8(+)T cell‐driven immune reactions by interfering with T‐cell stimulatory activity of dendritic cells through gap junctional intercellular communication, another pathway that we identify in our analysis.
Calcium signaling pathway was also identified in this analysis. Ca2 + signals regulate the activation of lymphocytes, their differentiation, and multiple effector functions as well as a variety of transcriptional programs. 25 After the engagement of immunoreceptors, such as T‐cell and B‐cell antigen receptors and the Fc receptors on mast cells and NK cells, the intracellular concentration of calcium ions is increased through the sequential operation of two interdependent processes. 26 Finally, a number of cancer‐related pathways were found to be involved in the microRNA–PBMC response to exercise. There is growing interest in the preventive effects of physical fitness in a variety of cancers, but the mechanisms of this effect remain enigmatic. 44 Simultaneously, microRNAs have become a target of intense investigation in cancer pathogenesis. 11 Our data suggest a possible connection between exercise alteration of PBMC microRNA profiles and the role of physical activity in modulation of malignancy.
Comparing microRNA responses to exercise in PBMCs and neutrophils
Our study permitted us to gain some initial perspectives on the patterns of microRNA response to exercise in different types of circulating leukocytes. As shown in Figure 2 , there was little overlap between the microRNAs that changed in response to the same exercise challenge in neutrophils (data from our earlier study 35 consisting of largely the same individuals) and those that changed in PBMCs in the current experiments. This is not surprising; as we reviewed our previous studies on gene expression in response to exercise, we found that only about 34% of exercise‐affected genes changed similarly in both neutrophils and PBMCs. Moreover, the intersecting analysis performed in neutrophils identified only three pathways, also distinctly different from the pathways we found in PBMCs using this in silico approach in this study. Thus, although brief exercise leads to an increase in the vast majority of leukocytes in the circulation, our data suggest that the ultimate genomic and epigenetic profile of the circulating cells depends to some extent on the type of leukocyte that is either directly or indirectly altered by exercise.
Figure 2.
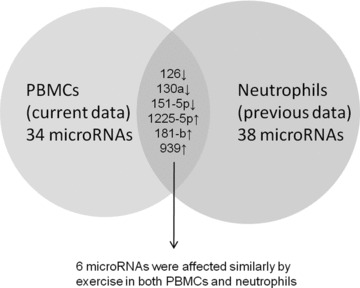
Comparison of microRNAs altered by exercise in PBMCs (this study) with those changed in neutrophils (earlier study). Of the 34 PBMC microRNAs, only six changed in the same manner in neutrophils as well.
Study challenges and limitations
Although hypothesis driven, this was an exploratory study. Because we did not a priori know the optimal conditions for isolating and identifying microRNAs from circulating PBMCs, we elected to focus our sample collection and preparation methods in a way that would optimize microRNA recovery. Consequently, we did not simultaneously measure both microRNAs and mRNAs in these subjects; rather, we relied on our previous data for the intersecting analysis. We also looked only at the PBMC microRNA expression changes immediately after the exercise. In future studies we plan to study more points in time and determine the duration of exercise‐induced effects on microRNA expression. We treated the PBMCs as one group of cells, and it will remain for future studies to determine whether the individual components of this heterogeneous collection of lymphocytes, monocytes, etc. reflect the overall microRNA response. Finally, this study was not designed to answer a key question that arises whenever one collects cells from the circulating blood—namely, are genomic or epigenetic changes observed in the circulating cells the direct result of exercise on those cells or, alternatively, do the differences derive from shifting genomically distinct populations of leukocytes as they migrate from marginal pools into the central circulation in response to exercise. 1 Nonetheless, either of these direct or indirect possibilities suggests an elegant, precise, and as of yet poorly understood mechanism that links exercise with immune cell function.
Summary
This study shows for the first time that exercise leads to an altered profile of microRNAs in PBMCs. The results complement our earlier findings in neutrophils. Many of the individual microRNAs that we found to be influenced by exercise are already known to play key mechanistic roles in regulating inflammatory function of PBMCs such as lymphocytes and monocytes. Moreover, the gene pathways likely to be influenced by microRNAs that we identified through our intersecting in silico analysis were ones that in some cases are clearly related to the broad adaptive mechanisms that link exercise with health and disease. These data in healthy human beings, along with the explosion of knowledge focused on microRNAs as “fine‐tuners” of immune function in diseases ranging from cancer to diabetes, 39 , 42 justify further exploration of the impact of exercise on microRNA regulation in each of the leukocyte subpopulations that comprise the PBMCs.
Acknowledgment
We thank Ms. Cheryl Nugas, and Ms. Georgia Bachman for their technical assistance. This work was supported in part by National Institutes of Health grants P01HD‐048721, and the UCI Institute for Clinical and Translational Science (CTSA grant) UL1RR031985.
Supporting information
Table S1. Association between microRNA and gene expression in response to brief exercise in human PBMCs and gene pathways that were significantly affected.
Supporting info item
Grants
References
- 1. Adams G, Zaldivar F, Nance D, Radom‐Aizik S, Kodesh E, Cooper DM. Exercise and leukocyte interchange among central circulation, lung, spleen, and muscle. Brain Behav Immun. 2011; 25(4): 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez‐Saavedra M, Antoun G, Yanagiya A, Oliva‐Hernandez R, Cornejo‐Palma D, Perez‐Iratxeta C, Sonenberg N, Cheng HY. miRNA‐132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011; 20: 731–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asirvatham AJ, Magner WJ, Tomasi TB. miRNA regulation of cytokine genes. Cytokine. 2009; 45: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008; 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008; 9: 839–845. [DOI] [PubMed] [Google Scholar]
- 6. Bopp T, Radsak M, Schmitt E, Schild H. New strategies for the manipulation of adaptive immune responses. Cancer Immunol Immunother. 2010; 59: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bottoms SE, Howell JE, Reinhardt AK, Evans IC, McAnulty RJ. Tgf‐Beta isoform specific regulation of airway inflammation and remodelling in a murine model of asthma. PLoS One. 2010; 5: e9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brannan JD, Turton JA. The inflammatory basis of exercise‐induced bronchoconstriction. Phys Sportsmed. 2010; 38: 67–73. [DOI] [PubMed] [Google Scholar]
- 9. Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011; 157: 163–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duursma AM, Kedde M, Schrier M, le SC, Agami R. miR‐148 targets human DNMT3b protein coding region. RNA. 2008; 14: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011; 223: 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franco M, Cooper RS, Bilal U, Fuster V. Challenges and opportunities for cardiovascular disease prevention. Am J Med. 2011; 124: 95–102. [DOI] [PubMed] [Google Scholar]
- 13. Garbacki N, Di VE, Huynh‐Thu VA, Geurts P, Irrthum A, Crahay C, Arnould T, Deroanne C, Piette J, Cataldo D, et al MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS One. 2011; 6: e16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, van Berkel TJ, Biessen EA. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010; 394: 792–797. [DOI] [PubMed] [Google Scholar]
- 15. Hoene M, Weigert C. The stress response of the liver to physical exercise. Exerc Immunol Rev. 2010; 16: 163–183. [PubMed] [Google Scholar]
- 16. Katz DL, Cushman D, Reynolds J, Njike V, Treu JA, Walker J, Smith E, Katz C. Putting physical activity where it fits in the school day: preliminary results of the ABC (Activity Bursts in the Classroom) for fitness program. Prev Chronic Dis. 2010; 7: A82. [PMC free article] [PubMed] [Google Scholar]
- 17. Kjaer M, Langberg H, Heinemeier K, Bayer ML, Hansen M, Holm L, Doessing S, Kongsgaard M, Krogsgaard MR, Magnusson SP. From mechanical loading to collagen synthesis, structural changes and function in human tendon. Scand J Med Sci Sports. 2009; 19: 500–510. [DOI] [PubMed] [Google Scholar]
- 18. Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR‐181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007; 129: 147–161. [DOI] [PubMed] [Google Scholar]
- 19. Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008; 29: 343–351. [DOI] [PubMed] [Google Scholar]
- 20. Matsakas A, Patel K. Skeletal muscle fibre plasticity in response to selected environmental and physiological stimuli. Histol Histopathol. 2009; 24: 611–629. [DOI] [PubMed] [Google Scholar]
- 21. Matzinger P. Friendly and dangerous signals: is the tissue in control? Nat Immunol. 2007; 8: 11–13. [DOI] [PubMed] [Google Scholar]
- 22. Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF‐kappa B activation through coordinated regulation of let‐7a and miR‐125b in primary human macrophages. J Immunol. 2010; 184: 5029–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nemet D, Oh Y, Kim HS, Hill MA, Cooper DM. The effect of intense exercise on inflammatory cyotkines and growth mediators in adolescent boys. Pediatrics. 2002; 110: 681–689. [DOI] [PubMed] [Google Scholar]
- 24. O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine‐tuners of Toll‐like receptor signalling. Nat Rev Immunol. 2011; 11: 163–175. [DOI] [PubMed] [Google Scholar]
- 25. Oh‐hora M. Calcium signaling in the development and function of T‐lineage cells. Immunol Rev. 2009; 231: 210–224. [DOI] [PubMed] [Google Scholar]
- 26. Oh‐hora M, Rao A. Calcium signaling in lymphocytes. Curr Opin Immunol. 2008; 20: 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oliveira M, Gleeson M. The influence of prolonged cycling on monocyte Toll‐like receptor 2 and 4 expression in healthy men. Eur J Appl Physiol. 2010; 109: 251–257. [DOI] [PubMed] [Google Scholar]
- 28. Ostrowski K, Rohde T, Asp S, Schjerling P, Pedersen BK. Pro‐ and anti‐inflammatory cytokine balance in strenuous exercise in humans. J Physiol. 1999; 515 (Pt 1): 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA‐21 and microRNA‐148a contribute to DNA hypomethylation in lupus CD4 + T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010; 184: 6773–6781. [DOI] [PubMed] [Google Scholar]
- 30. Pedersen BK, Febbraio MA. Muscle as an endocrine organ: focus on muscle‐derived interleukin‐6. Physiol Rev. 2008; 88: 1379–1406. [DOI] [PubMed] [Google Scholar]
- 31. Pedersen BK, Hoffman‐Goetz L. Exercise and the immune system: regulation, integration, and adaptation. Physiol Rev. 2000; 80: 1055–1081. [DOI] [PubMed] [Google Scholar]
- 32. Radom‐Aizik S, Zaldivar F, Jr. , Leu SY, Cooper DM. A brief bout of exercise alters gene expression and distinct gene pathways in peripheral blood mononuclear cells of early‐ and late‐pubertal females. J Appl Physiol. 2009; 107: 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radom‐Aizik S, Zaldivar F, Jr. , Leu SY, Cooper DM. Brief bout of exercise alters gene expression in peripheral blood mononuclear cells of early‐ and late‐pubertal males. Pediatr Res. 2009; 65: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Radom‐Aizik S, Zaldivar F, Jr. , Leu SY, Galassetti P, Cooper DM. Effects of 30 min of aerobic exercise on gene expression in human neutrophils. J Appl Physiol. 2008; 104: 236–243. [DOI] [PubMed] [Google Scholar]
- 35. Radom‐Aizik S, Zaldivar F, Jr. , Oliver S, Galassetti P, Cooper DM. Evidence for microRNA involvement in exercise‐associated neutrophil gene expression changes. J Appl Physiol. 2010; 109: 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ring S, Karakhanova S, Johnson T, Enk AH, Mahnke K. Gap junctions between regulatory T cells and dendritic cells prevent sensitization of CD8(+) T cells. J Allergy Clin Immunol. 2010; 125: 237–246. [DOI] [PubMed] [Google Scholar]
- 37. Robson‐Ansley P, Toit GD. Pathophysiology, diagnosis and management of exercise‐induced anaphylaxis. Curr Opin Allergy Clin Immunol. 2010; 10: 312–317. [DOI] [PubMed] [Google Scholar]
- 38. Rouas R, Fayyad‐Kazan H, El ZN, Lewalle P, Rothe F, Simion A, Akl H, Mourtada M, El RM, Burny A, et al. Human natural Treg microRNA signature: role of microRNA‐31 and microRNA‐21 in FOXP3 expression. Eur J Immunol. 2009; 39: 1608–1618. [DOI] [PubMed] [Google Scholar]
- 39. Sebastiani G, Vendrame F, Dotta F. MicroRNAs as new tools for exploring type 1 diabetes: relevance for immunomodulation and transplantation therapy. Transpl. Proc. 2011; 43: 330–332. [DOI] [PubMed] [Google Scholar]
- 40. Simpson RJ, Guy K. Coupling aging immunity with a sedentary lifestyle: has the damage already been done?–a mini‐review. Gerontology. 2010; 56: 449–458. [DOI] [PubMed] [Google Scholar]
- 41. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF‐kappaB‐dependent induction of microRNA miR‐146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006; 103: 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thai TH, Christiansen PA, Tsokos GC. Is there a link between dysregulated miRNA expression and disease? Discov Med. 2010; 10: 184–194. [PubMed] [Google Scholar]
- 43. Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al Modulation of miR‐155 and miR‐125b levels following lipopolysaccharide/TNF‐alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007; 179: 5082–5089. [DOI] [PubMed] [Google Scholar]
- 44. Walsh NP, Gleeson M, Shephard RJ, Gleeson M, Woods JA, Bishop NC, Fleshner M, Green C, Pedersen BK, Hoffman‐Goetz L, et al. Position statement. Part one: immune function and exercise. Exerc Immunol Rev. 2011; 17: 6–63. [PubMed] [Google Scholar]
- 45. Zaldivar F, Wang‐Rodriguez J, Nemet D, Schwindt C, Galassetti P, Mills PJ, Wilson LD, Cooper DM. Constitutive pro‐ and anti‐inflammatory cytokine and growth factor response to exercise in leukocytes. J Appl Physiol. 2006; 100: 1124–1133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Association between microRNA and gene expression in response to brief exercise in human PBMCs and gene pathways that were significantly affected.
Supporting info item


