SUMMARY
SUMOylation, the covalent attachment of a member of the SUMO (small ubiquitin-like modifier) family of proteins to lysines in target substrates, is an essential post-translational modification in eukaryotes. Microbial manipulation of SUMOylation recently emerged as a key virulence strategy for viruses and facultative intracellular bacteria, the latter of which only have been shown to deploy effectors that negatively regulate SUMOylation. Here, we demonstrate that the obligate intracellular bacterium, Anaplasma phagocytophilum, utilizes an effector, AmpA (A. phagocytophilum post-translationally modified protein A) that becomes SUMOylated in host cells and this is important for the pathogen’s survival. We previously discovered that AmpA (formerly APH1387) localizes to the A. phagocytophilum-occupied vacuolar membrane (AVM). Algorithmic prediction analyses denoted AmpA as a candidate for SUMOylation. We verified this phenomenon using a SUMO-affinity matrix to precipitate both native AmpA and ectopically expressed green fluorescent protein (GFP)-tagged AmpA. SUMOylation of AmpA was lysine-dependent, as SUMO-affinity beads failed to precipitate a GFP-AmpA protein when its lysine residues were substituted with arginine. Ectopically expressed and endogenous AmpA were poly-SUMOylated, which was consistent with the observation that AmpA colocalizes with SUMO2/3 at the AVM. Only late during the infection cycle did AmpA colocalize with SUMO1, which terminally caps poly-SUMO2/3 chains. AmpA was also detected in the cytosol of infected host cells, further supporting its secretion and likely participation in interactions that aid pathogen survival. Indeed, whereas siRNA-mediated knockdown of Ubc9 – a necessary enzyme for SUMOylation – slightly bolstered A. phagocytophilum infection, pharmacologically inhibiting SUMOylation in infected cells significantly reduced the bacterial load. Ectopically expressed GFP-AmpA served as a competitive agonist against native AmpA in infected cells, while lysine-deficient GFP-AmpA was less effective, implying that modification of AmpA lysines is important for infection. Collectively, these data show that AmpA becomes directly SUMOylated during infection, representing a novel tactic for A. phagocytophilum survival.
INTRODUCTION
Obligate intracellular bacterial pathogens are master cell biologists that utilize effector proteins to commandeer host cell pathways and reprogram the host cell environment to one that favors pathogen persistence and replication. Functionally characterizing such effectors is critical as it can lead to a better understanding of how these organisms facilitate their intracellular survival. Anaplasma phagocytophilum, a member of the family Anaplasmataceae, is a tick-transmitted obligate intracellular gram-negative bacterium that causes human granulocytic anaplasmosis (HGA), an emerging infection in the United States, Europe, and Asia. HGA presents as a febrile disease that can be accompanied by leukopenia, thrombocytopenia, elevated levels of serum transaminases, and raised susceptibility to potentially fatal secondary infections (Truchan et al., 2013). In vivo, the bacterium displays an unusual tropism for neutrophils, but in cell culture, promyelocytic and endothelial cell lines are useful models for studying the cell biology of A. phagocytophilum infection (Carlyon, 2012).
Following invasion, A. phagocytophilum resides within a host cell-derived vacuole that it actively remodels into a permissive niche (Truchan et al., 2013). Of the few identified bacterial effectors that localize to the A. phagocytophilum-occupied vacuolar membrane (AVM), only two – Ats1 (Anaplasma translocated substrate 1) and AptA (A. phagocytophilum toxin A) – have been ascribed functions (Huang et al., 2010b, Huang et al., 2010c, Sukumaran et al., 2011, Niu et al., 2010, Niu et al., 2012). We previously identified APH1387 as a 61.4-kDa effector that is induced within the initial hours of A. phagocytophilum infection and is strongly expressed during the bacterium’s intracellular replication phase. It has been detected on the AVM in neutrophils recovered from A. phagocytophilum infected mice as well as infected myeloid, endothelial, and tick cell lines (Huang et al., 2010c). Collectively, these phenomena imply the importance of the effector to A. phagocytophilum survival within diverse eukaryotic host cell types and warrant its further investigation. APH1387 carries three tandemly arranged direct repeats in its C-terminal portion at amino acids 180 to 272, 304 to 425, and 428 to 557 that together comprise 58% of the protein (Huang et al., 2010c). APH1387 has been referred to as P100 because when A. phagocytophilum infected whole cell lysates are resolved by SDS-PAGE, it migrates predominantly as a band having an apparent molecular weight of approximately 100 to 115 kDa along with less abundant, lower molecular weight bands that range from approximately 90 to 61 kDa (Storey et al., 1998, Huang et al., 2010c). Its aberrant electrophoretic migration pattern has been attributed to its acidic pI of 3.67. Other acidic proteins, such as the tandem repeat-containing proteins (TRPs) of the closely related Anaplasmataceae pathogen, Ehrlichia chaffeensis, have been shown to poorly bind SDS and, consequently, migrate with apparent molecular weights that are considerably higher than their actual molecular weights (Wakeel et al., 2010). Notably, however, recombinant APH1387 expressed in Escherichia coli migrates only as a single band (Huang et al., 2010c). The observed discrepancy that multiple isoforms are detected in lysates of A. phagocytophilum infected cells, but not in lysates of E. coli expressing recombinant APH1387 suggests that A. phagocytophilum-derived APH1387 may be post-translationally modified by the host cell during infection. Whether this occurs and, if so, the nature of the modification and its overall significance to the pathogen are unknown.
SUMOylation, the covalent attachment of a member of the SUMO (small ubiquitin-like modifier) family of proteins to lysine residues in targeted proteins, is an essential post-translational protein modification for all eukaryotic cells (Wilkinson et al., 2010). SUMOylation is a reversible process that is enzymatically analogous to, but functionally distinct from, the ubiquitination pathway. Over the past several years, this pathway has emerged as a major regulatory system of protein function that targets hundreds to thousands of substrates through direct SUMOylation and protein-protein interactions mediated by SUMOylation (Wilson, 2012). The wide range of cellular processes that SUMOylation modulates includes RNA processing, chromatin remodeling, genome maintenance, transcriptional regulation, mitosis, meiosis, differentiation, apoptosis, nucleocytoplasmic transport, regulation of ion channel activity, metabolic pathways, endocytic trafficking of receptors, and resistance to pathogens (Wilson, 2012, Wilkinson et al., 2010).
A SUMOylation consensus motif, ΨKxD/E, has been described where Ψ is a large hydrophobic residue and x represents any amino acid (Wilkinson et al., 2010). However, SUMOylation has been reported to occur outside of this consensus, and not all lysines found within such a motif are SUMO modified (Wilkinson et al., 2010). SUMO2/3 forms polymers on internal lysine residues of substrates both in vitro and in vivo (Tatham et al., 2001). While SUMO1 can directly conjugate proteins in vitro (Yang et al., 2006), in vivo it has only been observed to terminate SUMO2/3 polymers (Matic et al., 2008). The specific functional consequences of SUMOylation of individual substrates continue to be discovered and are difficult to predict. In general, SUMO modification of a target protein can result in one of three possible, even congruent, outcomes; the covalently attached SUMO moiety may function as an interface to recruit new interacting proteins to the substrate; it may prevent protein-protein interactions by occluding the binding site of a protein that would otherwise interact with the substrate; or it may cause a conformational change in the target protein that directly regulates its function (Wilkinson et al., 2010).
Given the pleiotropic effects of SUMOylation on eukaryotic cellular processes and the huge number of proteins that it regulates, it is not surprising that the manipulation of SUMOylation by microbial pathogens as a means for converting their host cells into replication-permissive niches has emerged as a key virulence strategy. It is established that viruses target host enzymes or encode their own enzymes to modulate the SUMOylation status of and thereby dysregulate host proteins involved in growth or pathogen resistance (Wilson, 2012). Also, viral proteins can be directly SUMOylated (Bekes et al., 2012, Wimmer et al., 2012). However, comparatively few examples exist of bacteria targeting SUMOylation.
Given their limited genetic repertoires, co-opting SUMOylation would be an especially advantageous strategy for obligate intracellular bacteria, as it could allow for maximal use of single effectors to commandeer batteries of host cell processes. A bacterial effector that is directly SUMOylated during infection has yet to be identified. In this study, we demonstrate that APH1387 is SUMOylated during infection, thereby revealing a novel method by which A. phagocytophilum exploits the host cell. We hereafter refer to APH1387 as AmpA (A. phagocytophilum post-translationally modified protein A).
RESULTS
In silico analysis predicts multiple AmpA SUMOylation sites
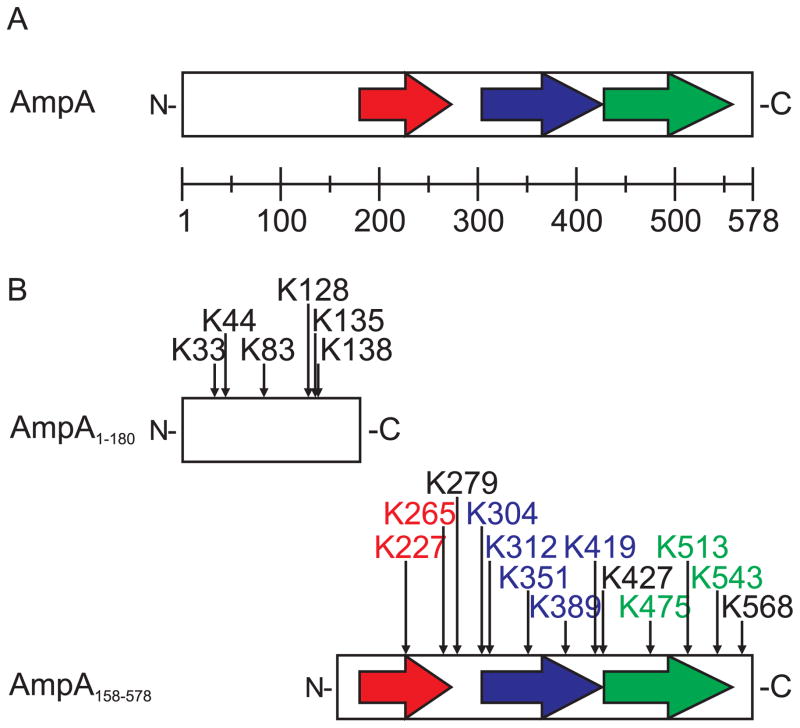
Based on our previous characterization of AmpA as a secreted effector and constituent of the AVM (Huang et al., 2010c), we examined the AmpA sequence for motifs that may implicate interactions with host cell factors. A striking feature was the abundance of lysine residues within the 578 amino acid length of AmpA: six lie within the non-repeat N-terminal region, and 13 within the tandem repeat-containing C-terminus for a total of 19 (Fig. 1). As lysines are frequent targets of SUMO post-translational modification, we examined AmpA using three SUMO prediction algorithms. Eight to 10 lysine residues were repeatedly predicted to be SUMOylated (Table 1) based on similarity to the ΨKxD/E consensus motif and to other residues known to flank SUMOylation sites (Ren et al., 2009, Teng et al., 2012). Of those predicted SUMOylation sites for AmpA, one lysine lies within the N-terminal non-repeat portion (K135) and two within the intervening spaces between and/or after tandem repeats (K427, K568). The remaining predicted SUMOylated lysines (K265, K304, K312, K389, K419, K513, and K543) are those that occur repetitively as part of the C-terminal tandem direct repeats. Because there had never been a published report of a bacterial protein being directly SUMOylated during infection, we set out to functionally verify whether AmpA is a SUMOylated effector.
Fig. 1.
Schematic diagrams of full-length A. phagocytophilum AmpA and truncated recombinant AmpA proteins used in these studies. A. Diagram of full-length AmpA. AmpA comprises an amino (N)-terminal region (amino acids 1 to 179), a tandem repeat region (amino acids 180 to 549), and a short carboxy (C)-terminal region (amino acids 550 to 578). The repeat region consists of 3 tandemly-arranged direct repeats (indicated by the colored arrows) consisting of 93 (red), 122 (blue), and 130 amino acids (green). The scale indicates 50 amino acid intervals. B. Diagrams of AmpA truncations. The AmpA C-terminal truncation AmpA1-180 (top) lacks the tandem repeat region and contains 6 lysine (K) residues within its 180 amino acid length. The AmpA N-terminal truncation AmpA158-578 (bottom) has the non-repeat region removed and contains 13 lysines within its 421 amino acid length. Each AmpA truncation was fused to GFP (not shown) on its N-terminus. Arrows above the diagram denote lysine (K) residues and their amino acid position within AmpA. Colored lysine positions coordinate to their locations within their respective tandem repeat regions (red, blue, and green arrows).
Table 1.
Predicted SUMOylation sites in AmpA
| K positiona | SUMOSp2.0b | SUMOplot | seeSUMO |
|---|---|---|---|
| 33 | |||
| 44 | |||
| 83 | |||
| 128 | |||
| 135 | Hc | H | Yd |
| 138 | |||
| 227e | |||
| 265 | H | H | Y |
| 279 | |||
| 304 | H | H | Y |
| 312 | M | H | Y |
| 351 | |||
| 389 | H | H | Y |
| 419 | L | L | Y |
| 427 | L | Y | |
| 475 | |||
| 513 | H | H | Y |
| 543 | L | L | Y |
| 568 | H | Y |
K position refers to the amino acid position in AmpA where each lysine (K) occurs.
SUMOSp2.0, SUMOplot, and seeSUMO denote the three SUMOylation prediction algorithms used to analyze AmpA.
H, high probability; M, medium probability; L, low probability, as scored by SUMOSp2.0 and SUMOplot.
Y, yes for SUMOylation; N, no for SUMOylation, as scored by seeSUMO.
Red, blue, and green colored lysine positions correspond to the first, second, and third tandem repeats, respectively, of AmpA as shown in Figure 1.
Ectopically expressed AmpA is SUMOylated on at least two lysine residues
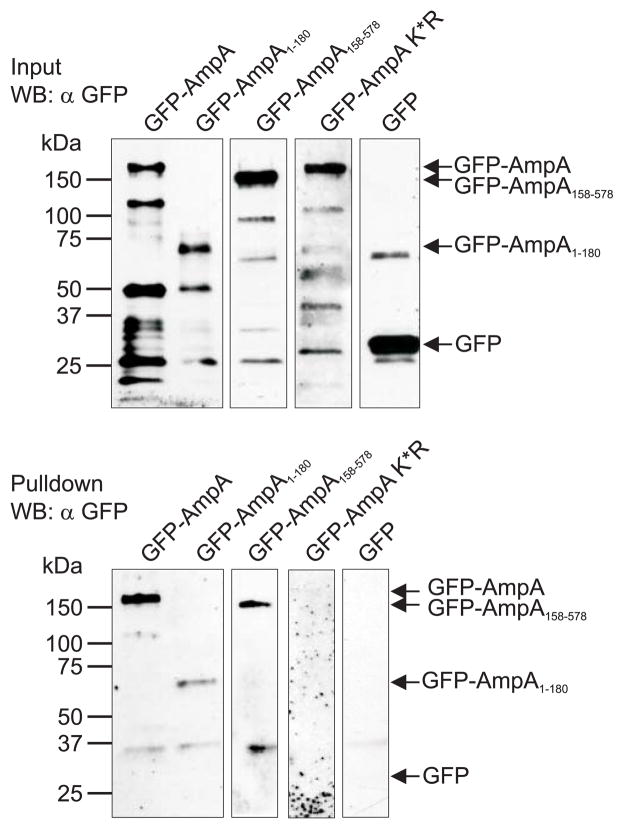
We next employed a SUMOylation pulldown system that uses SUMO interaction motifs (SIMs) coupled to beads to affinity purify mono- and poly-SUMOylated proteins from cell extracts. GFP-tagged full length AmpA (GFP-AmpA) or GFP alone was ectopically expressed in HeLa cells, which were employed for this purpose because they are much more amenable to transfection than the HL-60 and RF/6A cell lines that are commonly used to cultivate A. phagocytophilum. The host cells were lysed, incubated with the SIM-affinity beads, and captured SUMOylated proteins were screened by Western blot with GFP antibody. GFP-AmpA, but not GFP alone was precipitated despite the greater abundance of GFP alone relative to GFP-AmpA in input lysates (Fig. 2). To confirm that the nature of the AmpA pulldown was SUMO-specific, we also performed the assay using GFP-AmpA K*R in which all 19 of the AmpA lysines were substituted with arginine, which is structurally similar to lysine, but is SUMO intolerant. The SIM-affinity beads were unable to precipitate GFP-AmpA K*R (Fig. 2), thereby confirming both that the pulldown of AmpA was lysine dependent and that AmpA is SUMOylated.
Fig. 2.
Ectopically expressed AmpA is SUMOylated. SUMOylation pulldowns with ectopically expressed GFP-AmpA, GFP-AmpA1-180, GFP-Amp158-578, GFP-AmpA K*R, and GFP in HeLa cells. Western blots of protein input (top) and pulldown eluates (bottom) were screened with GFP antibody. Arrows denote the expected apparent molecular weights for each GFP-AmpA protein and the expected size for GFP. Blots are representative of 2–4 pulldowns performed for each GFP-AmpA protein.
In silico analyses predicted that up to 10 lysines within AmpA are potentially SUMOylated, with the vast majority of them occurring within the tandem repeat-containing C-terminal region (Table 1). In an attempt to narrow down which portion of ectopically expressed AmpA is SUMOylated, we created two AmpA truncations that were N-terminally fused to GFP. GFP-AmpA1-180 included the protein’s first 180 amino acids and contained six lysine residues, of which K135 was predicted as a SUMOylation site (Fig. 1B). GFP-AmpA158-578, which comprised the three C-terminal tandem repeats, harbored 13 lysine residues. Both truncations were ectopically expressed in HeLa cells; the cells were lysed and the lysates were incubated with the SIM-affinity beads. The SIM-affinity beads precipitated both GFP-AmpA1-180 and GFP-AmpA158-578 (Fig. 2). Thus, both the N- and C-terminal portions of AmpA become SUMOylated when ectopically expressed in eukaryotic cells and therefore at least two lysines within the whole of AmpA are SUMOylated. Even though GFP-AmpA, GFP-AmpA1-180, and GFP-AmpA158-578 in the input samples all migrated as multiple bands, the SIM-affinity beads primarily recovered the protein form exhibiting the highest apparent molecular weight for each. Thus, only a small portion of the total amount of ectopically expressed GFP-AmpA becomes SUMOylated.
Endogenous AmpA is SUMOylated during Anaplasma phagocytophilum infection of mammalian host cells
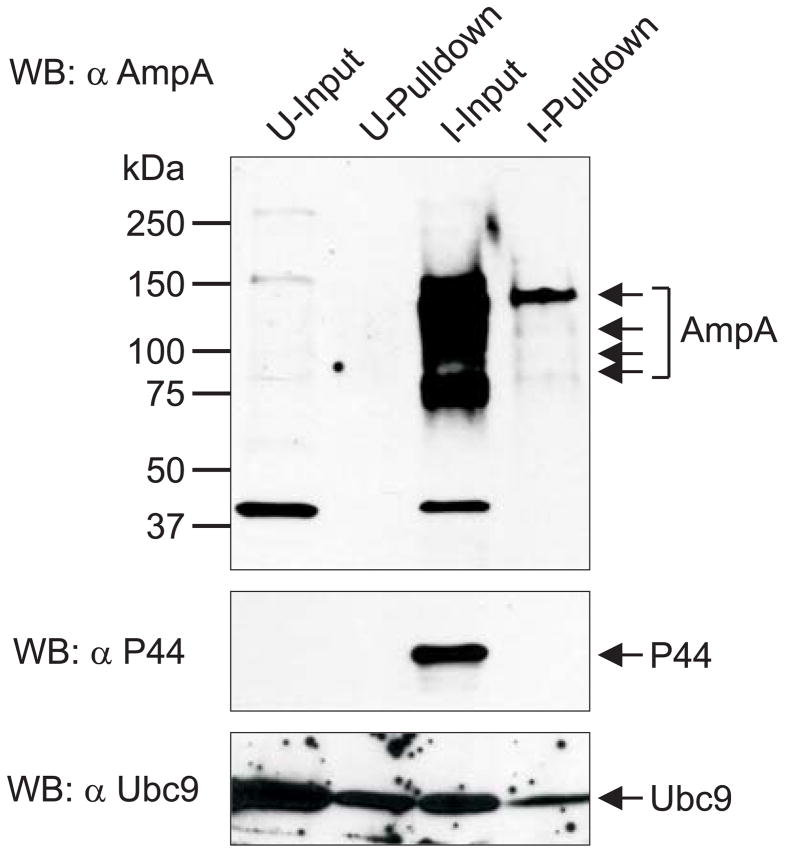
To determine if native AmpA is SUMOylated, we used AmpA antiserum to screen Western-blotted SUMOylated proteins that had been precipitated from lysates of uninfected and A. phagocytophilum-infected HL-60 cells. Consistent with previous reports (Storey et al., 1998, Huang et al., 2010c), multiple AmpA bands ranging in apparent molecular weight were detected in the infected sample input (Fig. 3). SIM-affinity beads recovered SUMOylated AmpA as a minor portion of the total AmpA protein content, migrating primarily as a single band with an apparent mobility of approximately 150 kDa. The E2 ligase of SUMOylation, Ubc9, is itself a target of SUMO modification (Wilkinson et al., 2010), and therefore served as a positive control. The A. phagocytophilum major surface protein P44 (Truchan et al., 2013), which is predicted to not be SUMOylated (data not shown), was used as a negative control. Ubc9 was detected in all input and pulldown samples, while P44 was only detected in the input from infected cells. These data demonstrate that, consistent with ectopically expressed GFP-AmpA, a small portion of the total AmpA protein content expressed by A. phagocytophilum during infection of human host cells is SUMOylated.
Fig. 3.
Native AmpA is SUMOylated in A. phagocytophilum infected HL-60 cells. Upper panel shows uninfected (U) and infected (I) HL-60 lysate input and pulldown lanes probed by Western blot analysis with AmpA antibody. Middle and lower panels show Western blotting of samples with antibodies against A. phagocytophilum P44 and Ubc9 as negative and positive controls for SUMOylation, respectively. Arrows denote the multiple isoforms of native AmpA and the expected sizes for P44 and Ubc9. Data presented are representative of four experiments with similar results.
Both ectopically expressed- and native AmpA are poly-SUMOylated
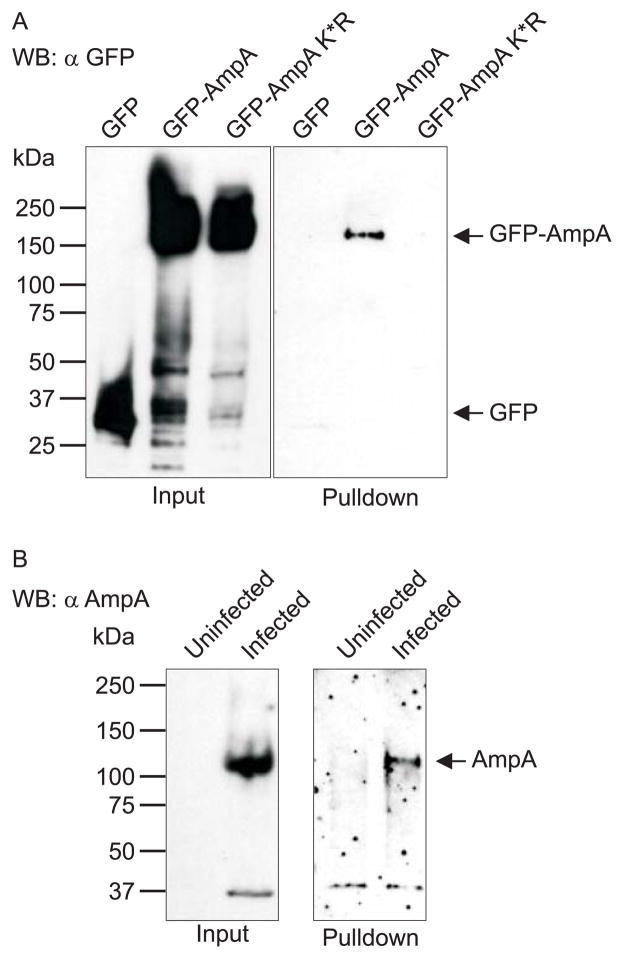
SUMO modification has several variations that can affect the function of its protein substrate. A protein can be mono-SUMOylated (one SUMO attached to a lysine residue), multiply mono-SUMOylated (several single SUMO moieties attached to individual lysines), poly-SUMOylated with a chain of SUMO2/3 moieties (Tatham et al., 2001), or a combination of two or more of these possibilities. Therefore, we next examined if AmpA is mono- or poly-SUMOylated using a poly-SUMO affinity matrix that is specific for SUMO2/3 chains and discriminates against free SUMO moieties and mono-SUMO-conjugates. Lysates of HEK-293 cells ectopically expressing GFP-AmpA, GFP-AmpA K*R, or GFP alone were incubated with the multi-SIM matrix. Following removal of unbound proteins by washing, bound poly-SUMOylated proteins were eluted, resolved by SDS-PAGE, Western blotted, and screened with GFP antibody. GFP-AmpA, but neither GFP-AmpA K*R nor GFP alone, was pulled down (Fig. 4A). Thus, the ability of GFP-AmpA to be precipitated in a poly-SUMOylation-dependent manner required the presence of lysine residues in the AmpA portion. HEK-293 cells were used in this assay to demonstrate that ectopically expressed AmpA becomes SUMOylated in another human cell type besides HeLa cells. Native AmpA from A. phagocytophilum infected HL-60 cells was also pulled down (Fig. 4B), emphasizing that AmpA is poly-SUMOylated during the course of infection.
Fig. 4.
Ectopically expressed and native AmpA are poly-SUMOylated. A. Input (left panel) and pulldowns (right panel) of HEK-293 cells transfected to express GFP-AmpA, GFP-AmpA K*R, or GFP alone. Western blots were probed with GFP antibody. Arrows denote the expected apparent molecular weights for GFP-AmpA, GFP-AmpA K*R, and GFP. B. Input (left panel) and pulldowns (right panel) of uninfected and A. phagocytophilum infected HL-60 cells. Western blots were probed with AmpA antibody. Arrow denotes the expected apparent molecular weight for native AmpA protein. Data shown are representative of two experiments with similar results.
AmpA is detected on the pathogen-occupied vacuole and in the cytosol of infected cells, and colocalizes with SUMO
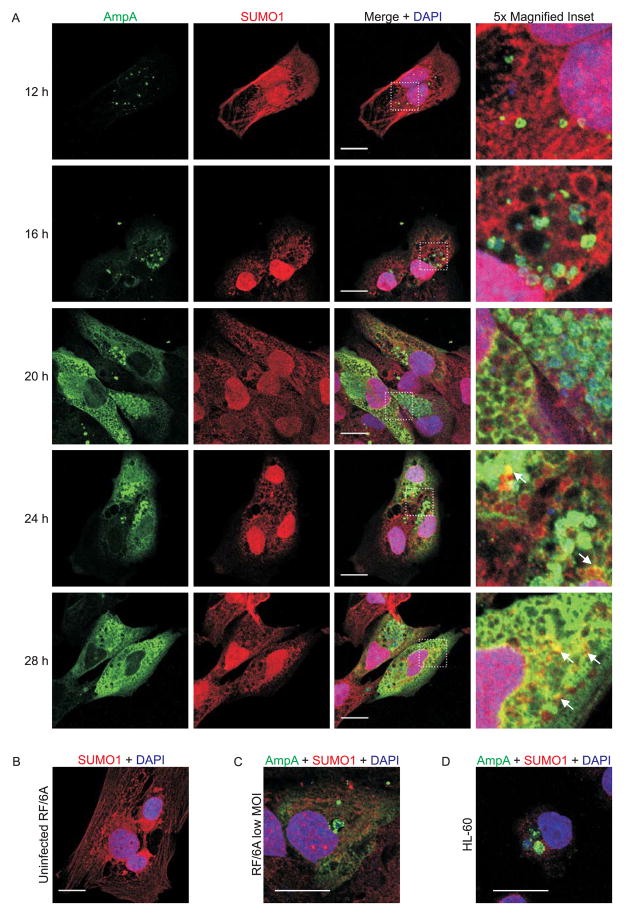
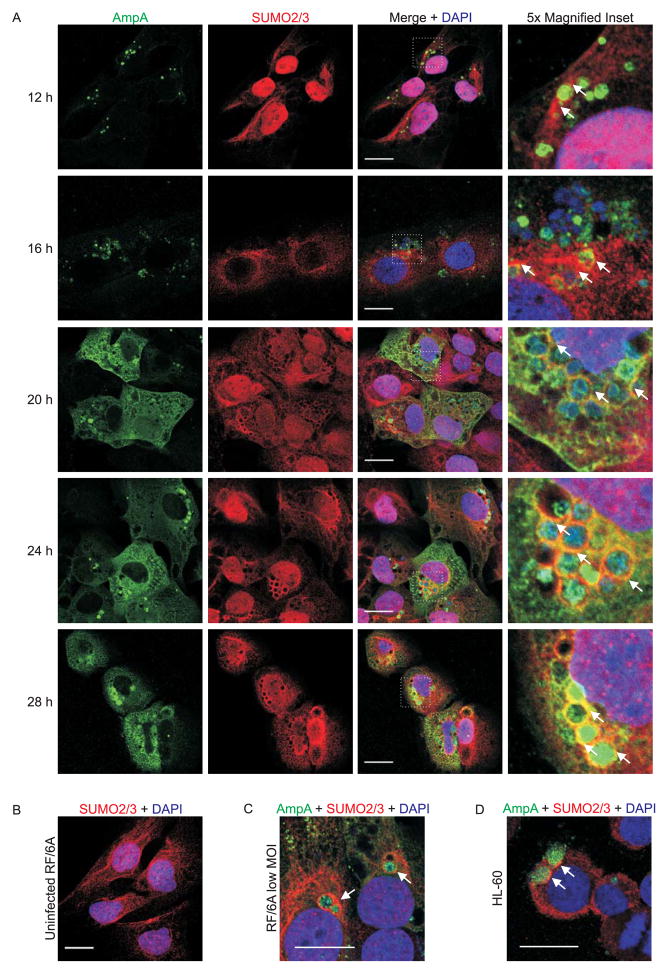
With the knowledge that AmpA both localizes to the AVM (Huang et al., 2010c) and acquires SUMO moieties during A. phagocytophilum infection, it stands to reason that the AVM might become decorated with SUMO moieties over the course of infection. To investigate this possibility, RF/6A cells were synchronously infected with A. phagocytophilum and the infection was allowed to proceed for 28 h, a period that would allow for the completion of a full infection cycle (Troese et al., 2009). RF/6A cells were used for the infection time course analysis because they are both permissive to A. phagocytophilum infection and are large and flat, which makes them ideal for imaging the A. phagocytophilum-occupied vacuole (ApV) (Munderloh et al., 2004, Sukumaran et al., 2011). The cells were examined by laser-scanning confocal microscopy (LSCM) for AmpA localization and for SUMO1 and SUMO2/3 recruitment to the ApV. As previously observed (Huang et al., 2010c), AmpA staining was detected on intravacuolar A. phagocytophilum organisms and the AVM beginning at 12 h, the time point at which the ApV was first easily discernible by immunofluorescence (Fig. 5A and Fig. 6A). At 12 and 16 h, aggregative SUMO2/3 staining was observed surrounding AmpA-positive ApVs (Fig. 6A). Beginning at 20 h and continuing throughout the remainder of the time course, aggregative SUMO2/3 signal colocalized with AmpA on the AVM (Fig. 6A). In contrast, SUMO1 signal colocalized with AmpA associated with the AVM beginning only at 24 h (Fig. 5A). Neither SUMO1 nor SUMO2/3 yielded aggregative staining patterns in uninfected RF/6A cells (Fig. 5B and Fig. 6B). Compared to SUMO2/3, the degree of SUMO1 labeling of the AVM was modest and not as discretely localized to the AVM. SUMO1 and SUMO2/3 staining remained on the cytosolic face of the AVM throughout infection and were not detected within the ApV lumen (Fig. 5A and Fig. 6A). Thus, SUMOylation only occurs at the host-pathogen interface of the AVM and SUMO moieties do not traffic inside the vacuole. Beginning at 20 h, AmpA signal was detected in the host cell cytosol and continually increased over the duration of infection (Fig. 5A and 6A), which suggests that, in addition to localizing to the AVM (Huang et al., 2010c), AmpA is also translocated into the cytosol. Recruitment of SUMO2/3, but not SUMO1, to the ApV was also observed in RF/6A cells infected with lower multiplicities of infection (Fig. 6C and 5C). Examination of A. phagocytophilum infected HL-60 cells at 24 h revealed that SUMO2/3 pronouncedly localized to the peripheries of ApVs and SUMO1 localization to ApVs was modest, at best, thereby validating that these phenomena occur during infection of both endothelial and myeloid host cells (Fig. 5D and 6D).
Fig. 5.
Localization of SUMO1 and AmpA in A. phagocytophilum infected mammalian host cells. A. Representative images of infected RF/6A cells at 12, 16, 20, 24, and 28 h post infection. Time points are marked at the left of each row. Cells were immunofluorescently stained to detect AmpA (green), SUMO1 (red), and DNA (DAPI, blue). Merged images are shown in the third column. Areas demarcated by white hatched boxes in the merged column are magnified five-fold in the fourth column. White arrows indicate representative areas where AmpA and SUMO1 signals colocalize. Scale bars, 20 μm. B. SUMO1 (red) and DNA (DAPI, blue) staining in an uninfected RF/6A cell. C and D. Merged confocal images of RF/6A cells infected with a low MOI of A. phagocytophilum (C) and A. phagocytophilum infected HL-60 cells (D) stained for AmpA (green), SUMO1 (red), and DNA (DAPI, blue) at 24 h post infection.
Fig. 6.
Localization of SUMO2/3 and AmpA in A. phagocytophilum infected mammalian host cells. A. Representative images of infected RF/6A cells at 12, 16, 20, 24, and 28 h post infection. Time points are marked at the left of each row. Cells were immunofluorescently stained to detect AmpA (green), SUMO2/3 (red), and DNA (DAPI, blue). Merged images are shown in the third column. Areas demarcated by white hatched boxes in the merged column are magnified five-fold in the fourth column. B. SUMO2/3 (red) and DNA (DAPI, blue) staining in an uninfected RF/6A cell. C and D. Merged confocal images of RF/6A cells infected with a low MOI of A. phagocytophilum (C) and A. phagocytophilum infected HL-60 cells (D) stained for AmpA (green), SUMO2/3 (red), and DNA (DAPI, blue) at 24 h post infection. White arrows indicate representative areas where AmpA and SUMO2/3 signals colocalize and/or SUMO2/3 accumulates on the AVM. Scale bars, 20 μm.
Pharmacologic inhibition of SUMOylation reduces the A. phagocytophilum load
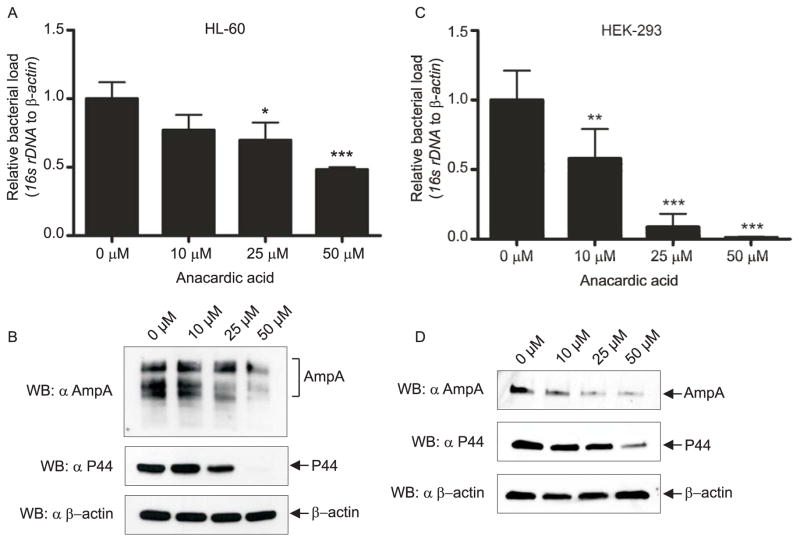
Because AmpA co-opts SUMOylation and the AVM accumulates SUMO moieties throughout infection, we next examined if pharmacologic inhibition of SUMOylation is detrimental to A. phagocytophilum intracellular survival. Anacardic acid is a small molecule inhibitor of SUMOylation that prevents formation of the E1-SUMO intermediate (Fukuda et al., 2009). Anacardic acid has been reported to exert antimicrobial activity against gram-positive bacteria, but to be ineffective against gram-negative organisms (Hemshekhar et al., 2011). Thus, any detrimental effect that anacardic acid might have on A. phagocytophilum would presumably result from its disruption of SUMOylation, rather than an antimicrobial effect on the bacterium. HL-60 or HEK-293 cells were pretreated with anacardic acid, after which they were infected with A. phagocytophilum organisms and subsequently maintained for 96 h. Media containing anacardic acid was replenished every 24 h. At the end of the time course, the anacardic acid and vehicle control treated infected cells were subjected to quantitative PCR (qPCR) and Western blot analyses. Anacardic acid had a dose-dependent inhibitory effect on A. phagocytophilum intracellular replication, as indicated by decreases in the A. phagocytophilum 16S rDNA-host cell β-actin ratio, and AmpA and P44 protein levels (Fig. 7). This inhibitory effect was more dramatic in HEK-293 cells as compared to HL-60 cells (Fig 7, A and C). Host cell β-actin levels were comparable among all samples and host cell viability was unaffected by anacardic acid treatment, as indicated by a lack of trypan blue uptake (data not shown).
Fig. 7.
Pharmacologic inhibition of SUMOylation reduces the A. phagocytophilum load. HL-60 (A and B) and HEK-293 (C and D) cells were treated with 0, 10, 25, or 50 μM anacardic acid for 4 h prior to infection by A. phagocytophilum. The infected cells were maintained for 96 h, with anacardic acid-containing growth media being replenished every 24 h. Cells were collected for qPCR (A and C) and Western blot analyses (B and D). For qPCR, relative DNA loads of A. phagocytophilum 16s rRNA gene were normalized to DNA loads of the human β-actin gene using the 2−ΔΔCT (Livak) method. Results shown are the means ± SD of triplicate samples and are representative of three independent experiments in HL-60 cells and two independent experiments in HEK-293 cells, with similar results. Statistically significant (*P < 0.05; **P < 0.005; ***P < 0.001) values relative to the 0 μM control are indicated. In B and D, 15 μg or 5 μg of whole cell lysate, respectively, was loaded per lane. Blots were probed with AmpA and P44 antibodies to examine pathogen burden, in comparison to the β-actin loading control. Data shown are representative of two independent sets of Western assays performed for each cell type.
Ubc9 expression is not altered by A. phagocytophilum infection, but knockdown of Ubc9 results in a modest increase in infection
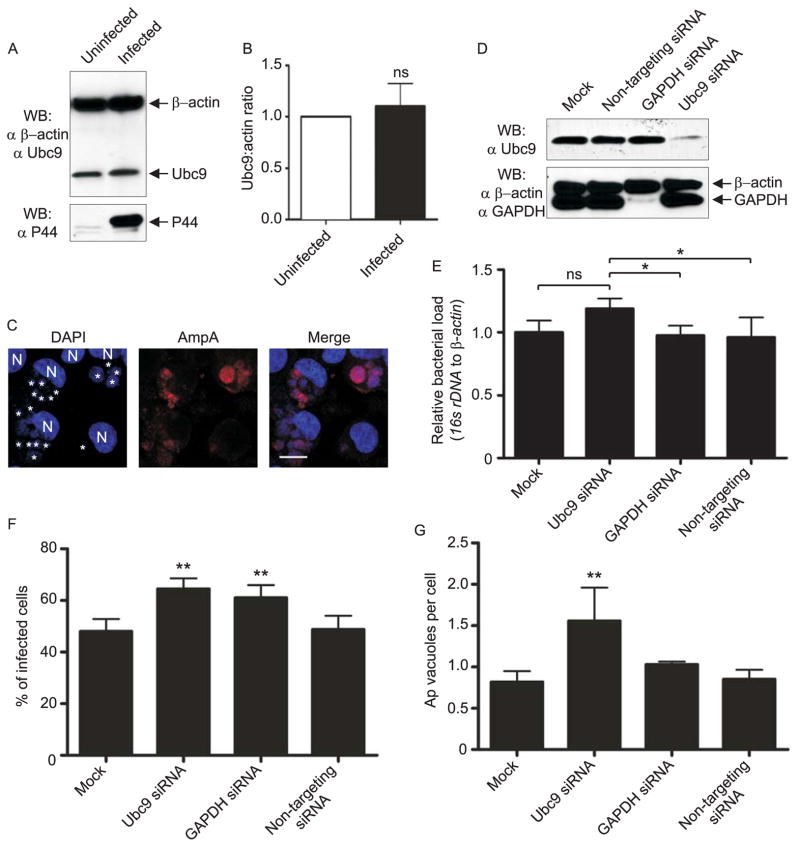
To further assess the role of SUMOylation in A. phagocytophilum infection, we focused on the SUMOylation E2 ligase, Ubc9. Because other intracellular bacterial pathogens mediate reduction of Ubc9 protein levels upon infection (Ribet et al., 2010, Kim et al., 2013, Kim et al., 2008), we assessed if this phenomenon occurs in A. phagocytophilum infected cells. Western blot and densitometry analysis of uninfected versus infected HL-60 cell lysates showed no difference in Ubc9 expression (Fig. 8, A and B). Next, we determined if siRNA-mediated knockdown of Ubc9 alters A. phagocytophilum infection. This experiment required host cells that would both support A. phagocytophilum intracellular growth and in which a high efficiency of transfection could be achieved. HeLa cells are highly amenable to transfection, but are not susceptible to A. phagocytophilum infection (data not shown). Therefore, we utilized HEK-293 cells because (1) we were able to routinely achieve transfection efficiencies of ≥75% using these cells versus less than 20% using RF/6A or HL-60 cells (data not shown); and (2) they are susceptible to A. phagocytophilum infection and support development of AmpA-positive ApVs (Fig. 8C). HEK-293 cells also support infection and intracellular replication of the closely related Anaplasmataceae family member, E. chaffeensis (Miura et al., 2011). HEK-293 cells were treated for 72 h with siRNA targeting Ubc9 or GAPDH, non-targeting siRNA, or transfection agent alone. Knockdown of protein expression was verified by Western blot (Fig. 8D). Treated cells were infected with A. phagocytophilum for 48 h, after which infection was assessed by qPCR. There was no significant difference in the A. phagocytophilum DNA load between Ubc9 knockdown and mock treated cells, but there was a significant, albeit modest, increase in the bacterial DNA load in Ubc9 siRNA treated cells relative to GAPDH and non-targeting siRNA treated cells (Fig. 8E). The percentages of infected cells and ApVs per cell were also enumerated using immunofluorescence microscopy (Fig. 8F and G). Whereas the percentages of infected cells in which Ubc9 or GAPDH had been knocked down were slightly higher than those for non-targeting siRNA-treated or mock-transfected cells (Fig. 8F), Ubc9 knockdown resulted in a significant increase in the number of ApVs per cell than all control treatments (Fig. 8G). Taken together, these results indicate that, unlike other intracellular bacteria, A. phagocytophilum does not manipulate Ubc9 expression and that Ubc9 knockdown results in a minor increase in A. phagocytophilum infection.
Fig. 8.
Ubc9 expression is not altered by A. phagocytophilum infection, but knockdown of Ubc9 results in a significantly increased bacterial load. A and B. A. phagocytophilum infection does not alter Ubc9 expression. A. Western-blotted whole cell lysates of uninfected and A. phagocytophilum infected cells were screened with β-actin and Ubc9 antibodies. B. Three pairs of infected and uninfected HL-60 lysates were examined in duplicate by Western blotting and the ratio of Ubc9 to β-actin analyzed by densitometry. “ns”, not significant. C. HEK-293 cells support A. phagocytophilum infection. HEK-293 cells that had been incubated with A. phagocytophilum were stained with DAPI (blue) to denote host cell nuclei (N) and bacteria (*). Cells were also stained with anti-AmpA (red) prior to visualization using LSCM. Scale bar, 10 μm. D–G. Effect of ubc9 knockdown on the A. phagocytophilum load in infected HEK-293 cells. HEK-293 cells were transfected with siRNA targeting ubc9 or gapdh, non-targeting siRNA, or transfection agent only (mock). At 72 h post transfection, the cells were boosted with a second siRNA transfection and infected with A. phagocytophilum. At 48 h post infection, the cells were resuspended and seeded on coverslips for immunofluorescent staining or collected for whole cell lysis and Western blotting analysis. D. Western blots demonstrating siRNA knockdown of Ubc9. Whole cell lysates from mock-, non-targeting siRNA-, gapdh siRNA-, and ubc9 siRNA-transfected cells were screened with antibodies targeting Ubc9 and GAPDH to confirm knockdown, and β-actin as a loading control. Blots are representative of more than four independent siRNA knockdown assays. E–G. The A. phagocytophilum load is modestly higher in host cells in which ubc9 has been knocked down. siRNA-treated and control host cells that had been infected with A. phagocytophilum were collected and processed for qPCR (E) or immunofluorescence microscopy (F and G). E. Relative loads of A. phagocytophilum 16S rRNA gene were normalized to human β-actin using the 2−ΔΔCT (Livak) method. Results shown are the means ± SD of triplicate samples and are representative of two independent experiments with similar results. Statistically significant (*P < 0.05) values relative to the ubc9 knockdown are indicated. F and G. Coverslips containing siRNA knockdown cells were processed for immunofluorescence and screened with anti-P44 to denote ApVs inside host cells. The mean ± standard deviations of percentages of infected HL-60 cells (F) and Ap vacuolar inclusions per HL-60 cell (G) were determined using immunofluorescence microscopy. Data presented are representative of two independent experiments. Statistically significant (*P < 0.05; **P < 0.005) values are indicated.
Ectopically expressed AmpA limits A. phagocytophilum growth in a lysine-dependent manner
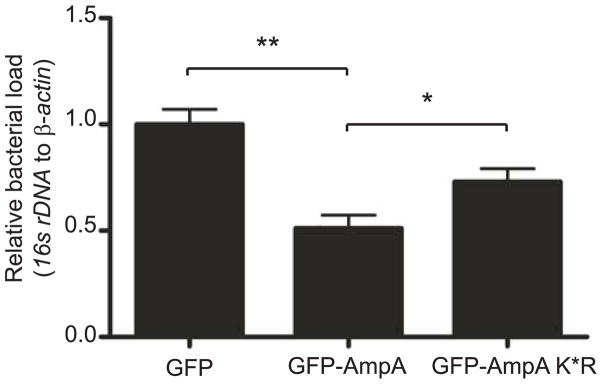
Given that AmpA is found at the AVM, in the host cytosol, and interfaces with host cell SUMOylation pathways, we rationalized that ectopically expressed AmpA would compete against bacterial-derived AmpA for access to host factors and thereby inhibit A. phagocytophilum intracellular development. Precedent for this approach is provided by the fact that ectopically expressed chlamydial effectors inhibit the growth of chlamdyiae within host cells (Cortes et al., 2007, Rockey et al., 1997). Furthermore, if lysine modification of AmpA by SUMOylation or another post-translational modification plays a role in AmpA interactions with host factors, then ectopically expressed AmpA K*R would be expected to exhibit a lesser inhibitory effect. To test our hypotheses, HEK-293 cells were transected to express GFP-AmpA, GFP-AmpA K*R, or GFP alone. Flow cytometric analyses confirmed that comparable transfection efficiencies for these constructs were achieved (data not shown). The transfected cells were infected with A. phagocytophilum and total DNA isolated from these cells at 24 h post infection was subjected to qPCR analysis for evaluation of bacterial load relative to host cell β-actin levels. As shown in Figure 9, overexpression of GFP-AmpA reduced infection levels by half when compared to GFP alone. GFP-AmpA K*R was unable to compete with native AmpA as efficiently as GFP-AmpA, implying the importance of the protein’s lysine residues in co-opting host cell factors to promote optimal A. phagocytophilum intracellular growth.
Fig. 9.
Ectopically expressed AmpA limits A. phagocytophilum growth in a lysine-dependent manner. Transfected HEK-293 cells expressing GFP-AmpA, GFP-Amp K*R, or GFP were infected with A. phagocytophilum. Genomic DNA was isolated and subjected to qPCR. Relative DNA loads of the A. phagocytophilum 16s rRNA gene were normalized to DNA loads of the human β-actin gene using the 2−ΔΔCT (Livak) method. Results shown are the means ± SD of triplicate samples and are representative of two independent experiments with similar results. Statistically significant (*P < 0.05; **P < 0.005) values relative to the GFP or GFP-AmpA are indicated. GFP vector alone was set as the control, equal to 1.0.
DISCUSSION
Numerous examples exist of pathogens targeting SUMOylation, many of which attempt to subdue SUMO modification in host cells. However, few intracellular bacteria are known to share this phenomenon. Xanthomonas campestris pathovar vesicatoria, the causative agent of bacterial spot of tomatoes and peppers, negatively regulates SUMOylation by decreasing overall host cell SUMOylation levels (Kim et al., 2013, Hotson et al., 2003, Kim et al., 2008, Roden et al., 2004). Listeria monocytogenes LLO (listeriolysin O), Clostridium perfringes PFO (perfringiolysin O), and Streptococcus pneumoniae PLY (pneumonlysin) impede global SUMO conjugation by inducing specific degradation of Ubc9 (Ribet et al., 2010). In contrast, A. phagocytophilum exploits host SUMO modification to decorate AmpA and does so without altering host Ubc9 levels. Although other means of pathogen SUMOylation modulation likely remain to be uncovered, A. phagocytophilum is the first intracellular bacterial pathogen demonstrated to exploit SUMOylation during infection of mammalian host cells as part of its intracellular survival strategy. Although a recombinant form of the E. chaffeensis effector, TRP120 was shown to be SUMOylated in an in vitro assay, and ectopically expressed TRP120 was SUMOylated (Dunphy et al., 2014), it has yet to be demonstrated whether native TRP120 is SUMOylated in E. chaffeensis infected host cells.
In addition to localizing to the AVM, AmpA is also secreted into the host cell cytosol, where it accumulates and is SUMOylated throughout the infection cycle. We hypothesize that SUMOylated AmpA could function to recruit interacting proteins to the ApV and/or may remain in the cytosol to modulate host cell processes that are critical for A. phagocytophilum survival. Both native and ectopically expressed AmpA are SUMO2/3-modified, as implied by the poly-SUMOylation assay results, but SUMO1 modification has not yet been excluded as a possibility. Most eukaryotic substrates can be conjugated by both SUMO1 and SUMO2/3 (Vertegaal et al., 2006), though the functional differences of SUMO1 versus SUMO2/3 have not been defined (Wilkinson et al., 2010). Regardless, SUMO1 and SUMO2/3 differ in both their conjugation dynamics and exhibit discrete patterns of localization in eukaryotic cells, indicating differential regulation of the SUMO paralogs (Wilkinson et al., 2010). Similarly, both AVM-localized and cytosolic AmpA colocalize with SUMO2/3 throughout the infection cycle, but only colocalize with SUMO1 at 24–28 h, around the time that A. phagocytophilum organisms transition from the replicative to infectious phase, egress from the host cell, and initiate the next round of infection (Troese et al., 2009). Thus, AmpA may be sequentially modified by SUMO2/3 and then SUMO1, a pattern that would be consistent with the fact that SUMO1 caps SUMO2/3 polymers on eukaryotic substrates in vivo (Matic et al., 2008). Although the functional roles of these SUMO variations is not always clear, evidence suggests that poly-SUMOylation can increase protein-protein interactions of targets with SIM-containing substrates, help maintain structural integrity of protein complexes, and even serve as ubiquitylation signals to promote proteasomal degradation of targets (Ulrich, 2008). Moreover, as there is a huge reserve of unconjugated SUMO2/3 but very little unconjugated SUMO1 in host cells (Saitoh et al., 2000), AmpA apparently taps into the available SUMO pool. SUMO2/3 modification of AmpA throughout the infection cycle is likely critical for bacterial intracellular survival, while terminal SUMO1 conjugation of AmpA may promote a late-stage infection cycle event. Though we observed AmpA colocalization with SUMO2/3 at the AVM, we cannot rule out the possibility that other SUMOylated bacterial and/or host proteins localize or are in close proximity to AmpA and the AVM.
Results from our pulldown assays and confocal studies evidence that only a small portion of the available AmpA pool, whether bacterial-derived or ectopically expressed, is actually SUMOylated at any given time. The “SUMO enigma” refers to the phenomenon observed in eukaryotic cells that only a small proportion of an available substrate protein need be SUMOylated to achieve maximal effect (Hay, 2005). For example, SUMO conjugation of the glutamate receptor subunit 6 (GluR6) results in its endocytosis from the plasma membrane. Even though only a small portion of SUMOylated GluR6 can be detected at any one time, a large proportion of GluR6 undergoes SUMO-induced endocytosis. Moreover, the spatial consequence of SUMOylation persists, as even after GluR6 has been deSUMOylated, it remains at the subcellular locale to where it trafficked, distinct from unmodified GluR6 (Martin et al., 2007, Wilkinson et al., 2010). The present study extends the “SUMO enigma” to a bacterial effector and underscores the ability of A. phagocytophilum to efficiently exploit SUMOylation.
The observed dose-dependent response of A. phagocytophilum infection to anacardic acid suggests that, despite potential off-target effects, pathogen load is decreased with increasing amounts of SUMOylation-inhibitory drug. Specific knockdown of the SUMO conjugating E2 enzyme, Ubc9, leads to a modest increase in infection and number of ApVs per cell. Thus, a balance appears to exist between the relevance of SUMOylation to A. phagocytophilum survival and the host cell anti-intracellular microbe response. In all likelihood neither the pharmacologic or siRNA-mediated treatment completely abolished SUMOylation in the cell. Indeed, a small amount of Ubc9 was still expressed after knockdown and the stability of anacardic acid over longer periods of time in cell culture is not known, which may have lead to cycling of SUMOylation inhibition and release with replacement of the drug every 24 h. Here, the “SUMO enigma” may also be playing a role, in that a small amount of SUMOylation is sufficient for A. phagocytophilum replication to persist. Complete inhibition of SUMOylation would likely have lethal effects on the host cell and, correspondingly, on the pathogen. Support for this premise is provided by the facts that deletion of Ubc9 in chicken DT40 lymphocytes ultimately leads to apoptosis (Hayashi et al., 2002) and Ubc9-deficient mouse embryos die early in development (Nacerddine et al., 2005). We complimented the anacardic acid and ubc9 knockdown approaches using ectopically expressed AmpA as a competitive agonist against A. phagocytophilum intracellular growth. As shown in other studies, ectopic expression of an effector in infected cells can either boost infection, as demonstrated for A. phagocytophilum Ats-1 (Niu et al., 2012), or hinder pathogen growth, as shown for Chlamydia caviae IncA and C. pneumoniae Cpn0585 (Cortes et al., 2007, Rockey et al., 1997). Ectopically expressed GFP-AmpA was an effective agonist that reduced the A. phagocytophilum load, presumably by sequestering host interacting partners away from native AmpA. Notably, ectopically expressed GFP-AmpA K*R was a less effective competitor, equating the loss of lysines with a likely loss in binding potential to host ligands. Although SUMOylation is not the only modification specific for lysines and the A. phagocytophilum AVM has been previously been shown to be decorated with monoubiquitin (Huang et al., 2012), the GFP-AmpA K*R data underscore the importance of AmpA lysine residues in its interaction with yet-to-be identified eukaryotic host factors.
Thematic homologies clearly exist between A. phagocytophilum AmpA and similar effectors of its fellow Anaplasmataceae family member, E. chaffeensis. Although lacking in sequence homology, AmpA and ehrlichial TRPs share the occurrence of repetitive sequences, localize to the pathogen-occupied vacuolar membrane, and are secreted into the host cytosol (Zhu et al., 2009, Huang et al., 2010c, Zhu et al., 2011, Luo et al., 2012). Yeast two hybrid analyses have revealed the great capacity of ehrlichial TRPs to interact with multiple host proteins with various functions, including transcriptional regulation, signal transduction, intracellular vesicular trafficking, translation, proteomic-mediated degradation, iron metabolism, and cytoskeletal organization. Many of these ligands localized to the E. chaffeensis-occupied vacuole (Luo et al., 2011, Wakeel et al., 2009, Zhu et al., 2011). In light of these prior reports and our data, the question is raised: how does one effector mediate interactions with so many different eukaryotic binding partners? Modifications such as SUMOylation may allow the effector to diversify its ligand repertoire. It is fathomable that different SUMO-modification patterns of an effector over the course of infection alter the pathogen’s interactome profile to facilitate events critical for each stage of infection. Such a use of SUMOylation would give obligate intracellular pathogens like A. phagocytophilum, which has a smaller genome and fewer annotated effectors than extracellular or facultative intracellular bacterial pathogens, the advantage of expanding its limited effector force to participate in a broader network of host modulatory activities. It is therefore imperative to map the modified lysine residues in AmpA and determine its binding partners in order to further our understanding of how this effector contributes to A. phagocytophilum intracellular survival.
EXPERIMENTAL PROCEDURES
In silico analyses
The AmpA sequence (APH1387; YP_505886.1) was analyzed for predicted SUMOylation sites using GPS-SUMO SumoSp Prediction of SUMOylation Sites (http://sumosp.biocuckoo.org/) (Ren et al., 2009); SUMOplot Analysis Program (http://www.abgent.com/sumoplot); and sequence-based evaluation of SUMOylation sites (seeSUMO)(http://bioinfo.ggc.org/seesumo/) (Teng et al., 2012).
Cell lines and cultivation of A. phagocytophilum
HL-60 cells were maintained and A. phagocytophilum strain NCH-1 was cultured in HL-60 cells as described (Huang et al., 2012). RF/6A monkey choroidal endothelial cells (CRL-1780; American Type Culture Collection [ATCC], Manassas, VA) were cultured and infected with A. phagocytophilum strain NCH-1 as described (Huang et al., 2012). HeLa 229 epithelial cells (CCL-1.2; ATCC) were grown in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, Sacramento, CA) at 37°C with 5% CO2. HEK-293 cells were cultured in Dulbecco’s Modified Eagle’s Medium with L-Glutamine, 4.5 g/L D-Glucose, and 100 mg/L sodium pyruvate (DMEM; Invitrogen) supplemented with 10% FBS, 1X MEM Non-Essential Amino Acids (Invitrogen), and 15 mM HEPES (Affymetrix, Cleveland, OH) at 37°C with 5% CO2. HEK-293 cells were infected with A. phagocytophilum organisms that had been naturally released from infected RF/6A cells into the culture media as follows. HEK-293 cells to be infected were seeded in a 25-cm2 flask and overlaid with 3 ml of media. Next, 2 ml of A. phagocytophilum laden media from heavily infected RF/6A cells was added for a final volume of 5 ml and the infected HEK-293 culture was incubated at 37°C with 5% CO2 for 48 h or longer.
Plasmid constructs
AmpA, AmpA1-180, and AmpA158-578 coding regions were amplified using primers listed in Table 2 and Platinum Pfx DNA polymerase (Invitrogen). Amplicons were purified using QIAquick PCR purification columns (QIAGEN, Valencia, CA) and cloned into pENTR/TEV/D-Topo (Invitrogen) as described previously (Huang et al., 2010c) to yield pENTR entry plasmids containing the DNA fragment of interest. DNA sequencing was used to verify insert integrity. Recombination of the insert sequence downstream of and in frame with the gene GFP in the pDest-53 plasmid (Invitrogen) was achieved using LR Clonase (Invitrogen) as described (Huang et al., 2010c). GFP-AmpA K*R was generated by gene synthesis of AmpA with the codons for all 19 lysine residues substituted with codons encoding arginine and flanked with EcoRI and SalI restriction sites (Biomatik, Ontario, Canada). AmpA K*R DNA was restriction cloned into the eGFP-C1 plasmid (Clontech, Palo Alto, CA) via the EcoRI/SalI sites and sequenced to verify insert fidelity.
Table 2.
Oligonucleotides used in this study
| Designation | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| pENTR-APH1387 | CACCa ATGTATGGTATAGATATAGAGCTAAG | CTAATAACTTAGAACATCTTCATCG |
| pENTR- | CACCTATGGTATAGATATAGA | CTAGTCTCAACTTCTTCAATT |
| APH13871-180 | GCTAAGTGATTACAGAATTGG | GCTGGTGCTTC |
| pENTR- APH1387158-578 | CACCTCGGGTGTAGATACGCAAGAAGAACAAG | CTAATAACTTAGAACATCTTCATCGTCAGGATCCTTTAACG |
| β-actin | AGAGGGAAATCGTGCGTGAC | CAATAGTGATGACCTGGCCGT |
| Ap 16S | TGTAGGCGGTTCGGTAAGTTAAAG | GCACTCATCGTTTACAGCGTG |
Underlined nucleotides correspond to a Gateway entry vector-compatible sequence.
Western blot analyses
Protein samples for Western blot analysis were resolved by SDS-PAGE, transferred to nitrocellulose or PVDF membrane, blocked in TBS containing 5% (vol/vol) non-fat dry milk, and probed as described (Carlyon et al., 2002). Primary antibodies were rabbit anti-AmpA (Huang et al., 2010c) used at a 1:5,000 dilution; mouse anti-β-actin (Santa Cruz Biotechnology, Dallas, TX) used at a 1:2,500 dilution; rabbit anti-GFP (Invitrogen) used at a 1:1,000 dilution; rabbit anti-Ubc9 (Santa Cruz) used at a 1:200 dilution; mouse anti-GAPDH (Sigma Aldrich, St. Louis, MO) used at a 1:10,000 dilution, and rabbit anti-P44 (Troese et al., 2011) used at a 1:5,000 dilution. Secondary antibodies were horseradish peroxidase (HRP)-linked goat anti-rabbit IgG and goat anti-mouse IgG, both of which were purchased from Cell Signaling Technology (Beverly, MA) and were used at a 1:10,000 dilution. Densitometry was performed using ImageJ (http://rsb.info.nih.gov/ij/).
In vivo SUMOylation assays
HeLa or HEK-293 cells were seeded in 6 well plates and transfected with 4.0 to 8.0 μg of endotoxin-free purified plasmid DNA (Maxi kit, QIAGEN) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s directions. Transfected HeLa or HEK-293 cells and A. phagocytophilum infected HL-60 cells were lysed in 50mM Tris-HCl, pH 7.5, 150mM NaCl, and 200mM iodoacetamide (Sigma) with protease inhibitor cocktail (Roche Diagnostics GmBH, Mannheim, Germany). Lysates (100 μg) were subjected to SUMO-Qapture-T pulldowns (Enzo Life Sciences, Farmingdale, NY) following the manufacturer’s protocol and analyzed by Western blotting. PolySUMOylated proteins were isolated and enriched from infected and transfected cells using the PolySUMO-Qapture kit (Enzo Life Sciences). Cells were lysed in 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and 200 mM iodoacetamide with protease inhibitor cocktail and processed according to the manufacturer’s directions using 200 μg of lysate. Eluates were analyzed by Western blotting.
LSCM analysis of AmpA colocalization with SUMO1 and SUMO2/3 in A. phagocytophilum infected cells
Coverslips of A. phagocytophilum infected RF/6A cells were processed at 12, 16, 20, 24, and 28 h post infection for LSCM as described (Huang et al., 2010c). Primary antibodies used were rabbit anti-SUMO1 (AbCam, Cambridge, MA) at a 1:250 dilution, and rabbit anti-SUMO2/3 (AbCam) at a 1:100 dilution, mouse anti-AmpA at a 1:200 dilution. To generate mouse polyclonal anti-AmpA serum, C3H/HeJ mice were immunized against glutahtione-S-transferase-tagged AmpA as previously described (Troese et al., 2011). Alexa Fluor 594-conjugated goat anti-rabbit IgG and Alexa Fluor 488-conjugated goat anti-mouse IgG (both from Invitrogen) were used as secondary antibodies prior to mounting with Prolong gold antifade plus 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and imaged with a Zeiss LSM 700 laser-scanning confocal microscope.
Assessment of the effect of ectopically expressed GFP-AmpA on A. phagocytophilum bacterial load
HEK-293 cells were transfected to express GFP, GFP-AmpA, or GFP-AmpA K*R. At 24 h, the transfected cells were incubated with host cell-free A. phagocytophilum organisms that had been naturally released from infected RF/6A cells as described above. The infection was allowed to proceed for 48 h, at which time total DNA was isolated from each culture using the DNeasy Blood and Tissue kit (QIAGEN). qPCR was performed on triplicate samples consisting of 100 ng genomic DNA, SsoFast Evagreen Supermix (Bio-Rad), and primers targeting genes encoding A. phagocytophilum 16S rDNA (APH_1000) and human β-actin (Table 2). Quantitative PCR assays were performed using the CFX96 real-time PCR detection system (Bio-Rad). Relative infection load levels were normalized to the DNA levels of the host cell β-actin gene using the 2−ΔΔCT (Livak) method (Livak et al., 2001) and the Prism 5.0 software package (Graphpad, San Diego, CA).
Pharmacologic inhibition of SUMOylation
Anacardic acid (EMD Millipore, Billerica, MA) dissolved in DMSO was used to pre-treat 3 × 106 HL-60 cells or 2 × 105 HEK-293 cells for 4 h at final concentrations of 10, 25, or 50 μM in primary growth media. HL-60 cells were synchronously infected with A. phagocytophilum (Troese et al., 2009) for one h, while HEK-293 cells were asynchronously infected with host cell-free A. phagocytophilum organisms that had been naturally released from infected RF/6A cells. All cells were subsequently maintained for 96 hours post infection with the anacardic acid/primary growth media replaced every 24 h. Cells were collected, washed with phosphate buffered saline, and processed for total DNA isolation for qPCR as above or whole lysis with RIPA buffer as described previously (Troese et al., 2011).
siRNA knockdown of ubc9 in A. phagocytophilum infected HEK-293 cells
To assess the role of SUMOylation on A. phagocytophilum infection, siRNA was used to knockdown expression of the SUMOylation E2 ligase, ubc9, in HEK-293 cells. 4 × 104 HEK-293 cells were seeded per well in 24 well plates or 2 × 105 cells per well of a 6-well plate. Cells were transfected 24 h later with ON-TARGETplus SMART pool siRNA (Thermo Fisher Scientific, Pittsburgh, PA) targeting human gapdh or ubc9 mRNA, or with a non-targeting siRNA using DharmaFECT 1 (Thermo Fisher Scientific). No siRNA (transfection agent only) served as a negative control. Seventy-two h post transfection, HEK-293 cells were transfected with a second round of siRNA and infected with A. phagocytophilum organisms that had been naturally released from infected RF/6A cell cultures as described above, and incubated an additional 48 h prior to processing. HEK-293 cells were suspended in culture media and re-seeded onto 12 mm coverslips for immunofluorescent staining with P44 antibody (Huang et al., 2010a) at a dilution of 1:500, collected for whole cell lysis in RIPA buffer and Western blotting analysis as described (Carlyon et al., 2002), or processed for qPCR as noted above. Coverslips were analyzed via spinning disk confocal microscopy using Slidebook software (Huang et al., 2010a) to denote A. phagocytophilum infected cells, which possessed one or more P44-positive ApVs, or uninfected cells, which did not. To determine the percentage of infected cells, the total number of infected host cells was divided by the total number of 400 cells counted per condition and multiplied by a factor of 100. Prism software was used to generate graphical data.
Statistical analyses
One-way analysis of variance (ANOVA) with Dunnett’s post test was performed using the Prism 5.0 software package to assess statistical significance (set at P < 0.05). For densitometry data, a one sample t test was used.
Acknowledgments
We thank Nathan L. Galloway and Bernice Huang for technical assistance, and Tim Lochmann and Shirley Taylor for providing HEK-293 cells. This study was supported by NIH grants R21 AI105364, R21 AI090170, and 2R01 AI072683 (to J.A.C.). A.R.B. is a fellowship recipient supported by VCU Institutional Research and Academic Career Development Award grant K12 GM093857. The Virginia Commonwealth University (VCU) Summer Student Program in Microbiology, Infectious Diseases, and Public Health Epidemiology provided support to L.J.M. LSCM was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, which is supported, in part, with funding from NIH-NINDS Center core grant 5P30NS047463.
References
- Bekes M, Drag M. Trojan horse strategies used by pathogens to influence the small ubiquitin-like modifier (SUMO) system of host eukaryotic cells. J Innate Immun. 2012;4:159–167. doi: 10.1159/000335027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyon JA. Establishing intracellular infection: modulation of host cell functions (Anaplasmataceae) In: Palmer GH, Azad A, editors. Intracellular Pathogens II: Rickettsiales. Washington, D. C: ASM Press; 2012. [Google Scholar]
- Carlyon JA, Chan WT, Galan J, Roos D, Fikrig E. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J Immunol. 2002;169:7009–7018. doi: 10.4049/jimmunol.169.12.7009. [DOI] [PubMed] [Google Scholar]
- Cortes C, Rzomp KA, Tvinnereim A, Scidmore MA, Wizel B. Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect Immun. 2007;75:5586–5596. doi: 10.1128/IAI.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy PS, Luo T, McBride JW. Ehrlichia chaffeensis Exploits Host SUMOylation Pathways To Mediate Effector-Host Interactions and Promote Intracellular Survival. Infect Immun. 2014;82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, et al. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- Hemshekhar M, Sebastin Santhosh M, Kemparaju K, Girish KS. Emerging Roles of Anacardic Acid and Its Derivatives: A Pharmacological Overview. Basic Clin Pharmacol Toxicol. 2011 doi: 10.1111/j.1742-7843.2011.00833.x. [DOI] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Molecular microbiology. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- Huang B, Hubber A, McDonough JA, Roy CR, Scidmore MA, Carlyon JA. The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes. Cell Microbiol. 2010a;12:1292–1307. doi: 10.1111/j.1462-5822.2010.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ojogun N, Ragland SA, Carlyon JA. Monoubiquitinated proteins decorate the Anaplasma phagocytophilum-occupied vacuolar membrane. FEMS immunology and medical microbiology. 2012;64:32–41. doi: 10.1111/j.1574-695X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, et al. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb Pathog. 2010b;49:273–284. doi: 10.1016/j.micpath.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Troese MJ, Ye S, Sims JT, Galloway NL, Borjesson DL, Carlyon JA. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect Immun. 2010c;78:1864–1873. doi: 10.1128/IAI.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Stork W, Mudgett MB. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell host & microbe. 2013;13:143–154. doi: 10.1016/j.chom.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, Mudgett MB. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in xanthomonas-infected tomato leaves. Plant Cell. 2008;20:1915–1929. doi: 10.1105/tpc.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. Ehrlichia chaffeensis TRP120 Interacts with a Diverse Array of Eukaryotic Proteins Involved in Transcription, Signaling, and Cytoskeleton Organization. Infection and immunity. 2011;79:4382–4391. doi: 10.1128/IAI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, McBride JW. Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infect Immun. 2012;80:2297–2306. doi: 10.1128/IAI.00154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Matsuo J, Rahman MA, Kumagai Y, Li X, Rikihisa Y. Ehrlichia chaffeensis induces monocyte inflammatory responses through MyD88, ERK, and NF-kappaB but not through TRIF, interleukin-1 receptor 1 (IL-1R1)/IL-18R1, or toll-like receptors. Infection and immunity. 2011;79:4947–4956. doi: 10.1128/IAI.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Lynch MJ, Herron MJ, Palmer AT, Kurtti TJ, Nelson RD, Goodman JL. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet Microbiol. 2004;101:53–64. doi: 10.1016/j.vetmic.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Developmental cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS pathogens. 2010;6:e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20800–20807. doi: 10.1073/pnas.1218674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, et al. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- Ribet D, Hamon M, Gouin E, Nahori MA, Impens F, Neyret-Kahn H, et al. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 2010;464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DD, Grosenbach D, Hruby DE, Peacock MG, Heinzen RA, Hackstadt T. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Molecular microbiology. 1997;24:217–228. doi: 10.1046/j.1365-2958.1997.3371700.x. [DOI] [PubMed] [Google Scholar]
- Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol Plant Microbe Interact. 2004;17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional Heterogeneity of Small Ubiquitin-related Protein Modifiers SUMO-1 versus SUMO-2/3. Journal of Biological Chemistry. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Storey JR, Doros-Richert LA, Gingrich-Baker C, Munroe K, Mather TN, Coughlin RT, et al. Molecular cloning and sequencing of three granulocytic Ehrlichia genes encoding high-molecular-weight immunoreactive proteins. Infect Immun. 1998;66:1356–1363. doi: 10.1128/iai.66.4.1356-1363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran B, Mastronunzio JE, Narasimhan S, Fankhauser S, Uchil PD, Levy R, et al. Anaplasma phagocytophilum AptA modulates Erk1/2 signalling. Cellular microbiology. 2011;13:47–61. doi: 10.1111/j.1462-5822.2010.01516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. The Journal of biological chemistry. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Teng S, Luo H, Wang L. Predicting protein sumoylation sites from sequence features. Amino Acids. 2012;43:447–455. doi: 10.1007/s00726-011-1100-2. [DOI] [PubMed] [Google Scholar]
- Troese MJ, Carlyon JA. Anaplasma phagocytophilum dense-cored organisms mediate cellular adherence through recognition of human P-selectin glycoprotein ligand 1. Infect Immun. 2009;77:4018–4027. doi: 10.1128/IAI.00527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Kahlon A, Ragland SA, Ottens AK, Ojogun N, Nelson KT, et al. Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense-cored cell. Infection and immunity. 2011;79:4696–4707. doi: 10.1128/IAI.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchan HK, Seidman D, Carlyon JA. Breaking in and grabbing a meal: Anaplasma phagocytophilum cellular invasion, nutrient acquisition, and promising tools for their study. Microbes and infection/Institut Pasteur. 2013;15:1017–1025. doi: 10.1016/j.micinf.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. The fast-growing business of SUMO chains. Mol Cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Vertegaal ACO, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and Overlapping Sets of SUMO-1 and SUMO-2 Target Proteins Revealed by Quantitative Proteomics. Molecular & Cellular Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Wakeel A, Kuriakose JA, McBride JW. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect Immun. 2009;77:1734–1745. doi: 10.1128/IAI.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeel A, Zhang X, McBride JW. Mass spectrometric analysis of Ehrlichia chaffeensis tandem repeat proteins reveals evidence of phosphorylation and absence of glycosylation. PLoS One. 2010;5:e9552. doi: 10.1371/journal.pone.0009552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VG. Sumoylation at the Host-Pathogen Interface. Biomolecules. 2012;2:203–227. doi: 10.3390/biom2020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer P, Schreiner S, Dobner T. Human pathogens and the host cell SUMOylation system. J Virol. 2012;86:642–654. doi: 10.1128/JVI.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Hsu CT, Ting CY, Liu LF, Hwang J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. The Journal of biological chemistry. 2006;281:8264–8274. doi: 10.1074/jbc.M510364200. [DOI] [PubMed] [Google Scholar]
- Zhu B, Kuriakose JA, Luo T, Ballesteros E, Gupta S, Fofanov Y, McBride JW. Ehrlichia chaffeensis TRP120 Binds a G+C-Rich Motif in Host Cell DNA and Exhibits Eukaryotic Transcriptional Activator Function. Infection and immunity. 2011;79:4370–4381. doi: 10.1128/IAI.05422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun. 2009;77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]