Significance
Actin is a monomeric protein that can polymerize into branched networks. Actin polymerization acts like an engine to drive cell movement and is regulated by multiple interacting proteins on the cell membrane. To understand the molecular details of how cells transmit signals from the membrane to the actin polymerization engine, we reconstituted this process in a test tube using seven purified proteins and membrane-coated glass beads. Using this “biomimetic” system, we discovered a sequence motif in the human protein Nck that activates a core component of the actin polymerization engine. This motif shares similarity with certain bacterial virulence factors that stimulate actin polymerization in infected human cells, suggesting that similar activation mechanisms have evolved in humans and bacterial pathogens.
Keywords: signal transduction, actin cytoskeleton, SH3 adapter, Nck, N-WASP
Abstract
Actin filament networks assemble on cellular membranes in response to signals that locally activate neural Wiskott–Aldrich-syndrome protein (N-WASP) and the Arp2/3 complex. An inactive conformation of N-WASP is stabilized by intramolecular contacts between the GTPase binding domain (GBD) and the C helix of the verprolin-homology, connector-helix, acidic motif (VCA) segment. Multiple SH3 domain-containing adapter proteins can bind and possibly activate N-WASP, but it remains unclear how such binding events relieve autoinhibition to unmask the VCA segment and activate the Arp2/3 complex. Here, we have used purified components to reconstitute a signaling cascade driven by membrane-localized Src homology 3 (SH3) adapters and N-WASP, resulting in the assembly of dynamic actin networks. Among six SH3 adapters tested, Nck was the most potent activator of N-WASP–driven actin assembly. We identify within Nck a previously unrecognized activation motif in a linker between the first two SH3 domains. This linker sequence, reminiscent of bacterial virulence factors, directly engages the N-WASP GBD and competes with VCA binding. Our results suggest that animals, like pathogenic bacteria, have evolved peptide motifs that allosterically activate N-WASP, leading to localized actin nucleation on cellular membranes.
Actin polymerization provides the force that drives membrane protrusion during cell motility as well as the propulsion of endocytic vesicles and intracellular pathogens. Branched actin networks are assembled on the surface of cellular membranes, where actin monomers are incorporated into the membrane-apposed ends of growing actin filaments (1–3). The formation of new branches is initiated by the Arp2/3 complex, which requires allosteric activation by membrane-associated nucleation-promoting factors. Neural Wiskott–Aldrich-syndrome protein (N-WASP) is an essential nucleation-promoting factor that integrates and transduces membrane-localized signals to the Arp2/3 complex.
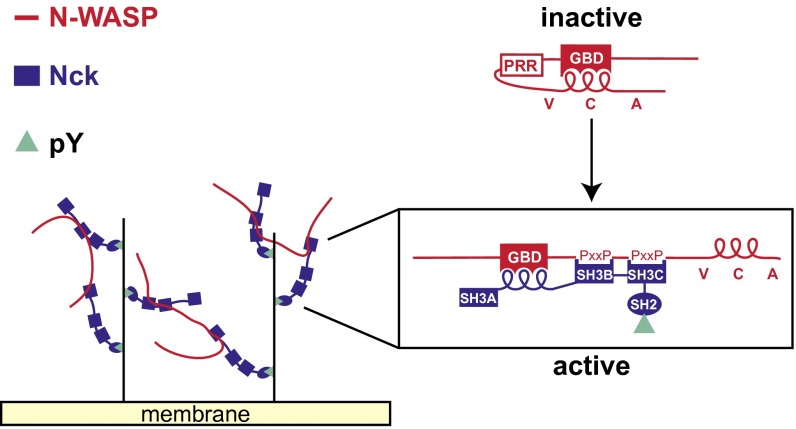
N-WASP constitutes a regulatory hub whose localization and activation state govern the spatiotemporal dynamics of actin network formation. Under resting conditions, N-WASP exists in an autoinhibited conformation in the cytoplasm. Signaling from tyrosine kinases, GTPases, and acidic phospholipids cooperatively activates N-WASP on the membrane (4–7). Two principal themes have emerged to describe N-WASP regulatory mechanisms: allosteric activation and oligomerization (8). Allosteric activation disrupts intramolecular autoinhibitory contacts between the C helix and the GTPase binding domain (GBD). These interactions maintain N-WASP in a closed conformation that sterically occludes its carboxyl-terminal verprolin-homology, connector-helix, acidic motif (VCA) segment (9, 10) (Fig. 3A). The small GTPase Cdc42 is the archetypal allosteric N-WASP activator. Cdc42 binds directly to the GBD and releases the VCA segment (11), which subsequently binds the Arp2/3 complex and promotes actin filament nucleation from the side of a preexisting actin filament. N-WASP oligomerization or clustering, mediated by signaling adapter proteins and acidic phospholipids, facilitates simultaneous interaction of two N-WASP molecules with one Arp2/3 complex. Simultaneous engagement of the constitutively inactive Arp2/3 complex by two VCA-type ligands is required for Arp2/3-mediated actin nucleation (12, 13).
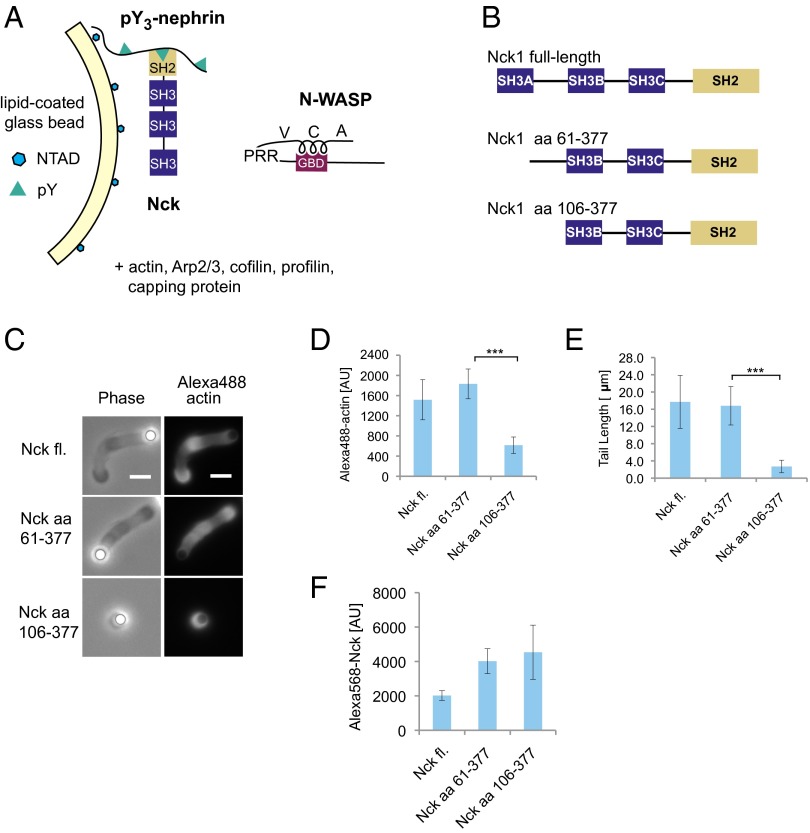
Fig. 3.
An inter-SH3 linker in Nck promotes actin network assembly by N-WASP. (A) Experimental strategy for localization of Nck to membranes containing pY3-nephrin. (B) Nck constructs used in the deletion analysis. (C) Representative images of actin tails formed by Nck deletion constructs. Motility reactions contained 200 nM N-WASP and 100 nM Nck. (Scale bar: 5 μm.) (D) Integrated fluorescence intensity of Alexa488 actin tails. (E) Actin tail length. (F) Integrated fluorescence intensity of bead-localized Alexa568 Nck constructs. (Mean ± SD; n > 20; ***P < 0.001.) fl., Full length; PRR, proline-rich region.
Src homology 3 (SH3) domain-containing adapter proteins have also been shown to activate N-WASP. Such signaling adapters often harbor multiple SH3 domains, each capable of binding a canonical polyproline motif (14). Genetic, cell biological, and biochemical evidence supports a role for the SH2/SH3 adapter protein Nck in the activation of N-WASP (15–22). Phosphorylated tyrosine residues on membrane receptors, such as the podocyte adhesion receptor nephrin and the vaccinia virus membrane protein A36R, localize Nck to the plasma membrane through its SH2 domain. Nck then directly binds N-WASP and the N-WASP–associated protein WIP, leading to localized actin polymerization (15, 20, 21, 23). Similar to Nck, Grb2 is an SH2/SH3 adapter that binds and activates N-WASP (24, 25), acting in concert with Nck to promote actin-dependent vaccinia virus motility (26). Other SH3 adapter proteins implicated in N-WASP activation include Crk-II (27), cortactin (28), Toca/CIP4 (29, 30), and Tks4/5 (31) (Fig. 1A). The mechanism by which SH3 adapters activate N-WASP is only partially understood. Although SH3-mediated oligomerization has been shown to increase N-WASP activity, these experiments were performed with a constitutively active N-WASP mutant lacking the GBD–VCA autoinhibitory interaction (32). Key unresolved questions are whether and how SH3 adapters counteract the autoinhibitory interactions that restrain access to N-WASP’s VCA segment.
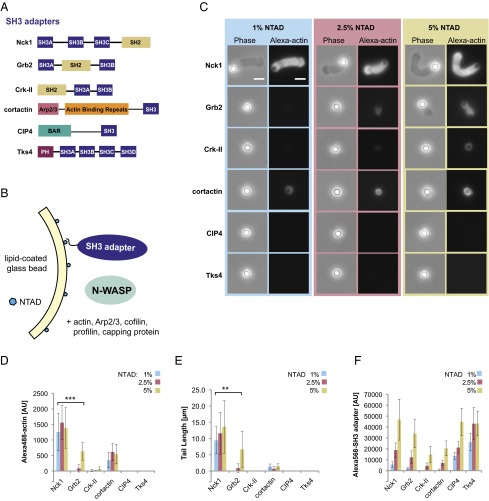
Fig. 1.
Membrane-associated actin networks assembled by SH3 adapters and N-WASP. (A) Domain organization of SH3 adapter proteins used in this study. (B) Experimental strategy for localization of His-tagged SH3 adapters using NTAD-doped membranes supported on silica microspheres. (C) Phase contrast and fluorescence images of lipid-coated beads (NTAD density: 1%, 2.5%, or 5%) incubated with the indicated SH3 adapter (250 nM), N-WASP (50 nM), Alexa488 actin, and actin regulatory components. After 15 min, reactions were fixed with glutaraldehyde. (Scale bar: 5 μm.) (D) Integrated fluorescence intensity of Alexa488 actin tails. (E) Actin tail length. (F) Integrated fluorescence intensity of bead-localized Alexa568 SH3 adapters (250 nM). (Mean ± SD; n > 20.) Asterisks indicate a significant difference between Nck1 and Grb2 at 1% and 5% NTAD: **P < 0.01; ***P < 0.001, respectively.
Here, we systematically compare the ability of distinct SH3 adapters to assemble actin and Arp2/3 networks while localized to a membrane surface. To this end, we have biochemically reconstituted the N-WASP/actin signaling cascade from pure components using a membrane bilayer supported on silica microspheres. We find that membrane-localized actin assembly varies dramatically depending on the SH3 adapter, with only Nck showing robust actin assembly. Structure–function analysis revealed a previously unidentified N-WASP activation motif embedded within a 45-aa linker that connects the first two SH3 domains of Nck. This conserved inter-SH3 domain linker binds directly to the N-WASP GBD and in concert with the SH3 domains, potently stimulates actin network assembly on membranes. Nck thus uses both allosteric and oligomerization-based mechanisms to activate N-WASP.
Results
Polarized Actin Network Assembly Promoted by SH3 Adapters and N-WASP.
We initially compared six full-length SH3 adapter proteins (Fig. 1A) using a well-established reconstituted motility system that contains purified N-WASP (amino acids 151–501; lacking the N-terminal EVH1 domain), Arp2/3 complex, actin, and the actin regulatory proteins profilin, cofilin, and capping protein (33, 34). As described in the Introduction, all six SH3 adapters have been implicated in cellular N-WASP activation and have been shown to bind the N-WASP proline-rich region. Because N-WASP–mediated actin assembly occurs on the cytosolic surface of cellular membranes, we used lipid bilayers supported on silica microspheres (7, 35, 36). Supported lipid bilayers were supplemented with a nickel-chelating lipid [1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (NTAD)] to localize His6-tagged versions of the SH3 adapter proteins (Fig. 1B). To test a range of SH3 adapter densities, we varied the molar fraction of NTAD in the lipid bilayers (1%, 2.5%, and 5% NTAD, mol%).
Lipid-coated beads were incubated in a solution containing the purified full-length SH3 adapter (250 nM), Alexa488-labeled actin, N-WASP, and the actin regulatory components used at concentrations that were previously shown to support bead motility (7, 33). After 15 min, reactions were fixed with glutaraldehyde, and the bead-associated actin structures were analyzed by wide-field epifluorescence imaging. Two measurements were used to quantify bead-associated actin: integrated fluorescence intensity of Alexa488 actin and actin tail length.
Nck was the only SH3 adapter that induced polarized actin tails on >95% of the beads at the lowest NTAD density tested (Fig. 1C and Fig. S1). Image analysis revealed that, in reactions driven by Nck, bead-associated actin intensity and tail length were already maximal at the lowest NTAD density (Fig. 1 D and E). Moreover, Nck-coated beads displayed robust actin-dependent motility when imaged live in a flow chamber containing N-WASP, Arp2/3 complex, and actin regulatory components (Movie S1). By contrast, Grb2 showed substantially weaker activity. Grb2-induced tails were not detected at 1% NTAD, and only diffuse actin “clouds” or short tails were observed at 2.5% and 5% NTAD, respectively (Fig. 1C, row 2). The Grb2-induced tails at 5% NTAD density contained one-half as much actin as the Nck-induced tails obtained at 1% NTAD and were significantly shorter (Fig. 1 D and E). A thin shell of bead-associated actin was visible in reactions containing cortactin (Fig. 1 C and D), consistent with its ability to directly activate Arp2/3 and bind actin filaments (37). However, cortactin did not promote the assembly of polarized actin tails. Reactions containing Crk-II, CIP4, or Tks4 were devoid of visible actin structures.
Fig. S1.
Low-magnification images of a motility reaction with His6-Nck. Epifluorescence images of a motility reaction using 1% NTAD lipids, 250 nM His6-Nck, and 50 nM N-WASP. Reactions were fixed with 3% glutaraldehyde after 15 min. Atto390-DOPE lipid is in magenta, Alexa488 actin is in green. (Scale bars: 30 µm.)
To ensure that the observed effects in the actin tail assembly assay were not caused simply by differential recruitment of the SH3 adapters, we used live imaging to quantify the fluorescence intensity of Alexa568-labeled adapters on the bead surface. Despite the common recruitment mechanism through hexahistidine tag and NTAD lipids, the adapters localized to different extents. At an NTAD density of 1%, the fluorescence intensities measured for CIP4 and Tks4 were significantly higher than for Nck, Grb2, Crk-II, and cortactin, all of which were used at a concentration of 250 nM (Fig. 1F, blue bars). We speculate that these differences arise from intrinsic lipid binding and/or a propensity to oligomerize on membrane surfaces. Most importantly, the Nck density at 1% NTAD was significantly lower than the density of the other five adapters at 5% NTAD. Despite achieving higher membrane densities than Nck, the other SH3 adapters were significantly less active or inactive. These results show that, of six SH3 adapters tested, Nck has by far the strongest ability to assemble polarized actin networks on the surface of supported membranes.
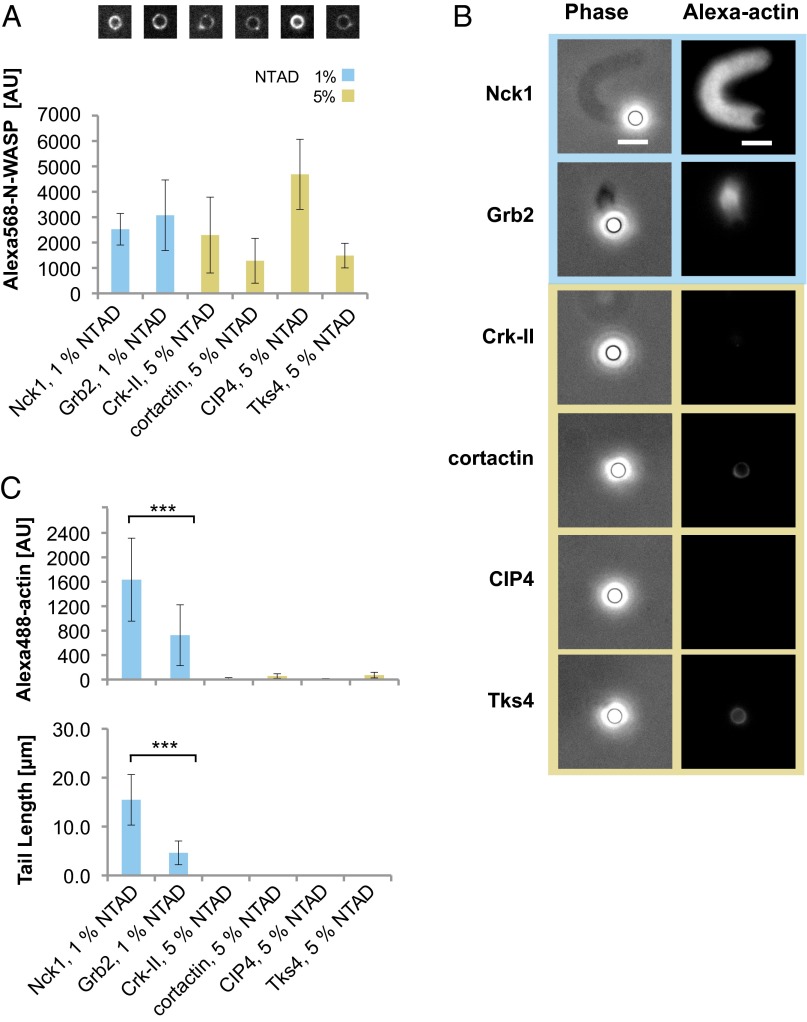
A potential explanation for Nck’s superior activity is that it recruits more N-WASP to the membrane compared with the other SH3 adapters. To test this, we measured the density of Alexa568-labeled N-WASP recruited to lipid beads in the presence of the His-tagged adapters. In initial experiments, Nck and Grb2 recruited similar levels of N-WASP, but the other SH3 adapters recruited substantially lower amounts. To compensate for these differences in apparent affinity, we increased the N-WASP concentration to 200 nM. Additionally, we increased the NTAD density to 5% for Crk-II, cortactin, CIP4, and Tks4, while maintaining 1% NTAD for Nck and Grb2. Under these conditions, N-WASP levels recruited by all of the SH3 adapters were within approximately twofold of each other (Fig. 2A). Despite the similar N-WASP density observed with all six SH3 adapters, Nck showed the strongest actin assembly activity (Fig. 2B). Tails assembled by Grb2 were approximately threefold shorter and incorporated approximately twofold less actin compared with Nck (Fig. 2C). Similar to our previous results (Fig. 1 C and D), Crk-II, cortactin, CIP4, and Tks4 did not assemble any detectable actin structures. These data indicate that the increased ability of Nck to assemble membrane-associated actin networks cannot be explained solely by enhanced N-WASP recruitment. We therefore considered the possibility that, among the SH3 adapters tested, Nck might deploy a unique N-WASP activation mechanism.
Fig. 2.
Nck is the most effective SH3 adapter in promoting N-WASP–dependent actin assembly. (A) Integrated fluorescence intensity and representative images of bead-localized Alexa568 N-WASP (200 nM) incubated with NTAD-doped lipid-coated beads and the indicated His-SH3 adapters (250 nM). To compensate for differences in N-WASP recruitment efficiency, the NTAD density was increased to 5% for Crk-II, cortactin, CIP4, and Tks4, whereas 1% NTAD was used for Nck1 and Grb2. (B) Representative phase contrast and fluorescence images of actin assembly reactions under conditions reported in A. (Scale bars: 5 μm.) (C) Integrated fluorescence intensity of Alexa488 actin tails and tail lengths (mean ± SD; n > 20). Asterisks indicate significant differences between Nck and Grb2: ***P < 0.001.
Discovery of an Inter-SH3 Domain N-WASP Activation Motif.
Nck is recruited to cellular membranes through its SH2 domain, which binds phosphorylated tyrosine residues on integral membrane receptors or membrane-associated adapter proteins. Nephrin is an Nck receptor on the plasma membrane of podocytes. Three phosphorylated tyrosine residues on nephrin bind the SH2 domain of Nck, and this interaction is essential for regulating podocyte actin dynamics in an N-WASP–dependent manner (20, 21).
To explore the mechanism of Nck-mediated N-WASP activation on membranes, we used a His-tagged, triphosphorylated nephrin peptide (His8-pY3-nephrin), which recruits untagged Nck to the surface of NTAD-containing membranes (38). Lipid-coated beads with 1% NTAD were preincubated with His8-pY3-nephrin and subsequently added to a solution containing untagged full-length Nck, N-WASP, and the actin regulatory components described above (Fig. 3A). Similar to His-tagged Nck recruited through NTAD lipids (Fig. 1C), untagged Nck recruited through pY3-nephrin promoted the assembly of actin tails on >95% of the lipid-coated beads (Fig. 3C and Fig. S2). Actin assembly promoted by untagged Nck was strictly dependent on the addition of His8-pY3-nephrin, and Nck mutants lacking the SH2 domain were inactive in this system.
Fig. S2.
Low-magnification images of a motility reaction with pY3-nephrin recruitment. Epifluorescence images of a motility reaction using 1% NTAD lipids, 100 nM Nck, and 200 nM N-WASP recruited to the beads by pY3-nephrin. Reactions were fixed with 3% glutaraldehyde after 15 min. Atto390-DOPE lipid is in magenta, Alexa488 actin is in green. (Scale bars: 30 µm.)
Using the pY3-nephrin system, we performed a deletion analysis of Nck (Fig. 3B). Whereas deletion of the amino-terminal SH3A domain had no effect, additional deletion of the inter-SH3AB linker led to a drastic reduction in actin tails (Fig. 3C). The few residual actin structures formed by this deletion construct contained threefold lower actin fluorescence, and the average tail length was reduced by 85% (Fig. 3 D and E). To exclude the possibility that decreased actin assembly was a trivial result of reduced membrane recruitment, we measured surface densities of the three Nck constructs in a separate experiment. Both deletion mutants were recruited similarly to one another and to a slightly higher extent than full-length Nck (Fig. 3F). Hence, the Nck mutant lacking the inter-SH3AB linker is recruited to pY3-nephrin but is specifically defective in promoting actin assembly (Fig. 3F). These experiments identify for the first time, to our knowledge, a 45-aa linker motif between the first two SH3 domains of Nck that strongly potentiates N-WASP–dependent actin assembly on membranes.
Direct Binding of the Inter-SH3 Linker to the N-WASP GBD.
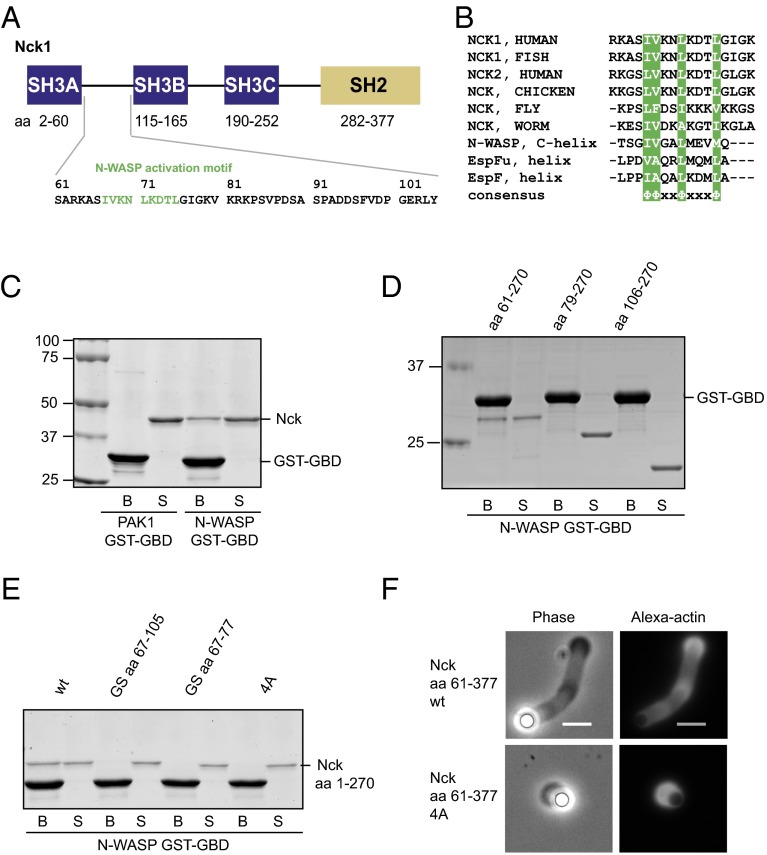
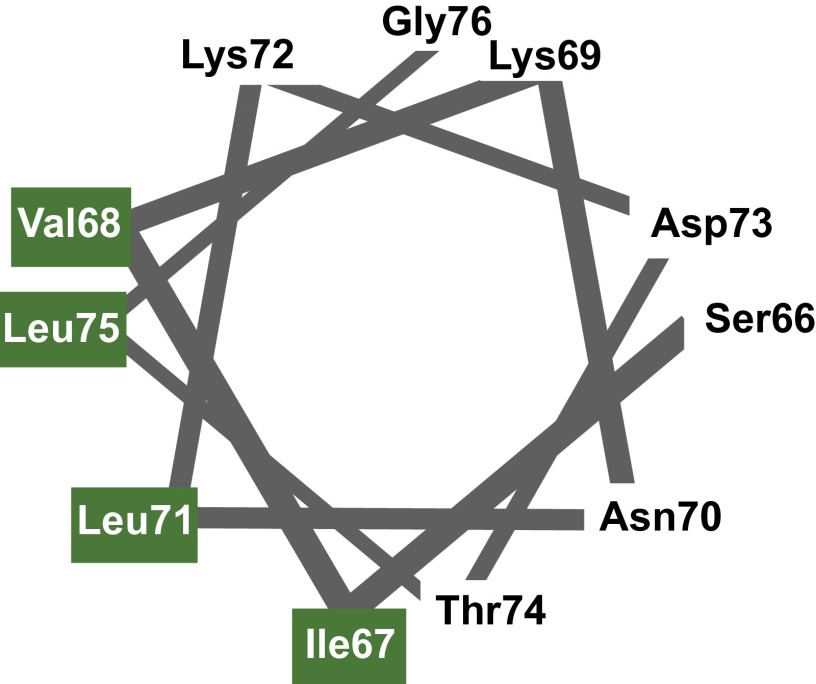
We hypothesized that the inter-SH3AB linker (Nck1 amino acids 61–105) might interact directly with N-WASP. Within this linker is a 9-aa motif (amino acids 67–75) that is highly conserved across species (Fig. 4 A and B). Moreover, the spacing of the hydrophobic residues suggested the possibility of forming an amphipathic helix (Fig. 4B and Fig. S3). Several amphipathic helical peptides have been shown to bind N-WASP through its GBD. In the autoinhibited state, the N-WASP GBD binds intramolecularly to the amphipathic C helix in the carboxyl-terminal VCA region (10) (Fig. 3A). Amphipathic helices can also activate N-WASP: helical motifs within the Escherichia coli virulence factors, EspFU and EspF, bind the GBD in competition with the inhibitory C helix (39–41). To our knowledge, such an activation mechanism has only been documented with bacterial pathogens and has not been found with endogenous eukaryotic activators.
Fig. 4.
The inter-SH3AB linker contains a conserved motif and binds directly to the N-WASP GBD. (A) Nck domain structure and sequence of the inter-SH3AB linker. (B) Sequence alignment comparing the Nck inter-SH3 linker from various organisms and previously known N-WASP GBD ligands. (C) Coomassie-stained gel of GST pull-down samples. Immobilized GST-GBD from N-WASP or PAK1 was used to pull down full-length Nck. (D) N-WASP GST-GBD was used to pull down the indicated Nck deletion constructs. (E) N-WASP GST-GBD was used to pull down Nck amino acids 1–270 WT and mutants. Samples comprising 1.25% of the unbound supernatant (S) and 12.5% of the bound fraction (B) were separated by SDS/PAGE and stained with Coomassie. GS aa 67–105, substitution of the entire SH3AB linker with a Gly-Ser linker; GS aa 67–77, substitution of the hydrophobic motif with a Gly-Ser linker. (F) Representative images of actin tails formed by Nck amino acids 61–377 WT and 4A mutant. Motility reactions contained 200 nM N-WASP and 100 nM Nck. (Scale bar: 5 μm.)
Fig. S3.
The hydrophobic residues of the inter-SH3 motif in Nck form a hydrophobic surface if arranged in an α-helical conformation.
To test whether the inter-SH3AB linker can directly interact with the N-WASP GBD, we first performed GST pull-down experiments with purified proteins. Full-length Nck bound to glutathione beads loaded with N-WASP GST-GBD but not to beads loaded with a structurally and functionally related GST-GBD derived from the PAK1 kinase (Fig. 4C). These results indicate a specific, direct, and previously undocumented interaction between Nck and the N-WASP GBD. Deletion of the SH2 and the SH3A domains from Nck did not affect the interaction (Fig. 4D). However, additional deletion of only 18 amino acids from the inter-SH3AB linker, including the putative amphipathic helix, completely abrogated binding to the GBD along with deletion of the entire linker (Fig. 4D). To further assess the importance of the inter-SH3AB linker, we used a construct lacking the SH2 domain (Nck amino acids 1–270). Replacing either the entire linker (amino acids 67–105) or only the hydrophobic motif (amino acids 67–77) with a Gly-Ser linker of equal length abolished the interaction (Fig. 4E). Finally, GBD binding was completely prevented by four alanine mutations at hydrophobic positions within the putative amphipathic helix [Ile67/Val68/Leu71/Leu75 (4A)]. These results strongly suggest that the inter-SH3AB linker, implicated above in promoting N-WASP–dependent actin assembly (Fig. 3 C–E), mediates direct interactions between Nck and the N-WASP GBD.
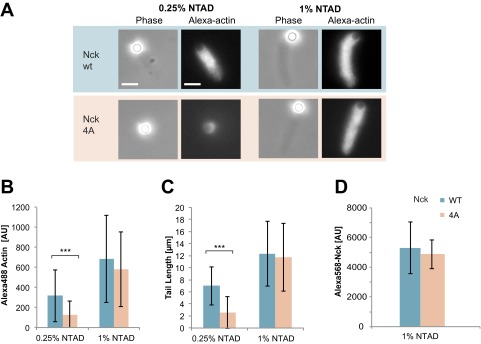
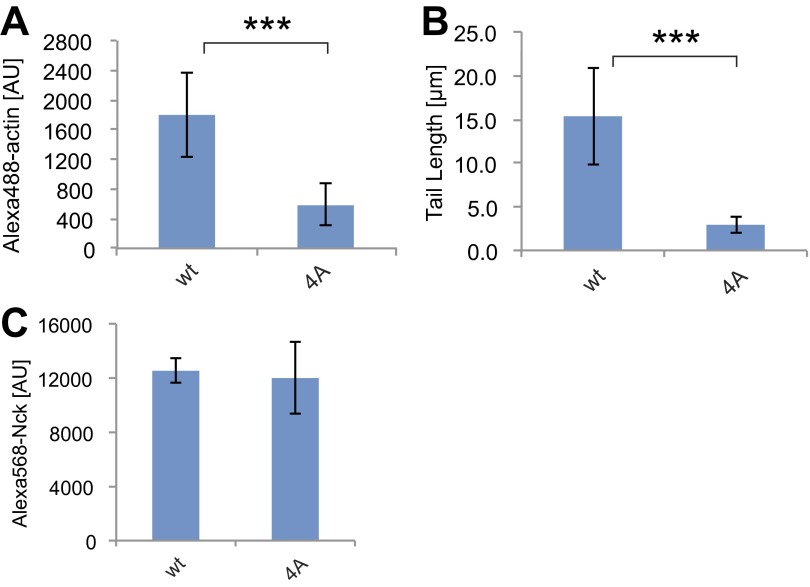
We next evaluated actin comet tail assembly by full-length Nck containing the four Ala mutations in the inter-SH3AB linker. In the presence of lipid-coated beads with a high NTAD density (1%), 4A Nck formed tails similar to the WT (Fig. S4). However, with a lower density of 0.25% NTAD, 4A Nck was defective, producing two- to threefold less actin than WT Nck (P < 0.001) (Fig. S4 A–C). We hypothesized that, at higher NTAD densities, polyvalent SH3-mediated interactions could potentially compensate for the defective linker in 4A Nck. To test this, we introduced the four Ala mutations in a construct lacking the SH3A domain (Nck amino acids 61–377). Here, the 4A mutant was strongly compromised in its ability to assemble actin tails, displaying a threefold reduction in polymerized actin and a fivefold decrease in tail length, even at 1% NTAD (P < 0.001) (Fig. 4F and Fig. S5 A and B). Collectively, these data establish a role for the amphipathic motif in actin comet tail formation, which becomes essential under conditions of low pY3-nephrin density or when the first SH3 domain is removed.
Fig. S4.
Comparison of full-length WT Nck with a 4A mutant hydrophobic motif. Motility assays were performed using 200 nM N-WASP and 100 nM Nck. Full-length Nck WT and 4A mutant were assayed. Nck was recruited to the membrane by pY3-nephrin. (A) Phase contrast and fluorescence images of motility reactions. (Scale bar: 5 μm.) (B) Integrated fluorescence intensity of Alexa488 actin tails. (C) Actin tail length. ***P < 0.001. (D) Alexa568 Nck recruited to the lipid-coated beads at 1% NTAD. Binding reactions contained 200 nM N-WASP and 100 nM Nck. Nck levels at the membrane were indistinguishable from background at 0.25% NTAD.
Fig. S5.
Bead motility reactions using Nck amino acids 61–377 (∆SH3A) WT and 4A mutant. Motility assays were performed using 200 nM N-WASP and 100 nM Nck amino acids 61–377 at 1% NTAD density. WT and the 4A mutant were assayed. Nck was recruited to the membrane by pY3-nephrin. (A) Integrated fluorescence intensity of Alexa488 actin tails. (B) Actin tail length. ***P < 0.001. (C) Alexa568 Nck recruited to the lipid-coated beads at 1% NTAD.
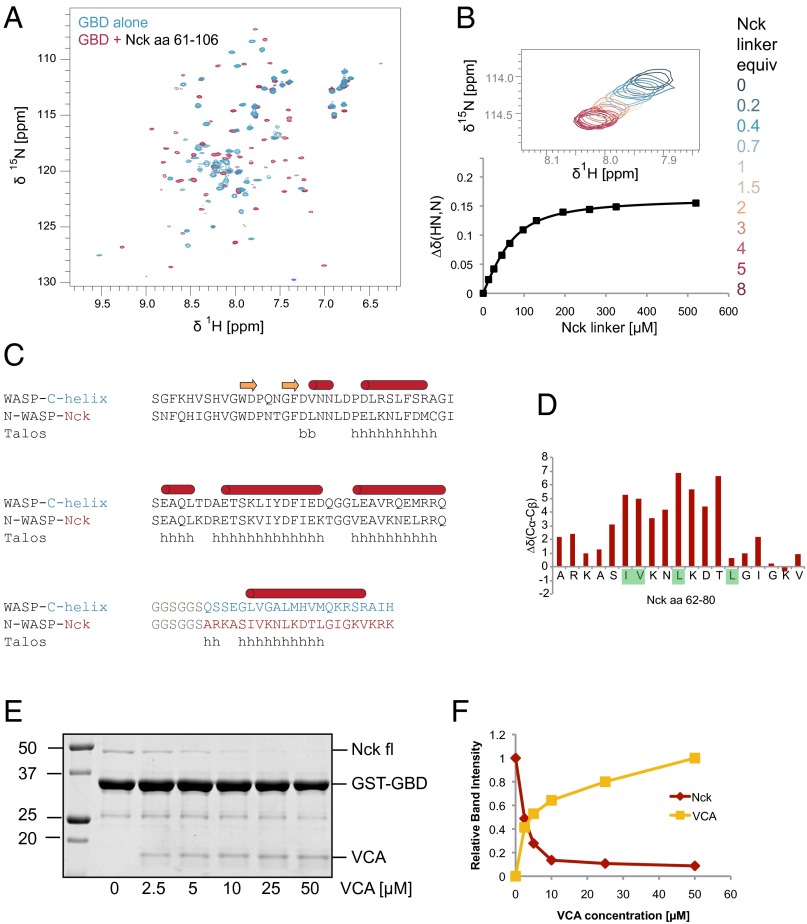
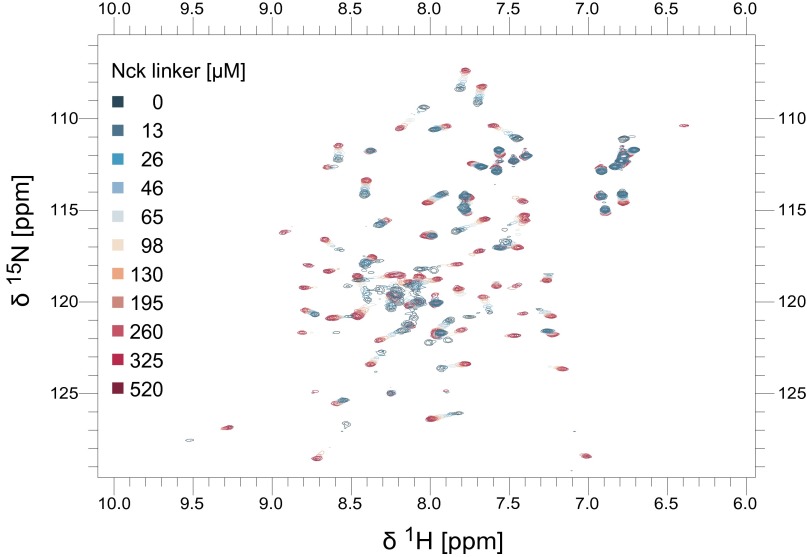
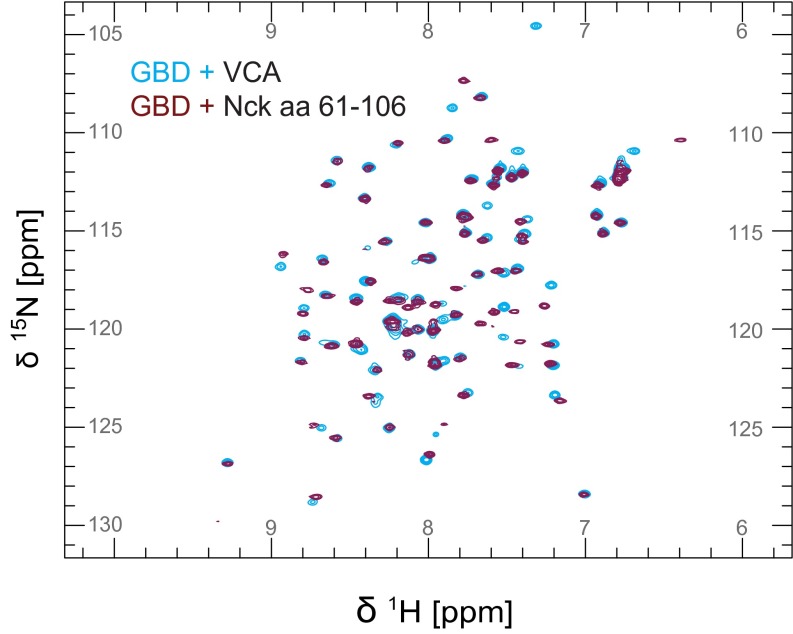
Structural studies of the WASP GBD (68% sequence identity to N-WASP GBD) have revealed a high degree of conformational plasticity. In the absence of a binding partner, the WASP GBD is partially folded or unstructured. Binding of the autoinhibitory C helix (10) or an activating EspFU-derived peptide (40) causes the GBD to adopt a mostly helical globular fold. To test whether the Nck linker behaves similarly, we recorded heteronuclear single-quantum coherence (HSQC) spectra of 15N-labeled N-WASP GBD in the presence and absence of the unlabeled Nck linker peptide (amino acids 61–106). The GBD spectrum in the presence of the Nck linker peptide revealed greater peak dispersion and more uniform peak intensities and line widths compared with the GBD alone (Fig. 5A), consistent with a more compact, folded domain. Based on chemical shift differences from NMR spectra recorded with increasing concentrations of the Nck peptide, we determined the equilibrium Kd to be 33 ± 8 µM (Fig. 5B and Fig. S6). We also recorded the N-WASP GBD spectrum with the VCA peptide and observed similar chemical shifts to those recorded in the presence of the Nck linker peptide: 80% of the peaks show a weighted chemical shift difference of less than 0.1 ppm (Fig. S7).
Fig. 5.
NMR analysis reveals direct binding of the Nck inter-SH3 linker and the N-WASP GBD. (A) [15N, 1H]-heteronuclear single-quantum coherence (HSQC) NMR spectra of 15N-labeled N-WASP GBD acquired in the presence and absence of unlabeled Nck linker peptide (amino acids 61–106). (B) NMR titration of 15N-labeled GBD (65 μM) with the Nck linker peptide. Chemical shift changes, along with the corresponding curve fit, are shown for a representative peak from the [15N, 1H]-HSQC spectrum. A binding Kd of 33 ± 8 μM was determined by fitting titration curves to a total of 17 peaks. (C) Sequence alignment of a WASP-GBD C-helix construct (10) and the N-WASP GBD Nck linker fusion protein. The secondary structure of the WASP–C helix complex (1EJ5.pdb) is indicated. (D) Secondary structure prediction of the N-WASP Nck fusion protein by Talos+ is shown. (E) Coomassie-stained gel from a competition pull-down experiment (bound fraction). N-WASP GST-GBD was used to pull down Nck in the presence of increasing N-WASP VCA. (F) Integrated band intensities from the Coomassie-stained gel showing Nck and VCA bound to GST-GBD.
Fig. S6.
[15N, 1H]-HSQC spectra of a titration of 65 µM 15N-labeled N-WASP GBD with unlabeled Nck linker peptide (0–8 eq).
Fig. S7.
Overlay of [15N, 1H] HSQC spectra of 15N-labeled N-WASP GBD complexed with unlabeled N-WASP VCA (light blue) or Nck linker (dark red) peptide.
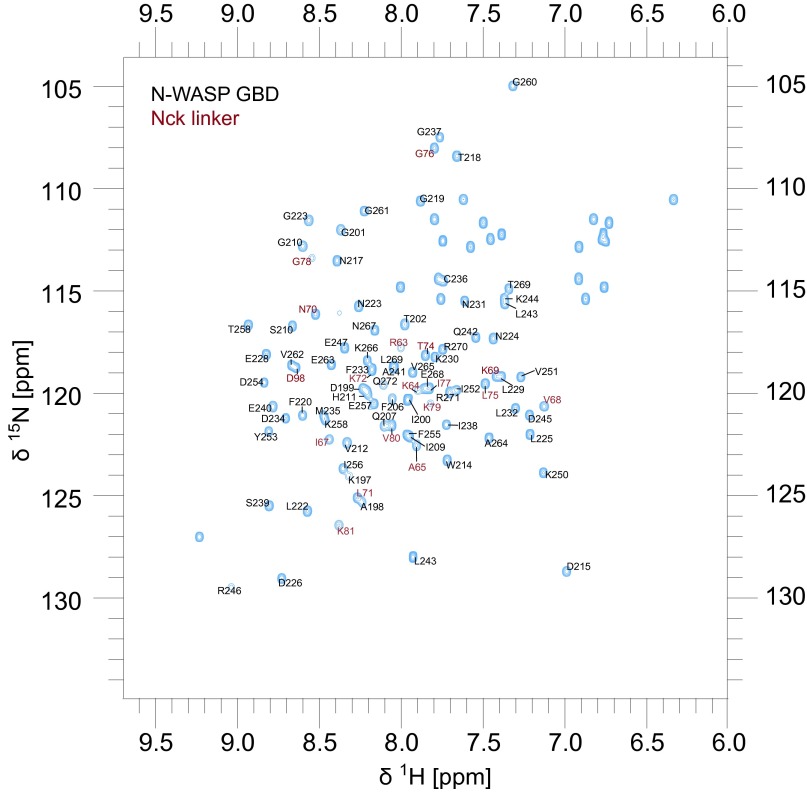
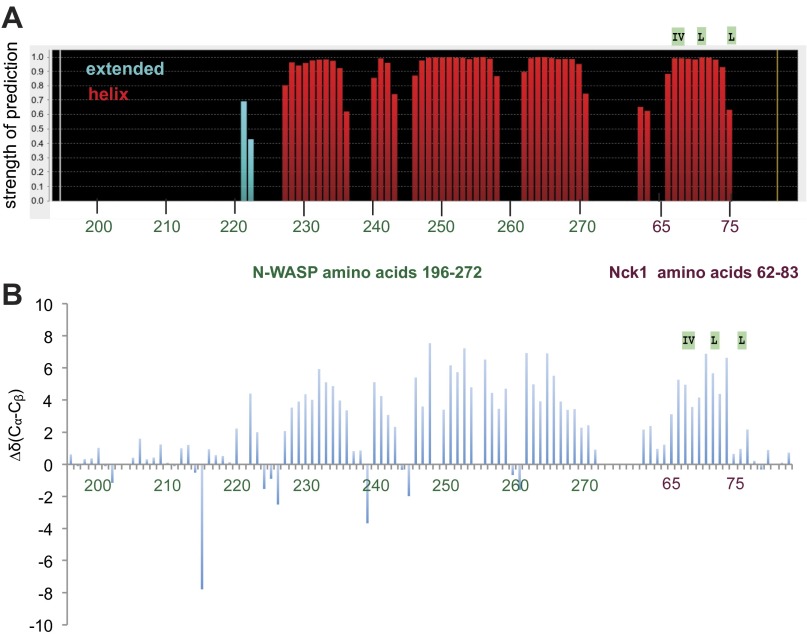
To characterize the GBD–Nck linker complex in greater detail, we used standard triple-resonance experiments to assign the backbone (86% completion) of a construct comprising the N-WASP GBD covalently linked to the inter-SH3AB segment from Nck (Fig. S8) (Biological Magnetic Resonance Data Bank accession no. 26643). Analysis of the HN, N, CO, Cα, and Cβ chemical shifts using Talos+ (42) indicates secondary structure similar to the WASP GBD in complex with the autoinhibitory C helix (Fig. 5C and Fig. S9A) (10). Importantly, the Talos+ analysis predicts a helical conformation for the Nck hydrophobic motif (amino acids 66–75). Moreover, the difference between the secondary Cα and Cβ chemical shifts for residues within the hydrophobic motif is positive (Δδ[Cα − Cβ] > 0) (Fig. 5D and Fig. S9B), consistent with a helical conformation (43). Collectively, these data suggest that the structure of the GBD–Nck linker complex is similar to that of the GBD–VCA complex. A prediction of this model is that binding of the C helix and the Nck linker to the GBD are mutually exclusive. Consistent with competitive binding, addition of increasing concentrations of VCA abrogated binding of full-length Nck to the GBD (Fig. 5 E and F). Based on these results, we conclude that Nck binds N-WASP through not only its SH3 domains but also, its inter-SH3AB linker. These interactions may act in a cooperative manner to destabilize autoinhibitory contacts between the GBD and VCA regions, resulting in allosteric activation of N-WASP (Fig. 6).
Fig. S8.
Assignment of the N-WASP–GBD–Nck linker complex. [15N, 1H]-HSQC of the N-WASP Nck fusion protein including N-WASP GBD (amino acids 196–272) and the Nck linker (amino acids 62–83). Peaks corresponding to Nck residues are labeled in red.
Fig. S9.
Secondary structure prediction. (A) Output of Talos+ secondary structure prediction based on HN, N, Cα, Cβ, and CO shifts. (B) Difference of Cα and Cβ secondary chemical shifts.
Fig. 6.
Proposed mechanism of N-WASP activation by Nck. In the autoinhibited conformation, the N-WASP C helix binds intramolecularly to the GBD. Nck binds to tyrosine-phosphorylated receptors on the membrane and activates N-WASP by competitively displacing the C helix from the GBD with its inter-SH3AB linker. The released VCA segment is available to activate the Arp2/3 complex. Polyvalent interactions between the SH3 domains of Nck and the proline-rich region (PRR) of N-WASP can lead to higher-order oligomer formation, increasing the local density of activated N-WASP molecules.
Discussion
In response to membrane-localized signals, N-WASP binds and activates the Arp2/3 complex, promoting the assembly of branched actin networks and the physical movement and deformation of cellular membranes. In addition to the canonical activators, Cdc42 and PIP2, membrane-localized SH3 adapter proteins have been implicated in N-WASP activation. It has remained unclear how SH3 adapters disrupt the intramolecular interactions that maintain N-WASP in an autoinhibited state.
Using purified components, we reconstituted N-WASP–dependent actin assembly on a lipid bilayer supported on silica microspheres, allowing side by side comparison of multiple SH3 adapters. We find that the SH2/SH3 adapter Nck activates N-WASP more potently than all of the other SH3 adapters, including Grb2, Crk-II, cortactin, CIP4, and Tks4. Although previous studies have focused on SH3/polyproline interactions to explain N-WASP activation by Nck, we identify here an activation motif in the linker between the first two SH3 domains (amino acids 67–75). This motif, which had no assigned function before this work, binds directly to the N-WASP GBD and competes with the autoinhibitory C helix in the VCA region, leading to allosteric activation of N-WASP.
The inter-SH3AB activation motif is conserved among Nck orthologs, suggesting that the N-WASP GBD interaction and activation mechanism is also evolutionarily conserved. N-WASP activation by the inter-SH3 linker is reminiscent of the mechanism used by the bacterial virulence factors, EspFU and EspF, to nucleate actin assembly in host cells. The activation motif in EspFU and EspF has the same pattern of hydrophobic residues as the Nck motif (Fig. 4B) (Φ-Φ-x-x-Φ-x-x-x-Φ; Φ is any hydrophobic residue and x is any residue) (39–41). Structural studies by multidimensional NMR have shown that the EspFU peptide adopts a helical conformation when bound to the WASP GBD (40). Likewise, our analysis of secondary chemical shifts strongly suggests that the Nck hydrophobic motif also adopts a helical conformation with the side chains of Ile67, Val68, Leu71, and Leu75 lying on one face of the helix (Fig. S3).
Similar to linker segments in many other proteins (44), the Nck inter-SH3AB linker is multifunctional. In addition to activating N-WASP through GBD binding, the inter-SH3AB linker has been shown to bind intramolecularly to the SH3B domain (45) and mediate Nck homo-oligomerization and phase separation (46). The N-WASP C-helix motif is similarly multifunctional, binding in a mutually exclusive manner to G actin and the Arp2/3 complex (47) in addition to mediating autoinhibition by intramolecular GBD interactions.
Our results, combined with previous studies, suggest that Nck can activate N-WASP by two complementary mechanisms. First, the Nck inter-SH3 linker can bind directly to the N-WASP GBD, thereby releasing the critical VCA segment. We note that the affinity of this interaction (Kd = 33 ± 8 µM) is probably insufficient to activate autoinhibited N-WASP on its own; cooperative binding of the linker, along with one or more SH3 domains, is likely required to disrupt N-WASP’s autoinhibitory interactions. Second, Nck can promote N-WASP oligomerization. When N-WASP binds to a poly-SH3 adapter that is itself oligomerized by a third multivalent adapter, phase separation can occur, enforcing a high density of N-WASP molecules (38, 48).
On recruitment to the membrane by pY3-nephrin/Nck complexes, N-WASP molecules can presumably equilibrate between autoinhibited and activated states. At high local densities of pY3-nephrin and Nck, the density of activated N-WASP molecules bound to the Arp2/3 complex may be sufficient to nucleate actin assembly without GBD engagement by the Nck hydrophobic motif. This model may explain our observation that, at high NTAD densities, the Nck 4A mutant (with a defective hydrophobic motif) assembles actin comet tails similar to the WT (Fig. S4). By contrast, a functional hydrophobic motif is essential at low NTAD densities or when the SH3A domain is deleted (Fig. 4F and Figs. S4 and S5). In the cellular context, a similar dependency on the Nck hydrophobic motif may occur under conditions where the local density of Nck is below the critical concentration required for phase separation (for example, when the balance of kinase and phosphatase activity is shifted to produce low phosphotyrosine levels on membrane-associated receptors that recruit Nck).
Consistent with previous observations (48), Nck and N-WASP phase separate in our hands when mixed together at micromolar concentrations. However, under the conditions of our bead motility assays (100–250 nM Nck and 50–200 nM N-WASP), we do not observe phase separation in solution. We also have not detected macroscopic phase separation on the lipid-coated beads in contrast to a recent study, in which macroscopic, phase-separated domains containing pY3-nephrin, Nck, and N-WASP were observed on planar lipid bilayers (38). Nevertheless, it is likely that membrane-associated Nck and N-WASP can form higher-order oligomers in our system, especially at high densities of NTAD (and pY3-nephrin); such oligomeric assemblies may be important for promoting localized actin nucleation (Fig. 6).
The quantitative relationships between Nck/N-WASP signaling inputs on the one hand—tyrosine kinase/phosphatase activities, phosphotyrosine valency, and phase separation—and actin network assembly and force production on the other hand remain to be elucidated. We anticipate that reconstituted signaling and actin assembly systems of even higher complexity will be essential for obtaining a complete mechanistic description of emergent phenomena at the intersection of signaling and cytoskeletal dynamics.
Materials and Methods
Protein Expression and Purification.
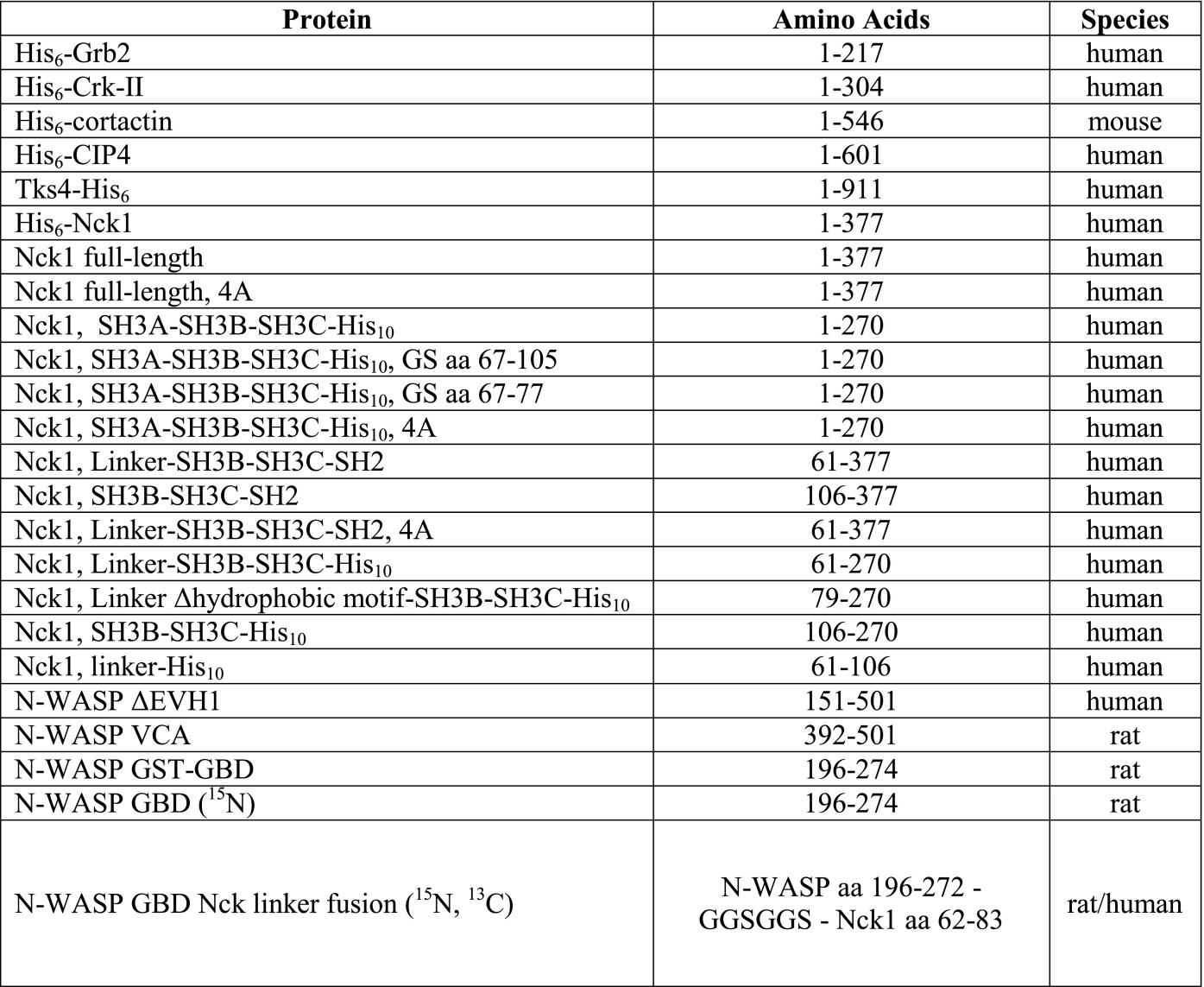
Expression plasmids, constructed from pET vectors, were transformed into E. coli BL21 (DE3) CodonPlus RIL (construct details are in Table S1). Cultures (2–4 L) were grown in LB at 18 °C overnight after induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside. For isotope labeling, M9 minimal medium was supplemented with 13C-glucose and/or 15NH4Cl, and bacteria were induced for 6 h at 30 °C.
Table S1.
Protein construct details
 |
Bacterial cell pellets were lysed using a microfluidizer (Microfluidics) in 20 mM Hepes, pH 7.5, 500 mM NaCl, 5–10% glycerol, and 4 mM β-mercapto-ethanol with protease inhibitor (4 mM Benzamidine or 1 mM di-isopropyl-fluorophosphate). For Nck and Grb2, 30 mM arginine was added to the lysis buffer. Lysates were cleared (24,000 × g for 40 min at 4 °C) and incubated in batch with Ni-nitriloacetic acid-agarose (Qiagen). The beads were washed with ATP buffer (lysis buffer supplemented with 10 mM MgCl2, 2 mM ATP, 30 mM KCl) and a high-salt buffer (lysis buffer with 1 M NaCl), and the protein was eluted with 400 mM imidazole.
Poly-His tags were cleaved from Nck, N-WASP (amino acids 151–501), cofilin, and profilin by incubation with tobacco etch virus (TEV) protease overnight. Tks4 and N-WASP were further purified with a heparin column (HiTrap; GE Healthcare), eluting with 50–1500 mM NaCl in storage buffer. N-WASP was stored in 20 mM Hepes, pH 7.5, 300 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). Tks4 was stored in 20 mM MES, pH 6.5, 250 mM NaCl, and 5% glycerol. All other proteins were further purified by size exclusion chromatography on S75 or S200 columns (GE Healthcare) equilibrated with storage buffer. The following storage buffers were used: Grb2 and Nck constructs (20 mM Hepes, pH 7.5, 500 mM NaCl, 0.5 mM TCEP, 10% glycerol), CIP4 (20 mM Hepes, pH 7.5, 150 mM NaCl), cortactin (50 mM Tris, pH 7.4, 100 mM NaCl, 10% glycerol), cofilin (20 mM Tris, pH 8, 50 mM NaCl, 1 mM DTT), and profilin (20 mM Tris, pH 8, 1 mM DTT, 1 mM EDTA).
GST-tagged N-WASP GBD and covalently linked GBD–Nck linker complex were purified on glutathione-Sepharose 4B (GE Healthcare). GST fusion protein was eluted with 30 mM glutathione (GSH) followed by dialysis in 20 mM Hepes, pH 7.5, 100 mM NaCl, and 1 mM DTT. For NMR experiments, the GST tag was cleaved by incubation with TEV protease followed by size exclusion chromatography.
To purify N-WASP VCA and Nck inter-SH3AB linker, cell pellets were lysed by incubation with 3 M guanidinium HCl overnight at room temperature (RT). The cleared lysate was applied to a Ni-chelating column (GE Healthcare) and washed with 1 M NaCl. Proteins were eluted with 0–400 mM imidazole followed by dialysis in storage buffer (Nck linker: 20 mM Hepes, pH 7.5, 200 mM NaCl; VCA: 20 mM Hepes, pH 7.5, 200 mM NaCl, 1 mM TCEP).
The following proteins were purified as previously described: actin (49), capping protein (50), and Arp2/3 complex (51), except that Arp2/3 complex was purified from frozen bovine thymus (Pel-Freez Biologicals).
Fluorescent Labeling.
Unless stated otherwise, proteins (5–50 μM) were labeled with 1 M eq Alexa568 maleimide (Invitrogen) on ice for 1 h. Crk-II was labeled overnight at 4 °C with 10 eq Alexa568 NHS (Invitrogen). Unreacted dye was removed by gel filtration. F-actin was labeled for 6 h on ice with 2 eq Alexa488-TFP (Invitrogen); unreacted dye was removed by pelleting the actin filaments followed by actin depolymerization. The extent of labeling (percentage of dye-labeled protein) was calculated by dividing the concentration of the protein-bound dye (determined by measuring the fluorescence emission intensity relative to a standard dilution series of the free dye) by the protein concentration. Protein concentrations were calculated from the absorbance at 280 nm after subtracting the dye contribution.
Preparation of Lipid-Coated Beads.
Chloroform solutions of 1,2-dioleoyl-sn-glycero-3-phosphocholine, NTAD (nickel salt; Avanti Polar Lipids), and Atto390-1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (Atto390-DOPE; ATTO-TEC) were mixed in molar ratios of 94:5:1, 96.5:2.5:1, and 98:1:1 of 1,2-dioleoyl-sn-glycero-3-phosphocholine:NTAD:Atto390-DOPE, evaporated in a stream of argon, and dried under high vacuum for at least 2 h. Lipids were resuspended in vesicle buffer (20 mM Hepes, pH 7.5, 100 mM NaCl, 330 mM sucrose) to a final concentration of 5 mM, sonicated for 20 s, subjected to five freeze–thaw cycles, and stored at −80 °C.
A suspension of glass beads (3.5 µL; 2.3-µm diameter; Total 2.8 × 107; Bangs Laboratories) was mixed with 5 µL lipid suspension and diluted to a final volume of 45 µL in vesicle buffer (6.2 × 108 beads mL−1). The suspension was sonicated (3 × 10 s) in a bath sonicator (Branson 2510) and incubated while rotating for 30 min at RT.
The beads were pelleted by pulse centrifugation, washed two times with 200 µL vesicle buffer, resuspended in 45 µL vesicle buffer, and used within 8 h.
Motility Assay with Different SH3 Adapters.
Lipid-coated beads (6.1 × 106 beads mL−1) containing 1%, 2.5%, and 5% NTAD were incubated with 0.25 µM His6-tagged SH3 adapter (20% Alexa568 labeled), 0.05 µM N-WASP, and motility components [9 µM actin (5% Alexa488 labeled), 0.075 µM Arp2/3, 0.05 µM capping protein, 2.6 µM profilin, 3.5 µM cofilin] in motility buffer (10 mM Hepes, pH 7.5, 2 mM MgCl2, 50 mM KCl, 50 mM NaCl, 1 mg/mL BSA, 2.5 mM ATP, 5 mM TCEP, 1.3 mM Dabco, 20 mM ascorbic acid, 0.2% methylcellulose). Samples were incubated for 15 min at RT while rotating. For quantitative image analysis, the reaction was diluted 1:1 in 3% glutaraldehyde (Ricca Chemical Company), spotted on a microscopy slide, covered with a coverslip, and imaged within 1 h.
Motility Reactions Using pY3-nephrin and Nck.
For each assay, lipid-coated beads (7.6 × 107 mL−1) were preincubated (15 min at RT) with 0.5 µM His8-pY3-nephrin (38) in buffer containing 10 mM Hepes, pH 7.5, 1 mg/mL BSA, and 50 mM KCl. Phosphonephrin-loaded beads were diluted eightfold in motility buffer containing 0.1 µM Nck (20% Alexa568 labeled), 0.2 µM N-WASP, and actin motility components as described above.
N-WASP and SH3 Adapter Density Measurements.
To compare the surface densities of SH3 adapters or N-WASP, we quantified the Alexa568 signal on the lipid-coated beads. Indicated amounts of Alexa568-labeled SH3 adapters or unlabeled SH3 adapter and Alexa568 N-WASP were incubated with lipid-coated beads in motility buffer for 15 min while rotating. The suspension (∼6 μL) was transferred to a flow chamber that had been passivated with –poly-(l-lysine)-PEG (3 mg/mL) (52) and imaged live as described below.
Microscopy and Image Analysis.
Epifluorescence and phase contrast images of fixed motility reactions were acquired with a 60× objective (N.A. 1.4) on an Olympus IX70 Inverted Microscope equipped with a Coolsnap HQ CCD Camera (Photometrics). Flat field-corrected images were analyzed using a CellProfiler Pipeline (53) to automatically locate beads, threshold images (Background Global Method), and determine the integrated fluorescence intensity of Alexa488 actin associated with each bead.
Live confocal images were acquired to quantify the density of N-WASP or SH3 adapters on lipid-coated beads. Images were obtained using a 100× objective (N.A. 1.49) on a Nikon ECLIPSE Ti Inverted Microscope equipped with a Yokoyama CSU-X1 Spinning Disk, a Hamamatsu EM CCD 9100–13 Camera, and 50-mW lasers [solid state, 405 nm; diode-pumped solid-state (DPSS), 561 nm]. For density measurements, flat field-corrected images were analyzed for bead fluorescence using a Mathematica script. The script identifies bead positions based on the Atto390-DOPE lipid fluorescence, quantifies bead fluorescence in the Alexa568 channel (SH3 adapter or N-WASP), and subtracts the median background value of a circular region outside of the bead. P values were calculated using an unpaired, two-tailed Student’s t test.
Pull-Down Assays.
GST-tagged N-WASP GBD (1.5 nmol; amino acids 196–274) or PAK1-GBD (1.5 nmol; amino acids 67–150; Cytoskeleton Inc.) was immobilized on GSH Sepharose 4B (20 µL per sample; GE Healthcare) for 15 min at 4 °C. The beads were washed two times with Nck storage buffer. Nck constructs (1.5 nmol total) were added to the beads and incubated for 2 h at a final concentration of 5 µM. The beads were washed two times, and the supernatant as well as the bead samples were subjected to SDS/PAGE. The Coomassie-stained gels were analyzed using a Licor Gel Imaging System. For competitive pull-down assays, N-WASP VCA (0–50 μM) was added to the binding reaction.
NMR Spectroscopy.
All NMR chemical shift assignment experiments were performed at 298 K on an Agilent DD2 600-MHz spectrometer equipped with a cold probe using NMRPipe for data processing (54) and NMRView for analysis (55). Backbone chemical shift assignments were obtained using standard 3D 15N-edited triple-resonance experiments including HNCACB, CBCA(CO)NH, and HNCO spectra (56). 13C-,15N-labeled N-WASP Nck fusion protein in 20 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.01% NaN3, and 5% D2O was used at a concentration of 550 µM. NMR titration data were acquired at 300 K on a Bruker DRX500 spectrometer (Topspin, version 1.3) equipped with a cryogenic probe. 15N-labeled N-WASP GBD (65 µM; in 20 mM Hepes, pH 7.5, 200 mM NaCl, 1 mM DTT, 0.01% NaN3, 5% D2O) was titrated with the Nck linker peptide (0–520 µM). Titration curves from 17 peaks were fit using CCPN Analysis (57). Weighted chemical shift differences were calculated using the following equation: Δδ = ((ΔδN × 0.2)2 + ΔδH2)1/2.
Supplementary Material
Acknowledgments
We thank Justin Farlow for help with image analysis, Sudeep Banjade and Michael Rosen for providing triphosphorylated nephrin peptide, and Sebastian Rumpf for critical reading of the manuscript. This research was supported by funds from Tobacco-Related Disease Research Program of the University of California Grant 19FT-0090 (to J.O.) and the Howard Hughes Medical Institute. NMR spectroscopy at University of Texas Southwestern Medical Center is supported by NIH instrumentation Grant 1S10OD018027-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (accession no. 26643).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510876112/-/DCSupplemental.
References
- 1.Le Clainche C, Carlier M-F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88(2):489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463(7280):485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290(5492):801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- 5.Papayannopoulos V, et al. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17(2):181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 6.Torres E, Rosen MK. Protein-tyrosine kinase and GTPase signals cooperate to phosphorylate and activate Wiskott-Aldrich syndrome protein (WASP)/neuronal WASP. J Biol Chem. 2006;281(6):3513–3520. doi: 10.1074/jbc.M509416200. [DOI] [PubMed] [Google Scholar]
- 7.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: Barbed end capture by N-WASP WH2 domains. Cell. 2007;128(5):901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391(6662):93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 10.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404(6774):151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 11.Abdul-Manan N, et al. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399(6734):379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 12.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108(33):E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ti S-C, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108(33):E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li SS-C. Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem J. 2005;390(Pt 3):641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frischknecht F, et al. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401(6756):926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 16.Gruenheid S, et al. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat Cell Biol. 2001;3(9):856–859. doi: 10.1038/ncb0901-856. [DOI] [PubMed] [Google Scholar]
- 17.Rohatgi R, Nollau P, Ho HY, Kirschner MW, Mayer BJ. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J Biol Chem. 2001;276(28):26448–26452. doi: 10.1074/jbc.M103856200. [DOI] [PubMed] [Google Scholar]
- 18.Campellone KG, et al. Clustering of Nck by a 12-residue Tir phosphopeptide is sufficient to trigger localized actin assembly. J Cell Biol. 2004;164(3):407–416. doi: 10.1083/jcb.200306032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera GM, Briceño CA, Takeshima F, Snapper SB, Mayer BJ. Inducible clustering of membrane-targeted SH3 domains of the adaptor protein Nck triggers localized actin polymerization. Curr Biol. 2004;14(1):11–22. doi: 10.1016/j.cub.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 20.Jones N, et al. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature. 2006;440(7085):818–823. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- 21.Verma R, et al. Nephrin ectodomain engagement results in Src kinase activation, nephrin phosphorylation, Nck recruitment, and actin polymerization. J Clin Invest. 2006;116(5):1346–1359. doi: 10.1172/JCI27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohamed AM, Boudreau JR, Yu FPS, Liu J, Chin-Sang ID. The Caenorhabditis elegans Eph receptor activates NCK and N-WASP, and inhibits Ena/VASP to regulate growth cone dynamics during axon guidance. PLoS Genet. 2012;8(2):e1002513. doi: 10.1371/journal.pgen.1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnelly SK, Weisswange I, Zettl M, Way M. WIP provides an essential link between Nck and N-WASP during Arp2/3-dependent actin polymerization. Curr Biol. 2013;23(11):999–1006. doi: 10.1016/j.cub.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.She HY, et al. Wiskott-Aldrich syndrome protein is associated with the adapter protein Grb2 and the epidermal growth factor receptor in living cells. Mol Biol Cell. 1997;8(9):1709–1721. doi: 10.1091/mbc.8.9.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlier MF, et al. GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex. J Biol Chem. 2000;275(29):21946–21952. doi: 10.1074/jbc.M000687200. [DOI] [PubMed] [Google Scholar]
- 26.Scaplehorn N, et al. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr Biol. 2002;12(9):740–745. doi: 10.1016/s0960-9822(02)00812-6. [DOI] [PubMed] [Google Scholar]
- 27.Tang DD, Zhang W, Gunst SJ. The adapter protein CrkII regulates neuronal Wiskott-Aldrich syndrome protein, actin polymerization, and tension development during contractile stimulation of smooth muscle. J Biol Chem. 2005;280(24):23380–23389. doi: 10.1074/jbc.M413390200. [DOI] [PubMed] [Google Scholar]
- 28.Kowalski JR, et al. Cortactin regulates cell migration through activation of N-WASP. J Cell Sci. 2005;118(Pt 1):79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- 29.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27(21):2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichot CS, et al. Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res. 2010;70(21):8347–8356. doi: 10.1158/0008-5472.CAN-09-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oikawa T, Itoh T, Takenawa T. Sequential signals toward podosome formation in NIH-src cells. J Cell Biol. 2008;182(1):157–169. doi: 10.1083/jcb.200801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padrick SB, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32(3):426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401(6753):613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 34.Mullins RD, Hansen SD. In vitro studies of actin filament and network dynamics. Curr Opin Cell Biol. 2013;25(1):6–13. doi: 10.1016/j.ceb.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacayo CI, et al. Choosing orientation: Influence of cargo geometry and ActA polarization on actin comet tails. Mol Biol Cell. 2012;23(4):614–629. doi: 10.1091/mbc.E11-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koronakis V, et al. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc Natl Acad Sci USA. 2011;108(35):14449–14454. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uruno T, et al. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3(3):259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- 38.Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife. 2014;3:e04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alto NM, et al. The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J Cell Biol. 2007;178(7):1265–1278. doi: 10.1083/jcb.200705021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng H-C, Skehan BM, Campellone KG, Leong JM, Rosen MK. Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U) Nature. 2008;454(7207):1009–1013. doi: 10.1038/nature07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sallee NA, et al. The pathogen protein EspF(U) hijacks actin polymerization using mimicry and multivalency. Nature. 2008;454(7207):1005–1008. doi: 10.1038/nature07170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spera S, Bax A. Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113(14):5490–5492. [Google Scholar]
- 44.Hsu WL, et al. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22(3):258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi K, Sun Z-YJ, Park S, Wagner G. Autoinhibitory interaction in the multidomain adaptor protein Nck: Possible roles in improving specificity and functional diversity. Biochemistry. 2010;49(27):5634–5641. doi: 10.1021/bi100322m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banjade S, et al. Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc Natl Acad Sci USA. 2015;112:E6426–E6435. doi: 10.1073/pnas.1508778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly AE, Kranitz H, Dötsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281(15):10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 50.Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155(2):251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egile C, et al. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146(6):1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bieling P, Telley IA, Hentrich C, Piehler J, Surrey T. Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 2010;95(2010):555–580. doi: 10.1016/S0091-679X(10)95028-0. [DOI] [PubMed] [Google Scholar]
- 53.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques. 2007;42(1):71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 54.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 55.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 56.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J Magn Reson B. 1994;103(3):203–216. [Google Scholar]
- 57.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.