Significance
A proportion of the variation in HIV-1 viral load in the infected population is influenced by host genetics. Using a large sample of infected individuals (n = 6,315) with genome-wide genotype data, we sought to map genomic regions that influence HIV viral load and quantify their impact. We identified amino acid positions located in the binding groove of class I HLA proteins (HLA-A and -B) and SNPs in the chemokine (C-C motif) receptor 5 gene region that together explain 14.5% of the observed variation in HIV viral load. Controlling for these signals, we estimated that an additional 5.5% can be explained by common, additive genetic variation. Thus, we demonstrate that common variants of large effect explain the majority of the host genetic component of HIV viral load.
Keywords: HIV-1 control, GWAS, heritability, infectious disease, genomics
Abstract
Previous genome-wide association studies (GWAS) of HIV-1–infected populations have been underpowered to detect common variants with moderate impact on disease outcome and have not assessed the phenotypic variance explained by genome-wide additive effects. By combining the majority of available genome-wide genotyping data in HIV-infected populations, we tested for association between ∼8 million variants and viral load (HIV RNA copies per milliliter of plasma) in 6,315 individuals of European ancestry. The strongest signal of association was observed in the HLA class I region that was fully explained by independent effects mapping to five variable amino acid positions in the peptide binding grooves of the HLA-B and HLA-A proteins. We observed a second genome-wide significant association signal in the chemokine (C-C motif) receptor (CCR) gene cluster on chromosome 3. Conditional analysis showed that this signal could not be fully attributed to the known protective CCR5Δ32 allele and the risk P1 haplotype, suggesting further causal variants in this region. Heritability analysis demonstrated that common human genetic variation—mostly in the HLA and CCR5 regions—explains 25% of the variability in viral load. This study suggests that analyses in non-European populations and of variant classes not assessed by GWAS should be priorities for the field going forward.
Upon infection with human immunodeficiency virus type 1 (HIV-1), there is substantial variability in viral control and rate of disease progression. After primary infection, characterized by high levels of viremia (HIV-1 RNA copies per milliliter of plasma) and transient loss of CD4+ T cells, most patients enter an asymptomatic period and maintain a relatively stable viral load off therapy. It has been well-established that this set point viral load (spVL) varies in the infected population and positively correlates with rate of disease progression (1). Thus, spVL is an easily measured and informative marker of clinical outcome.
Variability in spVL is influenced by host, viral, and environmental factors, including human genetic variation. Genome-wide association studies (GWAS) have consistently identified variation in the major histocompatibility complex (MHC) region on chromosome 6 as the major host determinant of HIV-1 viral load and disease progression (usually rate of CD4+ T-cell decline) (2–6). Similarly, studies of extreme phenotypes of HIV-1 progression [i.e., elite controllers (7, 8), long-term nonprogressors (9), and rapid progressors (10)] have underscored the primary role of the MHC in determining HIV-1 outcome. However, the GWAS of HIV-1—related phenotypes performed to date have been underpowered to identify the types of variants with modest effect sizes that have been observed to influence other complex human traits. To what extent additional host genetic factors contribute to HIV-1 control and the total variability in spVL explained by host genetics remain open questions.
Here, we report the results from the second phase of the International Collaboration for the Genomics of HIV-1 (for a complete list of contributors see SI Appendix, Note S1) (11), which has collected the majority of available genome-wide genotype data from HIV-1–infected patients with clinical follow-up. We tested ∼8 million variants, including single nucleotide polymorphisms (SNPs), short insertions and deletions (indels), classical human leukocyte antigen (HLA) alleles, and variable amino acids in HLA proteins for association with spVL in 6,315 HIV-1–infected individuals of European ancestry. We demonstrate that multiple independent signals exist at two genomic loci and implicate novel, potentially causal variants within these regions. Through heritability analysis, we estimate that the additive genetic contribution to spVL measurable through GWAS is 24.6%, the majority of which maps to variants in these two associated regions.
Results
Genome-Wide Association Analysis.
High-quality genotype data were obtained for 7,468 individuals of European ancestry from eight independent GWAS forming 10 genotype groups (SI Appendix, Table S1). The phenotypic endpoint most commonly shared between contributing centers was spVL, available for 6,315 individuals. After genome-wide genotype imputation, we tested ∼8 million common variants for association with spVL per group by linear regression and combined results using inverse-variance weighted metaanalysis. We observed significant associations on chromosomes 6 and 3, with several SNPs passing the threshold of genome-wide significance (P < 5 × 10−8) (Fig. 1). The strongest associated SNP on chromosome 6, rs59440261 (P = 2.0 × 10−83), lies in the MHC regions and is in strong linkage disequilibrium (LD) with the previously reported SNP rs2395029 (3) [r2 = 0.78, D′ = 1, minor allele frequency (MAF) rs59440261 = 0.06, MAF rs2395029 = 0.05]. The top chromosome 3 SNP, rs1015164 (P = 1.5 × 10−19), lies downstream of CCRL2, near an antisense transcribed sequence that overlaps chemokine (C-C motif) receptor 5 (CCR5) and is only weakly correlated to the CCR5Δ32 polymorphism known to impact HIV-1 disease progression (r2 = 0.03, D′ = 0.89, MAF rs1015164 = 0.30, MAF CCR5Δ32 = 0.10). Per-group analyses using the primary phenotypic endpoint (i.e., not necessarily spVL) (SI Appendix, Table S1) did not reveal any additional associated regions, and metaanalysis of these results was consistent with analysis of spVL (SI Appendix, Fig. S1).
Fig. 1.
Manhattan plot of genome-wide association results. After genotype imputation, ∼8 million common variants were tested for association with spVL in 6,315 individuals of European ancestry using linear regression. Per SNP -log10(P value) (y axis) are plotted by physical position (x axis). Genome-wide signals of association (P < 5 × 10−8, dotted line) were observed on chromosomes 6 and 3. The strongest associated SNPs per region were rs59440261 on chromosome 6 (P = 2.0 × 10−83) and rs1015164 on chromosome 3 (P = 1.5 × 10−19).
Additionally, we performed association analyses restricting the sample to extreme phenotypes of elite control (n = 887 HIV-1 controllers; n = 2,745 noncontrollers) or disease progression (n = 517 rapid progressors; n = 467 long-term nonprogressors). Association results were comparable with those obtained in the spVL analysis, with regions on chromosomes 6 and 3 being strongly associated (SI Appendix, Fig. S2). Thus, all further analyses were performed using the spVL phenotype.
Effect of Classical HLA Alleles on spVL.
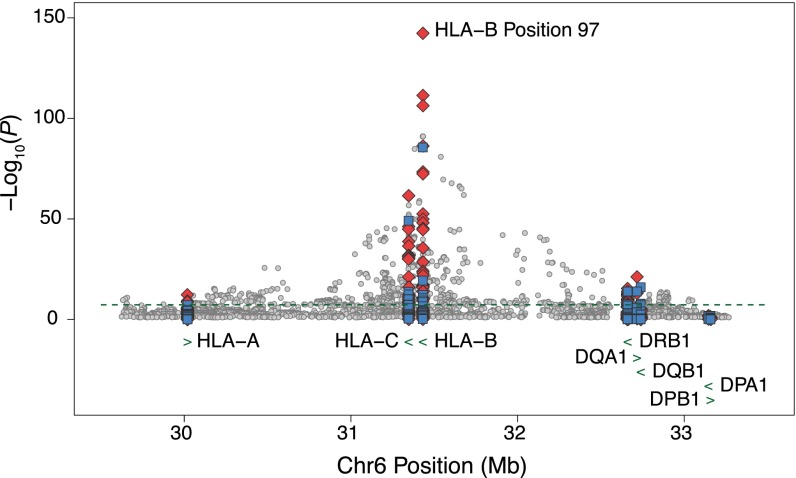
The SNP association signal on chromosome 6 centers on the class I HLA gene HLA-B, which is known to impact spVL (2, 8, 12). To gain a better understanding of functional variants in this region, we imputed classical class I and II HLA alleles, variable amino acid positions in HLA proteins, and additional single nucleotide variants. Association testing at these variants showed an increase in signal, with several variants having lower P values than those observed after genome-wide SNP imputation (Fig. 2).
Fig. 2.
Regional association plot of the chromosome 6 association peak. Association results, −log10(P value), for SNPs (gray circles), classical HLA alleles (blue boxes), and amino acids within HLA proteins (red diamonds). For biallelic markers, results were calculated by linear regression, including covariates. Association at amino acid positions with more than two alleles was calculated using a multi–degree-of-freedom omnibus test. The dashed line indicates genome-wide significance (P = 5 × 10−8). Amino acid position 97 (P = 4.6 × 10−143) in HLA-B showed the strongest association signal of any variant tested genome-wide.
Several classical HLA-A, HLA-B, and HLA-C alleles were associated with spVL, ranging in effect from strongly decreasing (notably HLA-B*57:01, effect size = −0.84) to strongly increasing (notably HLA-B*35:02, effect size = 0.36) (SI Appendix, Table S2). Given the presumed benefit of recognizing multiple viral epitopes through increased diversity at HLA alleles, we next tested for evidence of nonadditive effects at the HLA-B locus. Controlling for the additive effect at each allele, we observed evidence for a general heterozygote advantage across all HLA-B alleles that decreased spVL (P = 0.016, df = 1, effect size = −0.14) (SI Appendix, Fig. S3). Modeling per-allele nonadditive effects did not improve the fit over the general heterozygosity effect (P = 0.14, df = 13), and no single allele showed significant departure from additivity after accounting for multiple comparisons (SI Appendix, Table S3). Additionally, testing for a multiplicative effect between all pairs of HLA-B alleles did not uncover any significant interactions. These data confirm a protective role for general heterozygosity at HLA-B beyond the individual allelic additive effects.
Fine Mapping of MHC Association Signals.
Variable amino acid positions within the HLA class I proteins showed the strongest signal for association (Fig. 2). Notably, HLA-B position 97 (P = 4.6 × 10−143) was the strongest observed association study-wide, consistent with previous reports (7, 8). To determine which amino acid positions associated independently with spVL, we performed a forward conditional regression analysis. We identified (in order) positions 97, 67, and 45 in HLA-B and positions 77 and 95 in HLA-A as independently associated with spVL (Table 1). These positions fall within the peptide-binding groove of the respective protein (Fig. 3 A and B), and alleles at these positions had varying impact on spVL, ranging in effect from strongly decreasing to strongly increasing (Fig. 3C and SI Appendix, Table S4). Combining all alleles at these five positions explained 12.3% of the variance in spVL and accounted for the majority of the association signal at this locus (SI Appendix, Fig. S4). The relationship between these amino acid positions and classical HLA alleles is listed in SI Appendix, Table S5.
Table 1.
Independently associated amino acid positions in HLA proteins identified by stepwise forward conditional analysis
| Step | Position | Alleles* | Position P† | Model P‡ | Cumulative variance explained§ |
| 1 | HLA-B 97 | V/N/W/T/R/S | 4.6 x 10−143 | na | 0.102 |
| 2 | HLA-B 67 | Y/F/S/C/M | 3.7 x 10−112 | 3.2 x 10−15 | 0.112 |
| 3 | HLA-B 45 | E/T/K/M | 8.2 x 10−49 | 1.8 x 10−4 | 0.114 |
| 4 | HLA-A 77 | N/S/D | 1.8 x 10−12 | 9.4 x 10−12 | 0.122 |
| 5 | HLA-A 95 | L/I/V | 3.6 x 10−5 | 3.2 x 10−7 | 0.123 |
na, not applicable.
Per allele association statistics and frequencies are listed in SI Appendix, Table S4.
Position P values were calculated by a multi–degree-of-freedom omnibus test, including covariates and all alleles at that position.
Model P values were calculated by the likelihood ratio test comparing the model from the previous step to a model including the next position.
Cumulative variance explained was calculated by linear regression and represents the variance explained by including the positions identified at each step to the model from the previous step.
Fig. 3.
Location and effect of independently associated amino acids. Three-dimensional structures of (A) HLA-B (PDB ID code 2bvp) and (B) HLA-A (PDB ID code 4hwz) proteins. Conditional analysis identified five independent amino acid positions [positions 97, 67, and 45 in HLA-B and positions 77 and 95 in HLA-A (orange residues)] that line the peptide-binding groove and explain the majority of the association signal in the MHC. (C) Effect on spVL (i.e., change in log10 HIV-1 spVL per allele copy) of individual amino acid residues at each position. Results were calculated per allele using linear regression models, including allele dosage and principal components. Gray bars indicate the estimated change in spVL per amino acid allele at each position with standard error (whiskers). All identified positions accommodate >2 amino acid alleles, with allelic effects ranging from strongly protective (i.e., viral load decreasing) to deleterious (viral load increasing). Full association statistics and amino acid allele frequencies are listed in SI Appendix, Table S4.
Fine Mapping of CCR5 Region Association Signals.
The second highly significant signal of association centered over the CCR gene cluster on chromosome 3. Variation in the CCR5 gene is known to impact HIV-1 pathogenesis (13–16). The strongest known causal variant in this region is CCR5Δ32, which is known to reduce HIV-1 susceptibility and slow disease progression (13). Additionally, the CCR5 promoter haplotype P1 (Hap-P1) has been shown to associate with AIDS progression (15, 17). To account for these effects, we restricted the conditional analysis to 5,559 individuals for whom the CCR5Δ32 genotype was available and Hap-P1 carriage could be determined (Fig. 4). The top SNP association in this subset, rs4317138 (P = 7.7 × 10−22) (Fig. 4), is highly correlated to the top SNP identified in the analysis of the full sample (rs1015164, r2 = 0.97, D′ = 1, MAF rs4317138 = 0.31). Consistent with expectation, we observed a strong association between CCR5Δ32 and reduced spVL (P = 1.6 × 10−16, effect size = −0.28) and between CCR5 Hap-P1 haplotype and increased spVL (P = 1.8 × 10−19, effect size = 0.18).
Fig. 4.
Regional association plot of the chromosome 3 association peak. Association results for Mb 45.5–47 (Hg19) of chromosome 3 in a subset of individuals genotyped for CCR5Δ32 (n = 5,559). P values were calculated by linear regression, including covariates. The blue diamond, red square, and red diamond indicate the association strength of the top SNP (rs4317138, P = 7.7 × 10−22), Hap-P1 (P = 1.8 × 10−19), and CCR5Δ32 (P = 1.6 × 10−16), respectively. The dashed line indicates genome-wide significance (P = 5 × 10−8).
Conditioning on CCR5Δ32, 122 SNPs remained genome-wide significant (SI Appendix, Fig. S5). The top seven SNPs are in strong LD and fall within/near an antisense transcribed sequence RP11-24F11.2 (LOC102724297) that overlaps CCR5 (SI Appendix, Fig. S6). Conditioning on Hap-P1, these SNPs remained associated, with the strongest signal being rs1015164 (conditional P = 1.6 × 10−4). This SNP remained associated when conditioning on both Hap-P1 and CCR5Δ32 (P = 5.2 × 10−4) (Table 2). Interestingly, conditioning on rs1015164 explained the observed effect of Hap-P1 (conditional P = 0.09) but not CCR5Δ32 (P = 1.4 × 10−10), suggesting that this SNP tags additional, undescribed causal variants in this region. Taken together, these three variants explained 2.2% of the variance in spVL.
Table 2.
Conditional association results for variants in the CCR5 region
| Variant | Condition | |||||||||
| None | CCR5Δ32 | Hap-P1 | rs1015164 | CCR5Δ32 and Hap-P1 | ||||||
| Effect size | P value | Effect size | P value | Effect size | P value | Effect size | P value | Effect size | P value | |
| CCR5Δ32 | −0.28 | 1.6 x 10−16 | na | na | −0.22 | 1.4 x 10−10 | −0.22 | 1.4 x 10−10 | na | na |
| Hap-P1 | 0.18 | 1.8 x 10−19 | 0.15 | 1.4 x 10−13 | na | na | 0.06 | 0.09 | na | na |
| rs1015164(A) | 0.23 | 1.5 x 10−21 | 0.20 | 1.2 x 10−15 | 0.17 | 1.6 x 10−4 | na | na | 0.15 | 5.2 x 10−4 |
Effect size and P values were calculated using linear regression, including covariates to adjust for population structure and, where applicable, the variant/haplotype dosage (condition).
Assessing Narrow-Sense Heritability of HIV-1 spVL.
Combining the effects of the independently associated common variants in the HLA and CCR5 region explained 14.5% of the variability in spVL. We used genome-wide complex trait analysis (GCTA) (18) to address the extent to which additional, additive genetic factors may influence spVL and observed that genome-wide variation explains 24.6% [standard deviation (SD) = 3%] of the narrow-sense heritability (i.e., additive effects). We assessed the sensitivity of this estimate to potential overfitting by verifying that a randomly permuted phenotype vector (30 permutations) showed zero heritability. This genome-wide estimate decreased to 5.5% (SD = 3%) after controlling for the effects in the MHC/CCR5 regions. A series of analyses where we randomly selected two-thirds of all available samples supported this estimate (median 5% heritability, 6.9% interquartile range). Additionally, a complementary analysis using a polygenic score test demonstrated a similar lack of contribution from variants outside of the MHC and CCR5 regions (SI Appendix, Fig. S7). These results suggest that the identified common variants of large effect explain the majority of the host genetic component of HIV-1 spVL.
Discussion
Previous GWAS of HIV-1 control and disease progression lacked power to detect variants with modest effect sizes. By combining available genome-wide genotypes and clinical data from 6,315 HIV-1–infected individuals, we sought to get a more complete picture of the impact of common human genetic variation on HIV-1 disease across a range of effect sizes.
The MHC region demonstrated the strongest signal of association with spVL, with multiple, independent common variants of large effect mapping to this region. The long-range LD structure and high gene density (including many immunologically relevant genes) of the MHC make it impossible to definitively assign causality to any particular variant through purely statistical methods. However, the abundance of functional evidence and the centrality of the association signal in this study point to the class I HLA genes and, in particular, to HLA-B as being causal. Here, we observed strong associations between spVL and multiple alleles at HLA-A, -B, and -C over a broad range of effect sizes. Consistent with previous results (19), we observed evidence for a heterozygote advantage at the HLA-B locus. The comparatively weak statistical strength we report here may be due to methodological differences because (i) we control for additive effects at each allele and (ii) the larger sample size allows the consideration of an increased number of homozygous genotypes, reducing bias due to the low frequency (and thus increased proportion of heterozygosity) of strongly protective alleles. Thus, our results may more accurately reflect the true heterozygous effect.
By testing variant amino acid positions in classical HLA proteins, we confirmed the strong associations at positions 97 and 67 in HLA-B and observed additional signals at position 45 in HLA-B and positions 77 and 95 in HLA-A. The location of these amino acids in the peptide-binding groove of the respective proteins supports the hypothesis that the presentation of specific viral epitopes, directly dependent on the shape of the HLA peptide binding groove, is critical in determining the efficiency of the cytotoxic T-cell response. In addition to peptide presentation, HLA-C expression levels (20) and variation in non-HLA genes in the MHC region (21) have been proposed as impacting HIV-1 control. Detailed functional analyses of these effects will be required to fully understand the extent of the influence of MHC variation on the natural history of HIV-1 disease.
Although the impact of CCR5Δ32 on HIV-1 acquisition and disease progression has been well-described, this association has not been previously identified through GWAS. This lack of detection is likely due to the relatively limited LD between common SNPs and the CCR5Δ32 allele. Indeed, the top SNP identified on chromosome 3 in the full sample, rs1015164, is only weakly correlated to CCR5Δ32 (r2 = 0.03). Conditional analysis showed that several SNPs in this region were independently associated after controlling for the known effects of CCR5Δ32 and Hap-P1. These SNPs are located within/near an antisense transcribed sequence that overlaps CCR5 and thus may play a role in regulating its expression. Demonstration of causality of these variants and/or a silencing effect of the antisense transcribed sequence will require functional studies.
Measurable narrow-sense heritability attributable to non–genome-wide significant loci has been demonstrated for multiple complex traits (22, 23). Using genome-wide variants, we estimated that additive host genetic effects explain approximately one-quarter of the variance in HIV-1 spVL. However, after controlling for the genome-wide significant signals, the remainder of the genome explained only ∼5%. This limited residual heritability underscores the primary role of common variants of large effects in the MHC and CCR5 in HIV-1 control. Interestingly, analyses aimed at estimating the viral genetic component of heritability have been generally higher, ∼30–50%, than our estimated host component (24). However, it is difficult to disentangle these two values because host genetic variation, in particular the class I HLA region, exerts substantial pressure on the viral genetic sequence (25). Indeed, if the influence of host and viral genetics highly overlaps, up to an additional 70% of variability in spVL may remain unaccounted for. In addition to known nongenetic factors that impact spVL, such as age and sex, host genetic factors not measured by this study design (e.g., somatic recombination of T- and B-cell receptors, copy number variation, and rare variation) may also explain a substantial proportion of the variation. Comprehensive, joint analysis of the host and viral genetic components of spVL variation in large samples will also be of great interest due to the high sensitivity of HIV to reflect variation in the host environment.
For single variant analysis, this study had ∼80% power to detect common variants (at 10% frequency) that explain >0.5% of the variability in spVL. This level of sensitivity suggests that previous candidate gene studies that have claimed associations with spVL (of variants with relatively large effect size) are unlikely to be valid, given their lack of replication in the present study. This observation is consistent with previous GWAS that have directly examined (and failed to replicate) a number of these associations (2, 8).
The results presented herein combine the majority of genetic data available on untreated HIV-1–infected individuals of European ancestry. Because a substantial increase in sample size is unrealistic, because of current antiretroviral treatment guidelines (26), additional GWAS in this population are unlikely to provide further insight into the genetic architecture of HIV-1 control. Thus, studies in non-European populations, which heretofore have been underrepresented in GWAS, as well as investigations of other classes of genetic variation and genome-wide nonadditive and/or epistatic effects, should now be clear priorities in the field.
Methods
Ethics Statement.
All participants were HIV-1–infected adults, and written informed consent for genetic testing was obtained from all individuals as part of the original study in which they were enrolled (SI Appendix, Note S1). Ethical approval was obtained from institutional review boards for each of the respective contributing centers.
Samples and Contributing Centers.
DNA samples obtained from 21 individual cohorts or centers were genotyped as part of eight independent GWAS using various genotyping platforms (2, 5, 6, 8–10, 27, 28) and combined as part of the International Collaboration for the Genomics of HIV (SI Appendix, Table S1 and Note S1). All individuals were infected with HIV-1 and had phenotypic data relevant to viral control or disease progression. Primary phenotypes included spVL, long-term nonprogression, and elite control of HIV-1 viremia. The phenotype most commonly available was spVL (n = 6,315), which was used for the primary analysis. Additional analyses were performed on extreme phenotypes (elite control, long-term nonprogression, and rapid progression) and are presented in SI Appendix.
Genotype Quality Control and Imputation.
All quality control steps were performed per study using PLINK version 1.07 (29). Genotype data were combined based on geographic origin of the samples and/or genotyping platform, resulting in 10 genotype groups (SI Appendix, Table S1). Ancestry was inferred by principal components analysis using EIGENSTRAT (30), taking the HapMap 3 (31) sample as a reference. Only samples clustering with the HapMap Europeans were included. Study participants were excluded based on the following criteria: identity-by-descent of >0.125 (one individual per pair was removed), missingness of >2%, and inbreeding coefficients of <−0.1 or >0.1. SNPs were removed based on missingness of >5%, MAF of <1%, or Hardy–Weinberg equilibrium of P < 1 × 10−7.
Per group, genotypes for additional polymorphisms not directly assessed by the original genotyping platform were inferred using haplotype information (i.e., imputation of missing genotypes) (32) from the 1,000 Genomes Project Phase 1 v3 reference panel. Genotypes were prephased with mach v1 (33) and imputed using minimac (34). An additional imputation protocol using shapeit v2 (35, 36) and impute2 (37) was also implemented with highly concordant results. Imputed SNPs having a reported r2 score of <0.3 and minor allele frequency of <0.5% were excluded from downstream analysis.
Association Testing and Metaanalysis.
Single marker association tests were performed per genotype group regressing spVL on variant dosage using linear regression including principal components (PCs) to correct for population structure (30). In all cases, inclusion of PCs was sufficient to control for genomic inflation (lambda of ∼1) (SI Appendix, Table S1). Results were combined across genotype groups using inverse-variance weighted metaanalysis (38). In some cases, the primary endpoint for the original study was a binary trait (SI Appendix, Table S1). For these cohorts, we also tested the binary phenotype for association using logistic regression, including covariates as above and metaanalyzed across binary and quantitative endpoints using z-scores weighted by the group sample size. Power for detection of single variants was estimated using the genetic power calculator for quantitative traits (39).
Imputation and Association Testing in the MHC Region.
Classical HLA alleles, variant amino acids within HLA proteins, and additional SNPs in the MHC were imputed using the SNP2HLA pipeline, with a reference panel consisting of 5,225 individuals of European ancestry from the Type 1 Diabetes Genetics Consortium (40). Classical alleles and binary amino acid positions were individually tested for association using linear regression corrected for PCs and study-specific effects. Association was tested at multiallelic amino acid positions (i.e., three or more possible states) using a multi–degree-of-freedom omnibus test including covariates as above.
Testing for Nonadditive Effects of HLA-B Alleles.
Evidence of nonadditive effects at the HLA-B locus was assessed in a subset of individuals (n = 3,882) that carried two common alleles (minimum of five homozygous observations, n = 14 alleles). We first compared a model that included covariates (PCs and genotype group) and additive effects for each classical allele to a model that additionally included a heterozygosity effect; this approach is equivalent to having a general dominance term across all alleles. We similarly assessed the nonadditive effect of each allele individually. To estimate effect sizes of homozygote and heterozygote genotypes on spVL, we constructed additive models after excluding all homozygous individuals (for heterozygous effects) or excluding all heterozygous individuals (for homozygous effects). Interactions between specific alleles were assessed using models that contained additive terms for each allele and interaction terms between each pair of alleles.
Fine Mapping of Associated Regions.
To identify independent variants in associated regions, we used step-wise forward conditional testing, including covariates as above. In the MHC, due to the presence of multiallelic variants (i.e., >2 states), we used the likelihood ratio test (LRT). A position was considered independently associated if its addition to the model improved the fit after correcting for the total number of amino acids considered (LRT of P > 2 × 10−4). In the CCR5 region, conditional analysis was restricted to a subset of 5,559 participants genotyped for CCR5Δ32 and for whom the CCR5 Hap-P1 haplotype could be inferred (15, 17, 41). Variance explained by independently associated variants was calculated by comparing the adjusted r2 values from linear regression models, including covariates alone to one containing covariates and the selected variants.
Assessment of Narrow-Sense Heritability of spVL.
Heritability analysis was conducted with the GCTA software package (18) using common variants (MAF of >1%), which were accurately imputed in at least 99% of samples, pruned based on LD (r2 < 0.1). To avoid deflation of the total heritability estimate, the independently associated variants from the conditional analysis were also included. To reduce bias due to nonnormally distributed spVL measurements, cohorts enriched for HIV-1 controllers were removed. To empirically assess the error of the estimated variance component, we performed the analyses on 30 bootstrap replicates, by resampling the included individuals with replacement. To check for potential overfitting, we performed heritability analyses on 30 random assignments of the phenotypes to the genotypes. We assessed the effect of sample size by repeating the analysis over a grid of different sample sizes.
Supplementary Material
Acknowledgments
We thank Stuart Z. Shapiro (Program Officer, Division of AIDS, National Institute of Allergy and Infectious Diseases) and Stacy Carrington-Lawrence (Chair of Etiology and Pathogenesis, NIH Office of AIDS Research) for continued support. A portion of the computations were performed at the Vital-IT (www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514867112/-/DCSupplemental.
References
- 1.Mellors JW, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122(8):573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 2.Fellay J, et al. NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5(12):e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317(5840):944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelak K, et al. Infectious Disease Clinical Research Program HIV Working Group National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology (CHAVI) Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010;201(8):1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalmasso C, et al. ANRS Genome Wide Association 01 Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: The ANRS Genome Wide Association 01 study. PLoS One. 2008;3(12):e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Manen D, et al. Genome-wide association scan in HIV-1-infected individuals identifying variants influencing disease course. PLoS One. 2011;6(7):e22208. doi: 10.1371/journal.pone.0022208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaren PJ, et al. International HIV Controllers Study Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum Mol Genet. 2012;21(19):4334–4347. doi: 10.1093/hmg/dds226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereyra F, et al. International HIV Controllers Study The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limou S, et al. ANRS Genomic Group Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199(3):419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 10.Le Clerc S, et al. ANRS Genomic Group Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03) J Infect Dis. 2009;200(8):1194–1201. doi: 10.1086/605892. [DOI] [PubMed] [Google Scholar]
- 11.McLaren PJ, et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. 2013;9(7):e1003515. doi: 10.1371/journal.ppat.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97(6):2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene: Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez E, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA. 1999;96(21):12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin MP, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282(5395):1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 16.Smith MW, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression: Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277(5328):959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 17.McDermott DH, et al. Multicenter AIDS Cohort Study (MACS) CCR5 promoter polymorphism and HIV-1 disease progression. Lancet. 1998;352(9131):866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrington M, et al. HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283(5408):1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 20.Apps R, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340(6128):87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Clerc S, et al. Evidence after imputation for a role of MICA variants in nonprogression and elite control of HIV type 1 Infection. J Infect Dis. 2014;210(12):1946–1950. doi: 10.1093/infdis/jiu342. [DOI] [PubMed] [Google Scholar]
- 22.Gusev A, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95(5):535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser C, et al. Virulence and pathogenesis of HIV-1 infection: An evolutionary perspective. Science. 2014;343(6177):1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartha I, et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. eLife. 2013;2:e01123. doi: 10.7554/eLife.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Günthard HF, et al. International Antiviral Society-USA Panel Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312(4):410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 27.Herbeck JT, et al. Multistage genomewide association study identifies a locus at 1q41 associated with rate of HIV-1 disease progression to clinical AIDS. J Infect Dis. 2010;201(4):618–626. doi: 10.1086/649842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troyer JL, et al. Genome-wide association study implicates PARD3B-based AIDS restriction. J Infect Dis. 2011;203(10):1491–1502. doi: 10.1093/infdis/jir046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 31.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 36.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 37.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17(R2):R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 40.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8(6):e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkler CA, et al. Dominant effects of CCR2-CCR5 haplotypes in HIV-1 disease progression. J Acquir Immune Defic Syndr. 2004;37(4):1534–1538. doi: 10.1097/01.qai.0000127353.01578.63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.