Significance
Microtubules and their associated proteins are central to most cellular functions. They have been extensively studied at multiple levels of resolution; however, significant knowledge gaps remain. Structures of microtubule-associated proteins bound to microtubules are not known at atomic resolution. We used magic angle spinning NMR to solve a structure of dynactin’s cytoskeleton-associated protein glycine-rich (CAP-Gly) domain bound to microtubules and to determine the intermolecular interface, the first example, to our knowledge, of the atomic-resolution structure of a microtubule-associated protein on polymeric microtubules. The results reveal remarkable structural plasticity of CAP-Gly, which enables CAP-Gly’s binding to microtubules and other binding partners. This approach offers atomic-resolution information of microtubule-binding proteins on microtubules and opens up the possibility to study critical parameters such as protonation states, strain, and dynamics on multiple time scales.
Keywords: magic angle spinning NMR, microtubules, dynactin's CAP-Gly domain, structure determination, intermolecular interface determination
Abstract
Microtubules and their associated proteins perform a broad array of essential physiological functions, including mitosis, polarization and differentiation, cell migration, and vesicle and organelle transport. As such, they have been extensively studied at multiple levels of resolution (e.g., from structural biology to cell biology). Despite these efforts, there remain significant gaps in our knowledge concerning how microtubule-binding proteins bind to microtubules, how dynamics connect different conformational states, and how these interactions and dynamics affect cellular processes. Structures of microtubule-associated proteins assembled on polymeric microtubules are not known at atomic resolution. Here, we report a structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain of dynactin motor on polymeric microtubules, solved by magic angle spinning NMR spectroscopy. We present the intermolecular interface of CAP-Gly with microtubules, derived by recording direct dipolar contacts between CAP-Gly and tubulin using double rotational echo double resonance (dREDOR)-filtered experiments. Our results indicate that the structure adopted by CAP-Gly varies, particularly around its loop regions, permitting its interaction with multiple binding partners and with the microtubules. To our knowledge, this study reports the first atomic-resolution structure of a microtubule-associated protein on polymeric microtubules. Our approach lays the foundation for atomic-resolution structural analysis of other microtubule-associated motors.
Microtubules (MTs), polymeric assemblies of α/β tubulin, and their associated proteins are central to most cellular functions, including maintenance of the cytoskeleton, mitosis, differentiation, and intracellular transport of various cargos, including signaling molecules (1). Due to their critical role in cell division, MTs are targets of potent cancer therapeutics, including taxol, auristatin, and other MT-stabilizing/destabilizing drugs. Their clinical success, however, is also associated with dose-limiting toxicities, which has led to the development of new therapeutics against specific MT-associated proteins (2). On the other hand, mutations in, and/or dysfunction of, MTs and their associated proteins are invariably associated with disease. Specifically, point mutations in the CAP-Gly domain of the p150Glued subunit of the dynactin complex (the structure of the complex is shown in Fig. 1), which binds to MTs and the associated protein EB1, have been described in patients with Perry syndrome, distal spinal bulbar muscular atrophy, and amyotrophic lateral sclerosis (ALS) (3–6). Mutations in the cargo-binding domain and other regions of the dynein–dynactin motor complex lead to developmental defects and neurological diseases, such as ALS, Charcot–Marie–Tooth disease, and Huntington’s disease (7–10). Despite their prevalent and critical roles in cellular processes, there remains a significant gap, particularly at the atomic level, in understanding how MTs and their associated proteins function (generate directional force and maintain processivity), both in healthy and in diseased states.
Fig. 1.
(A) Overview of the structure of the dynactin complex. (Top Left) Overall structural organization of dynactin based on the recent cryo-EM structure (21). (Middle and Bottom Left) Domain structure of p150Glued microtubule-binding subunit of dynactin. The CAP-Gly domain (colored purple) is part of the p150Glued. (B) SDS/PAGE cosedimentation gel of CAP-Gly with microtubules. The lanes are labeled in order as follows: M, molecular mass markers; F, free CAP-Gly standard; P, pellet obtained by ultracentrifugation of CAP-Gly/microtubule solution; S, supernatant after ultracentrifugation of the pellet. (C) Transmission electron microscopy (TEM) images of microtubules (Top Left); U-13C,15N,2H-CAP-Gly/MT NMR sample (Top Right). The Bottom images were recorded before and after 2 wk of continuous multidimensional MAS NMR experiments.
Although impressive advances have been made using X-ray diffraction, electron microscopy, biochemistry, and cell biology methods (7, 11–20), including the recent 4.0-Å cryo-EM structure of the dynactin complex (21), essential information concerning the mechanisms of microtubule-based transport is lacking. There are no atomic-resolution structures of motor proteins assembled on polymerized microtubules. Structure determination of these assemblies is challenging due to their insolubility, large size, and lack of long-range order, precluding their analysis by the traditional atomic-resolution techniques, X-ray diffraction and solution NMR. More importantly, diffraction and EM methods do not report on dynamics, perhaps the most fundamental missing link needed to understand how MTs and MT motor protein produce force, how “information” is transmitted (allostery), and how mutations related to disease manifest themselves in terms of function. Magic angle spinning (MAS) NMR spectroscopy is uniquely poised to bridge this gap and provide atomic-resolution insights on microtubule-associated proteins bound to polymeric MTs (22–24). Recently, using a hybrid MAS NMR/molecular dynamics (MD) approach, we have gained a comprehensive view of the residue-specific conformational dynamics of CAP-Gly occurring over six decades of motional timescales (nano- to milliseconds), in its free form, assembled on MTs, and bound to EB1 (24). We discovered that loop regions of CAP-Gly are dynamic when both free and bound to MTs whereas their mobility is attenuated in complex with EB1.
In this report, we present a structure of CAP-Gly assembled with polymerized MTs, determined de novo by MAS NMR spectroscopy. We demonstrate that, by using three CAP-Gly/MT samples with three kinds of isotopic labels and numerous 13C-13C and 13C-15N distance restraints recorded from homo- and heteronuclear correlation spectra, in conjunction with torsion angle and hydrogen bonding restraints, the structure of CAP-Gly bound to MTs is solved to 1.9–2.5 Å equivalent resolution, with very tight NMR structural ensembles. To our knowledge, this study is the first atomic-resolution structure of any microtubule-associated protein bound to polymeric microtubules. For determination of intermolecular interfaces formed by CAP-Gly and MTs and/or other binding partners, we incorporated rotational echo double resonance (REDOR) and double-REDOR (dREDOR) filters into homo- and heteronuclear correlation experiments. This approach is critical when one of the binding partners cannot be isotopically labeled with magnetically active nuclei, such as is the case with mammalian microtubules. Our results reveal that the binding interfaces of CAP-Gly with MTs and with EB1 partly overlap. Finally, our study underscores the remarkable structural plasticity of CAP-Gly, permitting the protein to adopt different conformations depending on its binding partner.
More broadly, the MAS NMR approaches introduced in this work can be applied to determination of atomic-resolution structures of other proteins and their complexes with polymeric microtubules, actin, and other polymeric cytoskeleton structures.
Results
Biophysical Characterization of the CAP–Gly/MT Complex.
Fig. 1 illustrates a cosedimentation assay of a typical MAS NMR sample of the U-13C,15N-CAP-Gly/MT complex; CAP-Gly-MT binding assays performed under different conditions are also presented in SI Appendix. The results are fully consistent with our prior findings (22), indicating that CAP-Gly/MT binding is specific and that the binding affinity is estimated in the micromolar range. Of the 15.3 mg of hydrated CAP-Gly/MT complex in the NMR sample, ∼1–2 mg are CAP-Gly. Negatively stained transmission electron microscopy (TEM) was used for characterizing the morphology of MTs and CAP-Gly/MT assemblies. As shown in Fig. 1C and SI Appendix, Fig. S1, in the NMR sample, the microtubules are in a stable state (not a depolymerized dynamic state), and spinning the sample for 2 wk during MAS NMR experiments does not result in any degradation of the microtubules. Deuterated sample media do not have any effect on the MT morphology either.
Identification and Calibration of Distance Restraints in CAP-Gly Assembled on Polymerized Microtubules.
Hundreds of 13C-13C and 13C-15N medium- and long-range distance restraints are needed to obtain high-resolution protein structures by MAS NMR. The distance restraints require chemical shift assignments, which we performed de novo, as described in SI Appendix. 13C-13C restraints were obtained from combined R2-driven (CORD) spectra in U-13C,15N-CAP-Gly/MT (500 ms mixing time), U-15N/[2-13C-glucose]-CAP-Gly/MT (500 ms mixing time), and U-15N/[1,6-13C-glucose]-CAP-Gly/MT (500 and 200 ms mixing times). 13C-15N restraints were extracted from the proton assisted insensitive nuclei cross polarization (PAIN-CP) spectrum of U-15N/[2-13C-glucose]-CAP-Gly/MT. As shown in Fig. 2 and in SI Appendix, Figs. S3 and S6), the resolution and sensitivity of the datasets are excellent, and numerous meaningful distance restraints were recorded.
Fig. 2.
Expansions around the aliphatic regions of 2D CORD (A and B) and 2D PAIN-CP (C) spectra of sparsely labeled CAP-Gly bound to polymerized microtubules, demonstrating examples of long-range, medium-range, and sequential distance restraints used in the structure calculation. CORD spectrum of [1,6-13C-glucose,U-15N]-CAP-Gly/MT is shown in A; CORD spectrum of [U-13C,15N]-CAP-Gly/MT is shown in B; PAIN-CP spectra of [2-13C-glucose,U-15N]-CAP-Gly/MT is shown in C. In all datasets, the cross-peaks corresponding to the different distance restraint ranges are color coded as follows: long-range, green; medium-range, orange; sequential, black. Both ambiguous and unambiguous restraints were used in the structure calculation. All spectra were recorded at 20.0 T; the CORD mixing time was 500 ms.
To assign the cross-peaks in each spectrum, a semiautomatic protocol was used as detailed in SI Appendix. The autoassignment and manual check procedure assigned 1,496 peaks in total, of which 442 peaks were with multiple assignments. Five medium-range and six long-range distance restraints came from the PAIN-CP spectrum; these restraints proved to be essential in deriving the final high-quality structure. The majority of the assignments were unambiguous and these unambiguous restraints, together with the ambiguous restraints, were used for structure calculation in Xplor-NIH (25).
Structure of CAP-Gly Assembled on Polymerized Microtubules.
Fig. 3A shows the 3D structure of CAP-Gly assembled on microtubules calculated from the experimental MAS NMR restraints. The 10 lowest energy structures out of 500 calculated are depicted in SI Appendix, Fig. S4. As anticipated, the overall structural topology of the protein was retained upon binding to MTs, with the core of the protein comprised of four β-sheets and a short C-terminal α-helix. At the same time, as shown in Fig. 3D, loop regions in CAP-Gly underwent pronounced conformational changes upon the formation of a complex with MTs, compared with the free CAP-Gly and CAP-Gly/EB1 complex (15, 23, 26). These changes resulted in sidechains adopting different orientations in free and MT- and EB1-bound states (Fig. 3E). Such reorientations may be important for fine-tuning the CAP-Gly’s binding to EB1 and MTs. As discussed in Structural Plasticity of CAP-Gly, the loop regions are structurally well-defined, and the conformational rearrangements are related to CAP-Gly’s conformational plasticity reported by us (23, 24).
Fig. 3.
(A) The 3D structure of CAP-Gly bound to polymerized microtubules, calculated from MAS NMR restraints (PDB ID code 2MPX); the structure is an average from the ensemble of 10 lowest energy structures. The CAP-Gly residues are color coded according to the number (noted as “n”) of long-range distance restraints per residue detected in the various MAS NMR spectra. (B) A matrix representation of all intra- and interresidue contacts generated from the MAS NMR distance restraints. (C) The number of long-range distance restraints plotted vs. the residue number. The CAP-Gly secondary structure is shown on the top. (D) The structure of CAP-Gly bound to polymerized MTs (purple) shown in A superimposed with the average structure of free CAP-Gly (23) (orange; PDB ID code 2M02) and CAP-Gly in complex with EB1 (15) (light green; PDB ID code 2HKQ). (E) Loop conformations of CAP-Gly in free, MT-bound, and EB1-bound states corresponding to those in D: β3-β4 loop (Top) and β2-β3 loop (Bottom).
The statistics of distance and backbone torsion angle restraints used for the final structure refinement are presented in SI Appendix, Table S4. The structure was determined with a total of 1,194 nonredundant 13C-13C/15N-13C distance restraints (after excluding low-probability restraints), of which 408 and 237 are long- and medium-range restraints, respectively. We note that this number of restraints is significantly higher than that used for MAS NMR structure determination of CAP-Gly (917 total, 197 long-range, and 97 medium-range restraints) (23), as well as in the MAS NMR structures of similar-size proteins determined recently (27–29). To further evaluate the distance restraint quality, we summarized the results in a 2D contact map displayed in Fig. 3B, where a shaded square indicates one or more distance restraints (corresponding to a unique final assignment) between the corresponding residues. The map illustrates the importance of the numerous long-range distance restraints in establishing the spatial proximity between the various distant regions in the primary sequence. As shown in SI Appendix, Table S4, there are very few violations of energy terms in the final refinement: three NOE, one statistical torsion angle database potential (tDB), two dihedral angle (CDIH), and three van der Waals (VDW). The three violations of distance restraints reported by the final refinement calculation are highlighted in red in the contact map. They are all long-range restraints: V29Cγ1-V47C, I36Cα-V89Cγ1, and V33Cγ2-T43Cβ. In Fig. 3C, the number of long-distance restraints is plotted for each residue, illustrating that, on average, 11.5 long-range restraints per residue were derived from MAS NMR spectra. There are 14.6 long-range distance restraints per residue on average for residues in the regions corresponding to rigid secondary structure elements (β-sheets and α-helices), and 9.4 long-range restraints per residue for loop regions. This number of restraints per residue is much greater than that we recorded in free CAP-Gly (23): 9.8 and 2.8 restraints per residue for the rigid secondary structure elements and loop regions, respectively. This dramatic improvement in the restraint numbers for the CAP-Gly/MT complex is due to the use of several samples containing different sparse 13C labels, the acquisition of a 500-ms CORD spectrum in U-13C,15N CAP-Gly/MT, and the higher sensitivity of experiments at 20.0 T. This protocol results in well-defined conformations of the CAP-Gly’s loop regions.
As summarized in Table 1, the rmsd values within the ensemble of the above 10 structures are 0.36 ± 0.06 Å for all backbone heavy atoms (N, Cα, and Cο), and 0.27 ± 0.05 Å for backbone heavy atoms located in rigid secondary structures (α-helices and β-strands). The summary of the structure quality validation is presented in SI Appendix, Table S5. The equivalent resolution according to ProCheck-NMR (30) is 1.9 Å (with respect to average value), 2.4 Å (with respect to percentage of residues in helices, sheets, and loops), 2.6 Å (with respect to hydrogen bond energy SD), 1.6 Å (with respect to χ-1 pooled SD), and 1.0 Å (with respect to SD of χ-2 trans angle).
Table 1.
MAS NMR structure of CAP-Gly assembled on microtubules
| All residues, Å | In sheets and helices, Å | In loops, Å | Refs. | |
| Backbone* | 0.36 ± 0.06 | 0.27 ± 0.05 | 0.38 ± 0.06 | |
| vs. 2M02 | 2.22 | 1.02 | 2.49 | (23) |
| vs. 1TXQ | 1.78 | 0.76 | 1.94 | (26) |
| vs. 2COY | 1.74 | 0.87 | 1.87 | (42) |
| vs. 2HKQ | 1.79 | 0.78 | 1.96 | (15) |
| vs. 2HQH | 1.82 | 0.76 | 2.08 | (43) |
| vs. 3E2U | 1.76 | 0.78 | 1.99 | (39) |
Comparison with the previously determined structures of free CAP-Gly and CAP-Gly complexes with end-binding protein EB1 and the zinc-knuckle 2 domain of human CLIP-170 by MAS NMR, X-ray, and solution NMR.
Fit of the ensemble of the 10 lowest energy structures to the average structure, 2MPX.
Intermolecular Interface of CAP-Gly with Microtubules.
Determination of intermolecular interfaces formed by a biomolecule interacting with its binding partner by MAS NMR generally relies on (i) analysis of chemical shift perturbations or intensity changes observed for the biomolecule in the complex with respect to the free biomolecule (31, 32) and (ii) analysis of intermolecular dipolar-based heteronuclear correlations in differentially isotopically enriched samples, where one biomolecule contains a certain set of isotope labels (e.g., 13C) and its binding partner a different kind of labels (e.g., 15N) (33–35). For analysis of the CAP-Gly/MT interface, we could not use either approach because (i) the chemical shift perturbations are relatively small and present throughout the entire protein and (ii) isotopic labeling of MTs is a challenge. One approach is to prepare the U-13C,15N,2H-CAP-Gly/MT complex and use the nonexchangeable aliphatic 1H sites on MTs for selective magnetization transfers across the interface to establish correlations with heteronuclei (13C/15N) on CAP-Gly. However, this approach leads to artifacts associated with intramolecular transfers due to residual protonation in aliphatic sidechains (SI Appendix, Figs. S7 and S8).
We therefore turned our attention to the REDOR filter-based experiments, such as REDOR-HETCOR (35). Herein, a REDOR (36) filter is first used to selectively dephase 1H magnetization from hydrogens that are directly bonded to a heteronucleus (13C/15N), in a molecule that is U-13C/15N–labeled. The remaining 1H magnetization after the application of the REDOR filter belongs to protons that do not have any directly bonded 13C/15N sites. This magnetization selectively establishes intermolecular dipolar contacts across the interface through a 1H-13C/15N cross-polarization step. The resulting 2D (1H-13C)-REDOR-HETCOR spectrum contains a finite number of well-resolved intermolecular cross-peaks (Fig. 4B) REDOR filters have also been used for studies of water–protein interfaces, as implemented in a medium- and long-distance MELODI-HETCOR experiment (37, 38).
Fig. 4.
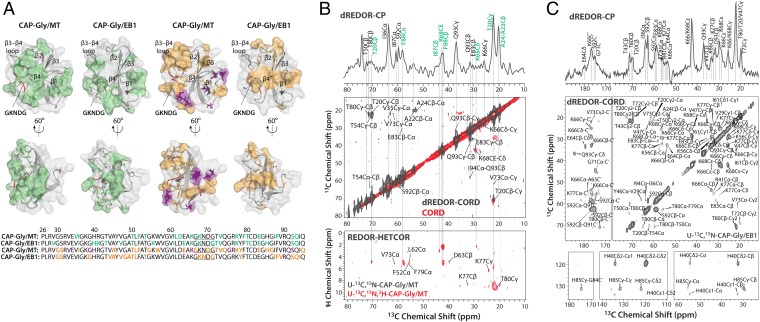
(A) Intermolecular interfaces of CAP-Gly with microtubules and end-binding protein EB1, derived from dREDOR (two Left panels) and chemical shift perturbations (two Right panels) in MAS NMR experiments, mapped onto the structure of CAP-Gly bound with MTs and in complex with EB1, respectively. The intermolecular interface of CAP-Gly with MT was obtained from dREDOR-based CPMAS and CORD experiments on U-13C,15N-CAP-Gly/MT (B, Top and Middle spectra, gray) and from CORD and 1H-13C-REDOR-HETCOR (35) experiments on U-13C,15N,2H-CAP-Gly/MT sample (B, Middle and Bottom spectra, red). Resonances obtained from both dREDOR-CORD and REDOR-HETCOR are labeled in green in B. (C) dREDOR-based CPMAS and CORD spectra of U-13C,15N-CAP-Gly/EB1. In A, the residues that belong to intermolecular interfaces of CAP-Gly with MT and EB1 are color coded as follows: with direct contacts in dREDOR- and HETCOR-based spectra (green); with large (> 1.0 ppm) and moderate (0.5–1.0 ppm) chemical shift perturbations (purple and orange, respectively). CAP-Gly residues at the interface with EB1 determined from chemical shift perturbations (23) are shown in orange. The primary sequence of CAP-Gly is depicted at the Bottom of A, and the residues are color coded as described above.
Because MT 1H assignments cannot be established at this time, we used double REDOR (dREDOR)-filtered 13C-13C correlation spectroscopy to assign the 13C CAP-Gly peaks. Here, 1H-13C/1H-15N REDOR is simultaneously applied to completely dephase CAP-Gly protons that are bonded to both 13C and 15N. 1H magnetization is dephased, resulting in no signal after the subsequent 1H-13C/15N cross-polarization of the U-13C,15N-CAP-Gly sample (SI Appendix, Fig. S9). On the contrary, the dREDOR–CPMAS spectrum of the U-13C,15N-CAP-Gly/MT complex contains a finite number of well-resolved 13C peaks that belong to the CAP-Gly residues at the interface with MTs (Fig. 4B). These signals arise from the 1H magnetization from the MTs transferred by 1H-13C cross-polarization across the intermolecular interface to the CAP-Gly residues. To assign these signals, we conducted a 2D dREDOR-CPMAS-CORD experiment. As shown in Fig. 4B, the spectrum contains multiple well-resolved diagonal and cross-peaks, which could be readily assigned, thus establishing the residues of CAP-Gly constituting the intermolecular interface with MTs. Notably, the same residues also gave rise to the cross-peaks in the REDOR-HETCOR spectrum of the U-13C,15N,2H-CAP-Gly/MT complex (Fig. 4B). Furthermore, in the REDOR-HETCOR spectrum, the K68 Cε, T20 Cγ, and K77 Cγ resonances correlated to multiple protons, which is a further indication that these residues are in spatial proximity to tubulin. In contrast, the CORD spectrum of U-13C,15N,2H-CAP-Gly/MT complex contained additional cross-peaks that belong to intramolecular correlations among CAP-Gly residues arising from residual protonation of the corresponding nonexchangeable sites. The residual proton signals were verified by the solution NMR spectra of the free U-13C,15N,2H-CAP-Gly (SI Appendix, Fig. S7).
The intermolecular interface of CAP-Gly with microtubules, derived on the basis of the above experiments, is illustrated in Fig. 4A and SI Appendix, Fig. S4B. The β3-β4 loop and the GKNDG motif within this loop constitute the primary interface with microtubules. This finding corroborates prior hypotheses, made on the basis of structural analysis of CAP-Gly complexes with EB1 and CLIP170, that the GKNDG motif likely constitutes a binding interface, with MT being a specific recognition sequence for EEY/F motifs ubiquitously present in CLIP170 and EB proteins, as well as in α-tubulin (26, 39). Our current results also clearly indicate that the bean-shaped CAP-Gly molecule binds to the microtubules with its flat surface comprised by its loops, the C-terminal β4-strand, and the short α-helix. These findings are in agreement with the recent cryo-EM results on the CAP-Gly(1-105)/MT and CAP-Gly(25-144)/MT complexes (40), which revealed the same flat side of CAP-Gly that forms the interface, including the proposed interaction of the GKNDG motif with the C terminus of tubulin. Of note, most of the surface-exposed, hydrophobic residues are on the flat side of CAP-Gly and are likely important for promoting the binding interaction.
Our results reveal that the intermolecular interfaces of CAP-Gly with microtubules and EB1 partly overlap (β3-β4 loop and S92-Q93 at helix turn). The binding interface of CAP-Gly with EB1 is known from previous studies (15, 26). The dREDOR-based spectra of the U-13C,15N-CAP-Gly/EB1 complex shown in Fig. 4C corroborate the previous findings and validate the dREDOR approach as applied to the CAP-Gly/MT complex.
As illustrated in Fig. 4A, chemical shift perturbations detected in CAP-Gly upon formation of the complex with MTs encompass multiple residues not found in the REDOR-based experiments. These residues are located at the N terminus, on the opposite side of the interface. Such perturbations are due to conformational changes in the protein residues upon binding to MTs but are not associated with the residues participating in the formation of the intermolecular interface with MTs. This finding underscores the fact that, when binding affinity is moderate, such as in CAP-Gly/MT, binding interface identification should not rely solely on chemical shift perturbations.
The CAP-Gly/MT interface contacts determined by MAS NMR are consistent with those predicted by us using molecular docking in ClusPro (41). We used the recent cryo-EM structure of MTs (12) and our MAS NMR structure of CAP-Gly bound to MTs. In the five top-ranked electrostatically driven models, CAP-Gly binds MT in the same orientation, at the C terminus of tubulin: The flat side (with β4 at the center) faces MT, and the GKNDG motif is oriented toward the minus end of MT (SI Appendix, Fig. S11). This predicted orientation and the binding contacts are in overall agreement with the interface determined from MAS NMR. We note that K77, F79, and T80 residues that, according to dREDOR, form direct contacts with MTs are on the opposite side of CAP-Gly and do not interact with tubulin in the docking model. This result is not surprising because the negatively charged C-terminal tail of tubulin (the E-hook), which is known to directly interact with CAP-Gly, is missing in the cryo-EM structure of MT, and the corresponding interactions are not captured in the model. Our MAS NMR experiments thus detect states invisible in cryo-EM due to conformational heterogeneity. On the basis of our results, we propose that the E-hook passes through the hydrophobic patch at the flat binding side and stretches to the β3-β4 loop of CAP-Gly to directly bind with the GKNDG motif.
Structural Plasticity of CAP-Gly.
CAP-Gly possesses conformational plasticity that is important for dynactin’s interactions with multiple binding partners (23). This conformational plasticity is intimately connected to CAP-Gly’s inherent mobility that spans many decades of motional timescales, from nano- to milliseconds (24). CAP-Gly’s motional profile is modulated by its environment, and, remarkably, both free and MT-bound proteins are dynamic in loops whereas CAP-Gly in complex with EB1 is considerably more rigid (24).
The atomic resolution of CAP-Gly bound to polymerized microtubules presented here provides an opportunity to gain additional insights into the conformational plasticity of the protein. To this end, we compared the MAS NMR structures of CAP-Gly assembled on MTs and free with the solution NMR structure of free CAP-Gly, and the X-ray structures of CAP-Gly/EB1 and CAP-Gly/ZnCLIP complexes. Table 1 contains the summary of the results in the form of rmsds between the different structures. The results indicate that, interestingly, the backbone rmsd between the MAS NMR-derived structures of CAP-Gly/MT complex and free CAP-Gly is the largest. The backbone rmsd between the MAS NMR structure of CAP-Gly/MTs and the solution NMR structure of free CAP-Gly (1.74 Å for all residues) has similar overall value as the rmsd for CAP-Gly complexes with EB1 or ZnCLIP (1.76–1.82 Å for all residues), but significantly larger rmsd in rigid secondary structure elements (0.87 Å vs. 0.76–0.78 Å). It is important to note that the overall structure of the core β2-β4 sheets superimposes very well in the different structures: free CAP-Gly, CAP-Gly assembled microtubules, and CAP-Gly in complex with EB1, as illustrated in Fig. 3D. At the same time, the β1 conformation shows relatively large differences in the structures of free CAP-Gly and CAP-Gly assembled on MTs. Not surprisingly, the largest differences among these structures are found for the loop regions spanning residues D63 to G86 and containing the GKNDG motif. In the free CAP-Gly structure, the long loop extends farther away from the core formed by β2-β4. The conformation of free CAP-Gly is more open. In contrast, upon binding to the microtubules or EB1, CAP-Gly adopts a more closed conformation. This result is also consistent with the fact that CAP-Gly is locked into a single conformation upon binding to EB1 or microtubules (23). Taken together, the results of this investigation and our prior studies indicate that the CAP-Gly loops reorganize for optimal interactions with microtubules and the multiple dynactin’s binding partners.
Discussion
Recently, a cryo-EM study reported the structure of several CAP-Gly constructs on polymerized MTs at 9.7–10.2 Å resolution (40). Although atomic resolution was not achieved in this study, several important observations were made: (i) The interface of CAP-Gly with MT includes the GKNDG motif, which interacts with the C terminus of tubulin; (ii) the tubulin E-hooks interact with the basic patches 1–25 and 106–144 of the protein, possibly permitting it to diffuse laterally to the next binding site; (iii) the orientation of CAP-Gly on MTs depends on the length of the construct used; and (iv) the recognition sites for CAP-Gly and EB1 on tubulin are nonoverlapping. Interestingly, electron density could not be detected for CAP-Gly(25-105), presumably due to the weaker association with MTs compared with CAP-Gly(1-105) and CAP-Gly(25-144) and the dynamic nature of the resulting CAP-Gly(25-105)/MT complex. Furthermore, owing to its flexibility the CAP-Gly domain and the entire dynactin’s shoulder projection are not visible in the high-resolution EM maps of the dynactin complex (21), precluding their structural analysis. Indeed, as we reported recently (24), CAP-Gly(19-107) bound to polymeric MTs is dynamic on the timescales spanning nano- to milliseconds, and the lack of electron density in the cryo-EM studies is consistent with our results. In contrast to the EM-based studies, MAS NMR experiments permit detection of dynamic complexes, as shown in our current work. Despite the dynamic nature of CAP-Gly(19-107)/MT, the atomic-resolution structure of CAP-Gly in this complex, including the intermolecular interface, could be determined through MAS NMR.
Our study revealed that CAP-Gly interacts with MTs through its loops (including the GKNDG motif), as well as residues on the C-terminal β4-strand and α-helix. Although we could not assign the tubulin residues interacting with CAP-Gly because at present we have not isotopically labeled mammalian tubulin, the topology of CAP-Gly’s interface derived on the basis of our MAS NMR data suggests that there needs to be a flexible region of tubulin “wrapping around” the CAP-Gly, indirectly corroborating the cryo-EM conclusion that tubulin’s E-hook must be involved in the formation of the intermolecular interface. In the future, it will be advantageous to use a hybrid approach combining MAS NMR with cryo-EM and computational analysis to gain atomic-resolution structural information on both the MT-associated proteins and their orientations on the MTs.
Concluding Remarks
The atomic-resolution structure of CAP-Gly on polymerized microtubules solved by MAS NMR establishes the foundations for our understanding of the biological function of microtubule-associated proteins assembled with polymerized microtubules. As demonstrated in this work, MAS NMR enables investigations of dynamic complexes between motor proteins and MTs, which cannot be addressed by other structural biology techniques. The structure of dynactin’s CAP-Gly on polymerized MTs revealed structural plasticity of the protein that is necessary for dynactin’s interaction with multiple binding partners. In the future, we envision that hybrid MAS NMR/cryo-EM/computational approaches will become essential to gain atomic-level understanding on the motor protein assemblies with microtubules.
Materials and Methods
Samples of isotopically labeled CAP-Gly(19-107) bound to polymerized MTs were prepared as reported previously (22, 24). Cosedimentation assays were performed, and negatively stained transmission electron micrographs were acquired (SI Appendix). Details of MAS NMR experiments, spectral analysis, and structural calculation are described in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Grant R01GM085306 from the National Institute of General Medical Sciences. We acknowledge the support of the National Science Foundation Grant CHE0959496 for the acquisition of the 850-MHz NMR spectrometer and of NIH Grants P30GM103519 and P30GM110758 for the support of core instrumentation infrastructure at the University of Delaware.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.T. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2MPX) and the Biological Magnetic Resonance Data Bank (BMRB code 25005).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509852112/-/DCSupplemental.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112(4):467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28(1-2):197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 3.Farrer MJ, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41(2):163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puls I, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33(4):455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 5.Puls I, et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57(5):687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka F, Ikenaka K, Yamamoto M, Sobue G. Neuropathology and omics in motor neuron diseases. Neuropathology. 2012;32(4):458–462. doi: 10.1111/j.1440-1789.2011.01281.x. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed S, Sun S, Siglin AE, Polenova T, Williams JC. Disease-associated mutations in the p150(Glued) subunit destabilize the CAP-gly domain. Biochemistry. 2010;49(25):5083–5085. doi: 10.1021/bi100235z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen XJ, Xu H, Cooper HM, Liu Y. Cytoplasmic dynein: A key player in neurodegenerative and neurodevelopmental diseases. Sci China Life Sci. 2014;57(4):372–377. doi: 10.1007/s11427-014-4639-9. [DOI] [PubMed] [Google Scholar]
- 9.Eschbach J, et al. Mutations in cytoplasmic dynein lead to a Huntington’s disease-like defect in energy metabolism of brown and white adipose tissues. Biochim Biophys Acta. 2011;1812(1):59–69. doi: 10.1016/j.bbadis.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Eschbach J, et al. Dynein mutations associated with hereditary motor neuropathies impair mitochondrial morphology and function with age. Neurobiol Dis. 2013;58:220–230. doi: 10.1016/j.nbd.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldaz H, Rice LM, Stearns T, Agard DA. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature. 2005;435(7041):523–527. doi: 10.1038/nature03586. [DOI] [PubMed] [Google Scholar]
- 12.Alushin GM, et al. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157(5):1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berezuk MA, Schroer TA. Dynactin enhances the processivity of kinesin-2. Traffic. 2007;8(2):124–129. doi: 10.1111/j.1600-0854.2006.00517.x. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463(7280):485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honnappa S, et al. Key interaction modes of dynamic +TIP networks. Mol Cell. 2006;23(5):663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14(11):713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roll-Mecak A, Vale RD. Making more microtubules by severing: A common theme of noncentrosomal microtubule arrays? J Cell Biol. 2006;175(6):849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siglin AE, et al. Dynein and dynactin leverage their bivalent character to form a high-affinity interaction. PLoS One. 2013;8(4):e59453. doi: 10.1371/journal.pone.0059453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallee RB, Williams JC, Varma D, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58(2):189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 20.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303(5658):676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 21.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347(6229):1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S, Siglin A, Williams JC, Polenova T. Solid-state and solution NMR studies of the CAP-Gly domain of mammalian dynactin and its interaction with microtubules. J Am Chem Soc. 2009;131(29):10113–10126. doi: 10.1021/ja902003u. [DOI] [PubMed] [Google Scholar]
- 23.Yan S, et al. Three-dimensional structure of CAP-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy: Conformational plasticity and interactions with end-binding protein EB1. J Mol Biol. 2013;425(22):4249–4266. doi: 10.1016/j.jmb.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan S, et al. Internal dynamics of dynactin CAP-Gly is regulated by microtubules and plus end tracking protein EB1. J Biol Chem. 2015;290(3):1607–1622. doi: 10.1074/jbc.M114.603118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19(4):449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 27.Fasshuber HK, et al. Structural heterogeneity in microcrystalline ubiquitin studied by solid-state NMR. Protein Sci. 2015;24(5):592–598. doi: 10.1002/pro.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morag O, Sgourakis NG, Baker D, Goldbourt A. The NMR-Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope. Proc Natl Acad Sci USA. 2015;112(4):971–976. doi: 10.1073/pnas.1415393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demers J-P, et al. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nat Commun. 2014;5:4976. doi: 10.1038/ncomms5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 31.Zech SG, Olejniczak E, Hajduk P, Mack J, McDermott AE. Characterization of protein-ligand interactions by high-resolution solid-state NMR spectroscopy. J Am Chem Soc. 2004;126(43):13948–13953. doi: 10.1021/ja040086m. [DOI] [PubMed] [Google Scholar]
- 32.Schütz AK, et al. The amyloid-Congo red interface at atomic resolution. Angew Chem Int Ed Engl. 2011;50(26):5956–5960. doi: 10.1002/anie.201008276. [DOI] [PubMed] [Google Scholar]
- 33.Asami S, Rakwalska-Bange M, Carlomagno T, Reif B. Protein-RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52(8):2345–2349. doi: 10.1002/anie.201208024. [DOI] [PubMed] [Google Scholar]
- 34.Weingarth M, Baldus M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc Chem Res. 2013;46(9):2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Tasayco ML, Polenova T. Magic angle spinning NMR experiments for structural studies of differentially enriched protein interfaces and protein assemblies. J Am Chem Soc. 2008;130(17):5798–5807. doi: 10.1021/ja711304e. [DOI] [PubMed] [Google Scholar]
- 36.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson. 1989;81(1):196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Yao XL, Schmidt-Rohr K, Hong M. Medium- and long-distance 1H–13C heteronuclear correlation NMR in solids. J Magn Reson. 2001;149(1):139–143. [Google Scholar]
- 38.Li S, Su Y, Luo W, Hong M. Water-protein interactions of an arginine-rich membrane peptide in lipid bilayers investigated by solid-state nuclear magnetic resonance spectroscopy. J Phys Chem B. 2010;114(11):4063–4069. doi: 10.1021/jp912283r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisbrich A, et al. Structure-function relationship of CAP-Gly domains. Nat Struct Mol Biol. 2007;14(10):959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Crevenna AH, Kunze I, Mizuno N. Structural basis for the extended CAP-Gly domains of p150(glued) binding to microtubules and the implication for tubulin dynamics. Proc Natl Acad Sci USA. 2014;111(31):11347–11352. doi: 10.1073/pnas.1403135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20(1):45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 42.Saito K, et al. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure. 2004;12(9):1719–1728. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi I, Plevin MJ, Ikura M. CLIP170 autoinhibition mimics intermolecular interactions with p150Glued or EB1. Nat Struct Mol Biol. 2007;14(10):980–981. doi: 10.1038/nsmb1299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.