Significance
Rhythmic gene regulation in mouse liver results from an intertwined relationship between feeding cycles and the circadian clock. Significant efforts have been made to understand this interaction but a complete picture of the resulting diurnal transcription–translation processes is still missing. Through the simultaneous quantification of temporal transcription, accumulation, and translation of mRNA in the liver, we investigated the regulatory landscape of mice with intact or deficient circadian clock subjected to different feeding regimens. We showed that circadian clock and feeding rhythms coordinate rhythmic transcription to drive downstream rhythmic mRNA accumulation and translation. However, a subset of genes harboring 5′-Terminal Oligo Pyrimidine tract or Translation Initiator of Short 5′-UTR elements encoding proteins involved in translation and mitochondrial activity, respectively, present a transcription-independent rhythmic translation mainly regulated by feeding.
Keywords: circadian rhythms, ribosome profiling, mRNA translation, 5′-TOP sequences, TISU motifs
Abstract
Diurnal oscillations of gene expression are a hallmark of rhythmic physiology across most living organisms. Such oscillations are controlled by the interplay between the circadian clock and feeding rhythms. Although rhythmic mRNA accumulation has been extensively studied, comparatively less is known about their transcription and translation. Here, we quantified simultaneously temporal transcription, accumulation, and translation of mouse liver mRNAs under physiological light–dark conditions and ad libitum or night-restricted feeding in WT and brain and muscle Arnt-like 1 (Bmal1)-deficient animals. We found that rhythmic transcription predominantly drives rhythmic mRNA accumulation and translation for a majority of genes. Comparison of wild-type and Bmal1 KO mice shows that circadian clock and feeding rhythms have broad impact on rhythmic gene expression, Bmal1 deletion affecting surprisingly both transcriptional and posttranscriptional levels. Translation efficiency is differentially regulated during the diurnal cycle for genes with 5′-Terminal Oligo Pyrimidine tract (5′-TOP) sequences and for genes involved in mitochondrial activity, many harboring a Translation Initiator of Short 5′-UTR (TISU) motif. The increased translation efficiency of 5′-TOP and TISU genes is mainly driven by feeding rhythms but Bmal1 deletion also affects amplitude and phase of translation, including TISU genes. Together this study emphasizes the complex interconnections between circadian and feeding rhythms at several steps ultimately determining rhythmic gene expression and translation.
Living organisms on Earth are subjected to light–dark cycles caused by rotation of the Earth around the sun. To anticipate these changes, virtually all organisms have acquired a circadian timing system during evolution that allows a better adaptation to their environment. As a consequence, most aspects of their physiology are orchestrated in a rhythmic way by the circadian clock (from the Latin circa diem, meaning “about a day”), an endogenous and autonomous oscillator with a period of around 24 h (1, 2). Not surprisingly, perturbations of this clock in mammals lead to pathologies including psychiatric, metabolic, and vascular disorders (1, 3, 4). At the organismal scale, the oscillatory clockwork is organized in a hierarchal manner. Within the suprachiasmatic nuclei (SCN) of the hypothalamus, the “master clock” receives light input via the retina and communicates timing signals to “enslave” oscillators in peripheral organs (1, 2). The molecular oscillator consists of interconnected transcriptional and translational feedback loops, in which multiple layers of control, including temporal posttranscriptional and posttranslational regulation, play important roles (5). These additional layers of regulation are largely coordinated by systemic cues originating from circadian clock and/or feeding-coordinated rhythmic metabolism, allowing the adjustment of the molecular clockwork with the metabolic state of the cell (6). During the last decades, efforts have been made to produce a comprehensive knowledge of the transcriptional regulation orchestrated by the circadian clock (7–9) and feeding rhythms (10, 11). More recently, additional layers of regulation at the translational (12, 13) and posttranslational (14) levels have been described, increasing the complexity of the phenomenon involved in the establishment of the rhythmic mRNA expression and protein synthesis landscapes.
To gain more insight into potential rhythmic coordination at all stages of mRNA regulation, we monitored simultaneously mRNA transcription, accumulation, and translation using total RNA sequencing and ribosome profiling (15, 16).We describe here a modified protocol of this technique that allows deep sequencing of ribosome-protected mRNA fragments rapidly and with high throughput from mouse tissues. We measured transcription, accumulation, and translation of mouse liver mRNAs under different feeding rhythms and in clock-deficient animals. In this way, we showed that rhythmic transcription drives predominantly rhythmic mRNA accumulation and translation for a majority of genes. The study of WT and brain and muscle Arnt-like 1 (Bmal1) KO mice shows that the interconnection between circadian clock and feeding rhythms has broad impacts on rhythmic genes expression. Surprisingly, Bmal1 deletion seems to alter posttranscriptional level more importantly than transcription. In addition, it seems that global mRNA accumulation drives translation whereas a small subset of genes presents a diurnal change in their translation efficiency. These genes are involved in translation or mitochondrial activity and harbor a 5′-Terminal Oligo Pyrimidine tract (5′-TOP) or Translation Initiator of Short 5′-UTR (TISU) motif, respectively. Their rhythmic translation efficiency is mainly driven by feeding rhythms and food restriction increases their amplitudes and temporal coordination. However, Bmal1 deletion also affects amplitudes and phases of mRNA translation, notably for TISU genes. By measuring simultaneously all of the aspects of mRNA regulation, this study shows for the first time to our knowledge the role of circadian and feeding rhythms in the establishment of rhythmic mRNA and protein synthesis.
Results
Ribosome Profiling Around the Diurnal Cycle in Mouse Liver.
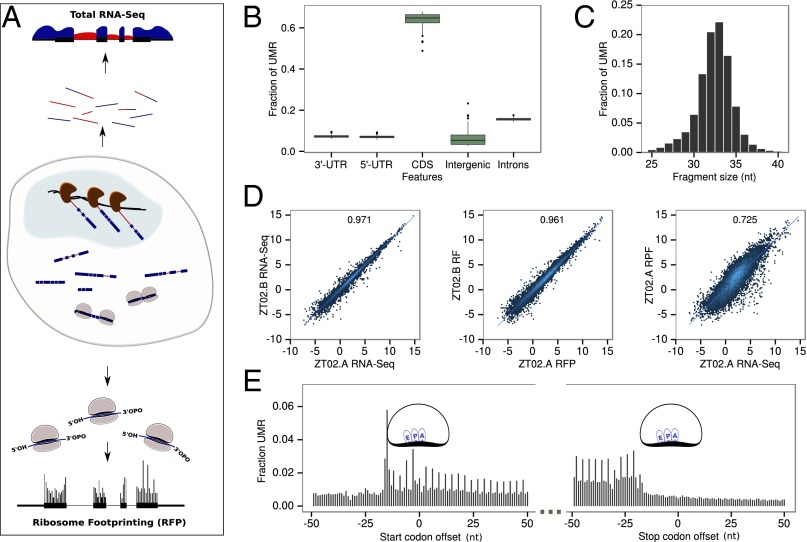
To monitor temporal mRNA transcription, accumulation, and translation, we extracted total RNA and ribosome-protected mRNA fractions from livers of individual mice every 2 h under ad libitum feeding (ALF). In parallel, the same experiment was performed every 4 h in WT and Bmal1 KO animals under a night-restricted feeding (RF) regimen. In total, 84 samples were subjected to RNA sequencing (RNA-Seq) and ribosome profiling (Ribo-Seq) (Fig. 1A). For RNA-Seq, intronic reads provide a good proxy for transcription rate and exonic reads for mRNA abundance (17). Owing to the large number of samples, we developed a simplified and faster protocol for ribosome profiling library generation, similar to that in ref. 18. Briefly, ribosome footprints (RFPs) ends were modified with polynucleotide kinase to generate libraries using 5′- and 3′- mRNA adapters. The quality and reproducibility of the resulting Ribo-Seq libraries were high (Fig. S1), showing a low percentage of PCR duplicates and minor sequence-dependent ligation (Fig. S2).
Fig. 1.
Ribosome profiling around the diurnal cycle in mouse liver. (A) Schematic representation of the experiment. Total mRNAs are directly sequenced (top) and ribosome loaded-mRNA are purified (ribosome footprinting) before sequencing (bottom). RNA-Seq from total RNA (genomic sequence is represented by a black line, surrounded by RNA-Seq signal) is quantified in intronic (red) and exonic (blue) regions to estimate pre-mRNA and mRNA levels. Ribosome density is quantified by modified ribosome profiling protocol. (B) The overall fraction of uniquely mapped reads (UMRs) of size between 28 and 34 bp up to one mismatch from the 84 Ribo-Seq samples in protein CDSs and nonprotein coding regions. (C) Size distribution of RFPs for UMR up to one mismatch. (D) Reproducibility of biological replicates. (Left) Log2 of total normalized counts (reads per kilobase per million, RPKM) for exonic RNA-Seq. (Middle) Ribo-Seq. (Right) Comparison of RPKM for Ribo-Seq and exonic RNA-Seq for each annotated protein coding gene (only UMR). R2 indicates Pearson correlation at the top of each panel. (E) Density of 5′ ends of 32-nt ribosome footprints at the starts and ends of ORFs shows three nucleotides periodicity. Positions of the related E, P, and A sites of the ribosomes are indicated.
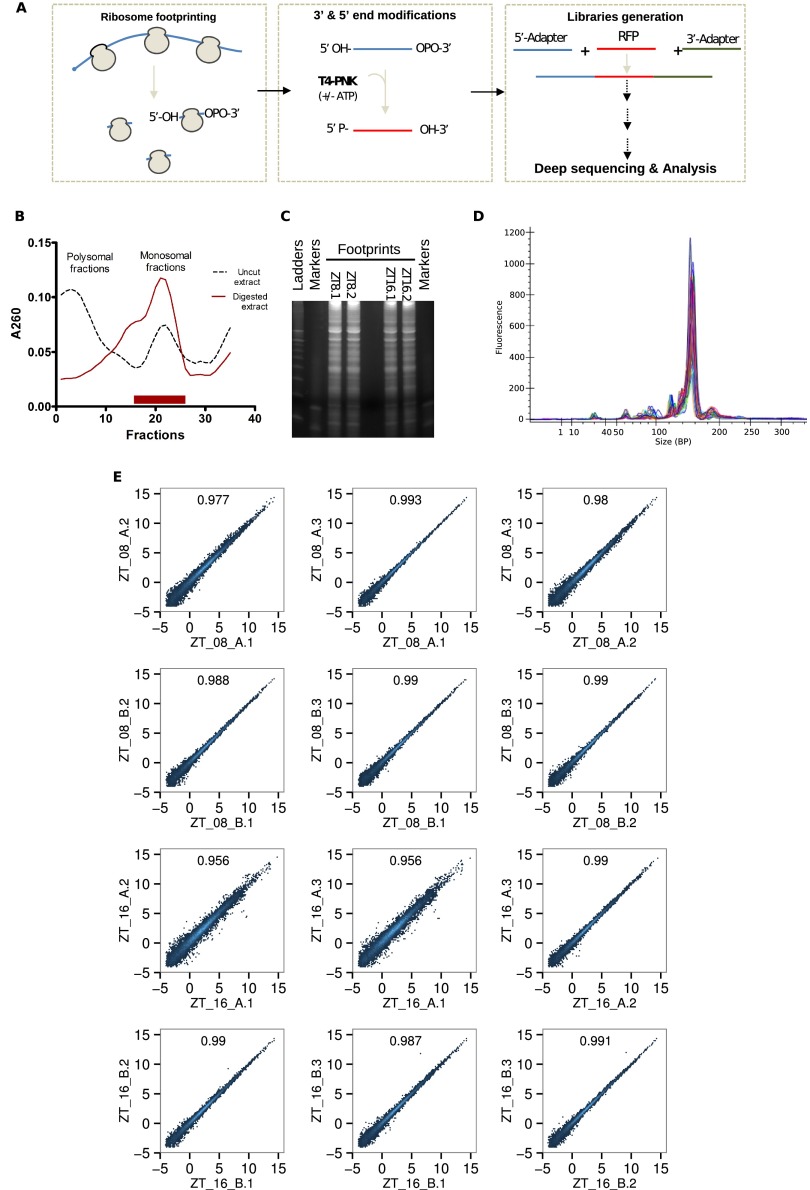
Fig. S1.
Technical validation of ribosome profiling experiments. (A) Simplified representation of the modified ribosome profiling method. RNase I digestion leaves a 5′-OH and a 3′-cyclophosphate. T4 polynucleotide kinase treatment performed in the absence of ATP converts the 3′-cyclophosphate into a 3′-OH whereas phosphorylating the 5′-OH ends in presence of ATP. Libraries are then generated as described in Materials and Methods. (B) Example of 260-nm optical density profile from sucrose gradient fractions. RNase I-digested lysates (red line) harbor clear monosome enrichment, whereas untreated controls (dark dashes) exhibit a standard polysome-enriched profile. Monosomal RNA was then extracted from fractions delimited with a red bar. (C) Extracted RNAs were resolved on 15% TBE-urea-PAGEs to select 28- to 36-nt-long RNA fragments. (D) Homogenous size distribution of libraries with a strong and sharp library peak at the expected size (∼150 bp). (E) Technical reproducibility of technical replicates for each individual gene (log2 RPKM). R2 indicates Pearson correlation at the top of each panel. Four samples (two per time point) were treated as described before. Then, fragmented RNAs were split, resolved on three distinct 15% TBE-urea-PAGEs, and analyzed as described in Materials and Methods.
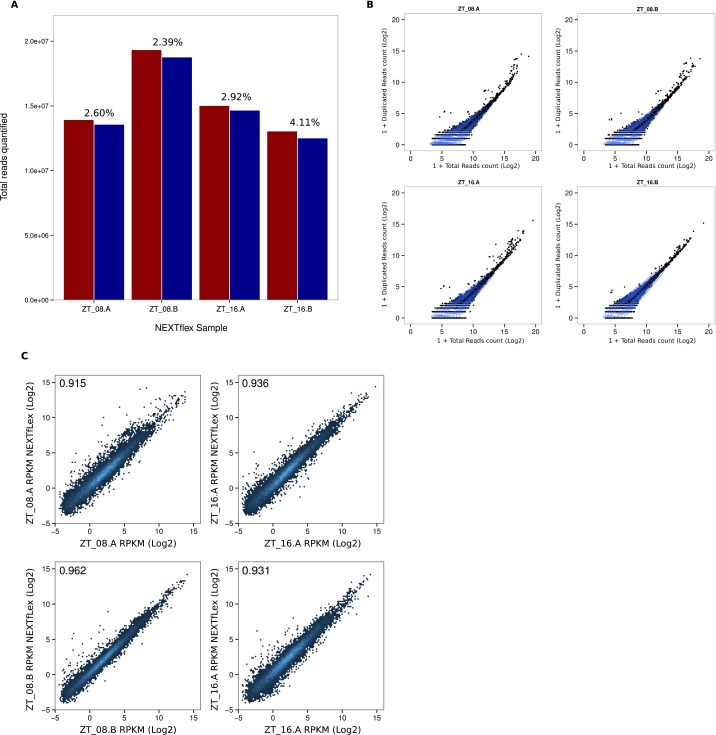
Fig. S2.
Validation of Ribo-Seq libraries (NextFlex). (A) NEXTflex libraries were generated from four different samples with randomly barcoded adapters to identify duplicated reads and possible adapters bias. Total numbers of reads quantified with (red) or without (blue) duplicated sequences are represented. For each sample, percentages of duplicated reads are indicated. (B) Number of duplicated reads sequences in function reads counts (log2) for individual genes. Blue lines represent densities of genes and are aggregated around minimal duplicated values. (C) Reproducibility of RFP signals (RPKM log2) obtained with NEXTflex and TrueSeq protocols. Correlation scores are represented in each plot.
Overall, Ribo-Seq reads mapped mainly on coding DNA sequences (CDSs) (Fig. 1B) with a mean footprint size surrounding 32 nucleotides (Fig. 1C), consistent with previous results obtained in mammalian cells or tissues (18–21) as well as with ribosome size. Biological reproducibility for both RNA-Seq and Ribo-Seq was high (Fig. 1D; R2 of 0.97 and 0.96, respectively). Moreover, mRNA abundance and total ribosome density still exhibited fairly high correlation (R2 = 0.73), highlighting both the overall proportionality between number of translating ribosomes and number of transcripts, as well as gene-specific translational kinetics (initiation or elongation). Finally, densities of ribosome footprints at the starts and ends of CDSs in protein-coding transcripts exhibited the expected trinucleotide repeat, reflecting the main ORF and the three-nucleotide movement of the ribosomes (Fig. 1E). Together, these results emphasize that this technique allows us to obtain with high throughput a high-quality dataset that can be used to monitor simultaneously transcription, mRNA abundance, and translation in different conditions.
Rhythmic Ribosome Footprint Signal Is Mainly Driven by Rhythmic mRNA Accumulation.
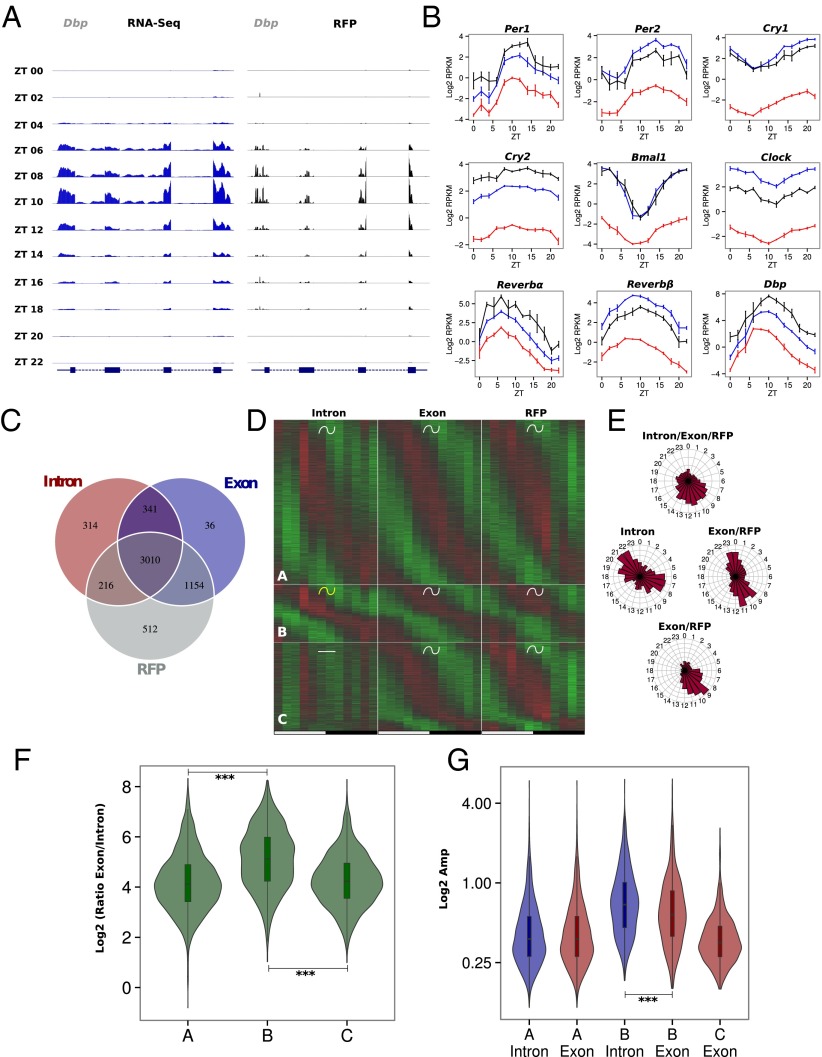
We first investigated gene expression in ALF mice. Considering some well-studied circadian clock-regulated genes (e.g., Dbp) confirmed that our data are of high quality and showed the expected rhythmicity for both RNA-Seq and Ribo-Seq (Fig. 2A). As anticipated, circadian clock genes show rhythms at all stages, from their respective pre-mRNAs to the accumulation of the mRNA and downstream mRNA translation (Fig. 2B), confirming that translational regulation has a minor impact upon clock gene expression (12). To assess and quantify rhythmic regulations at the different steps, we developed computational models and classified genes depending on the rhythmic or constant behavior at each measured level (pre-mRNA, mRNA, and ribosome footprints) (Dataset S1). For a great majority of genes, rhythmically translated mRNAs are subjected to rhythmic mRNA accumulation and a significant part of those (72%) exhibit rhythmic transcription (Fig. 2C).
Fig. 2.
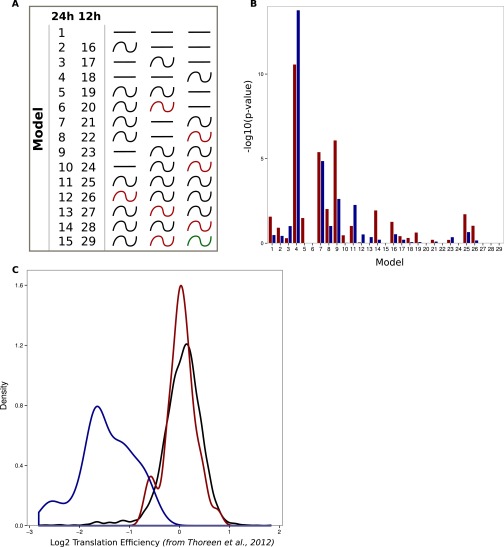
Rhythmic ribosome footprint signals are mainly driven by rhythmic mRNA accumulation. Model selection to assess rhythmicity is applied on WT ad libitum datasets combining intronic, exonic, and RFP signal. Harmonic regression is used with a period of 24 h and 12 h. Genes are assigned to one of the 29 models described in Fig. S5A. An arbitrary threshold of 0.4 is set on the BIC weight. Genes with log2 RPKM >0 at the exon and RFP levels are selected. (A) Exon RNA-Seq (Left) and RFP (Right) signals for the circadian clock-regulated Dbp gene. The two signals synchronously peak at ZT10. (B) Pre-mRNA (red), mRNA (blue), and RFP (black) signals in log2 RPKM for circadian clock core genes and clock-controlled genes. (C) Number of genes with rhythmic pre-mRNA, mRNA, and RFPs. The largest group shows rhythms on all three levels. (D) Three groups of genes showing identical mRNA and RFP rhythms. Standardized relative expression is indicated in green (low) and red (high). White and black boxes represent light and dark periods, respectively. (E) Phase distribution of the three groups described in D. (F) Distribution of mRNA (Exon)/pre-mRNA (Intron) ratios for the three groups of genes. An increased ratio suggested a more stable mRNA and longer half-life. A Welch′s t test indicates that this ratio is significantly higher in group B compare with groups A (P = 1⋅10−62) and C (P = 1⋅10−49). (G) Distribution of pre-mRNA (blue) and mRNA (red) amplitudes for the three groups. Group B harbors decreased amplitude in exons compare with introns (paired t test: P = 5⋅10−13) as a consequence of long half-lived transcripts, despite a general trend for higher amplitude.
We evaluated the impact of rhythmic translation on rhythmic protein levels by comparing mRNA and RFP levels with recently published protein levels (14). As described for rhythmic mRNAs, the majority of rhythmic RFPs encoded nonrhythmic proteins, likely as a consequence of long protein half-lives (Fig. S3). However, these observed flat profiles in protein abundance do not necessarily indicate flat activity because, for example, newly synthesized proteins can be more active than old oxidized proteins (22–24). Therefore, total protein level quantified by mass spectrometry may not always reflect rhythmic activity of newly synthesized protein originating from rhythmic RFP accumulation. However, rhythmic RFP signals typically show no significant delay with mRNA accumulation, whereas an average delay of around 6 h is observed between RFP signals and protein accumulation, as expected for relatively long-lived proteins (Fig. 3C) (14).
Fig. S3.
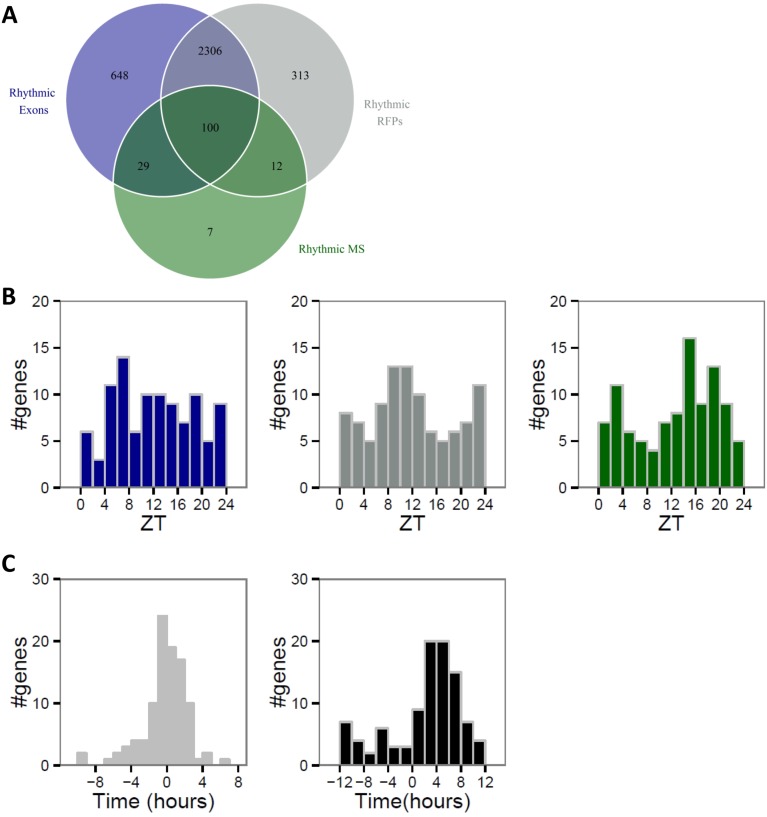
mRNA accumulation mainly drives protein rhythm. Proteins identified and quantified for at least eight time points in published MS data (14) were analyzed and compared with corresponding exonic and RFP reads obtained in our ribosome profiling data. Rhythmicity was assessed by performing a multiple linear regression for each relative time as described in ref. 14. Exonic, RFP, and MS signals were considered rhythmic for corrected P value (Benjamini–Hochberg method, ref. 82) below 0.25. (A) Venn diagram showing number of rhythmic proteins (green), mRNAs (blue), and rhythmic RFPs (gray). (B) Phase distributions of the 100 rhythmic proteins (green) encoded by rhythmic mRNA (blue) and RFP (gray). (C) Phase delays between mRNA and RFPs (Left) for group B show that phase of translation follows mRNA accumulation phase. Protein accumulation occurs mainly 4–6 h after the peak of RFP signal (Right).
Fig. 3.
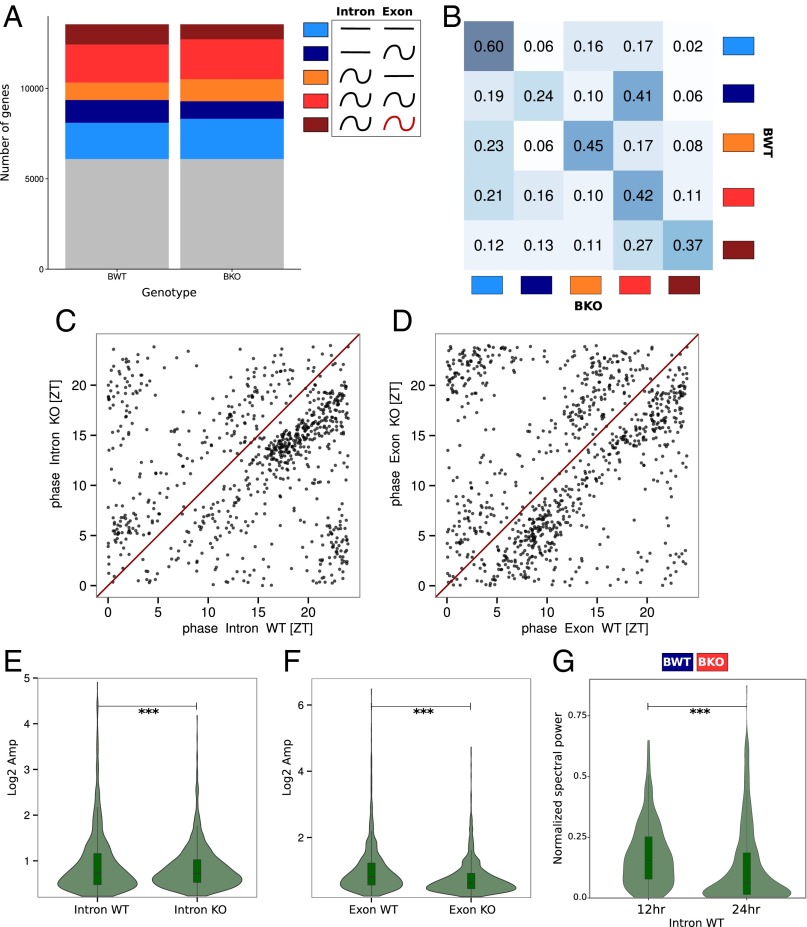
Effects of Bmal1 deletion on rhythmic mRNA accumulation. Model selection to assess rhythmicity is applied on Bmal1 WT and KO RF dataset combining intronic, exonic, and RFP signal. Harmonic regression is used with a period of 24 h. Genes are assigned to one of the 877 models generated by the six conditions. A threshold of 0.1 is set on the BIC weight. Genes with log2 RPKM >0 at the exon and RFP levels in WT condition are selected. (A) Genes are clustered in the six groups depending of the model they were assigned and grouped in function of their rhythmic pattern at the intronic and exonic level: gray, genes not assigned in any group; light blue, constant pre-mRNA and mRNA levels; dark blue, constant pre-mRNA and rhythmic mRNA; orange, rhythmic pre-mRNA and constant mRNA; red, rhythmic pre-mRNA and mRNA; brown, rhythmic pre-mRNA and mRNA with different rhythmic parameters. (B) Fraction of genes belonging in one of the five clusters in KO compared with WT mice. Intensity of the blue color in each box represents the conservation degree of behavior of the genes between WT and KO, with darkest blue corresponding to higher conservation. (C) Phase distribution of rhythmic pre-mRNA in WT and KO mice. (D) Phase distribution of rhythmic mRNA in WT and KO mice. (E) Distribution of pre-mRNA amplitudes in WT and KO mice. KO mice present slightly increased amplitude compare with WT (paired t test: P = 1⋅10−4). (F) Distribution of mRNA amplitudes in WT and KO mice. KO mice present decreased amplitude compare with WT (paired t test: P = 1⋅10−32). (G) Fourier transform is applied for genes belonging to the dark blue cluster in WT and red in KO mice. Distribution of amplitude density for the 24-h and 12-h harmonics are computed for WT animals. The high component of the 12-h harmonic indicates a predominantly 12-h rhythmic pre-mRNA level (paired t test: P = 2⋅10−6).
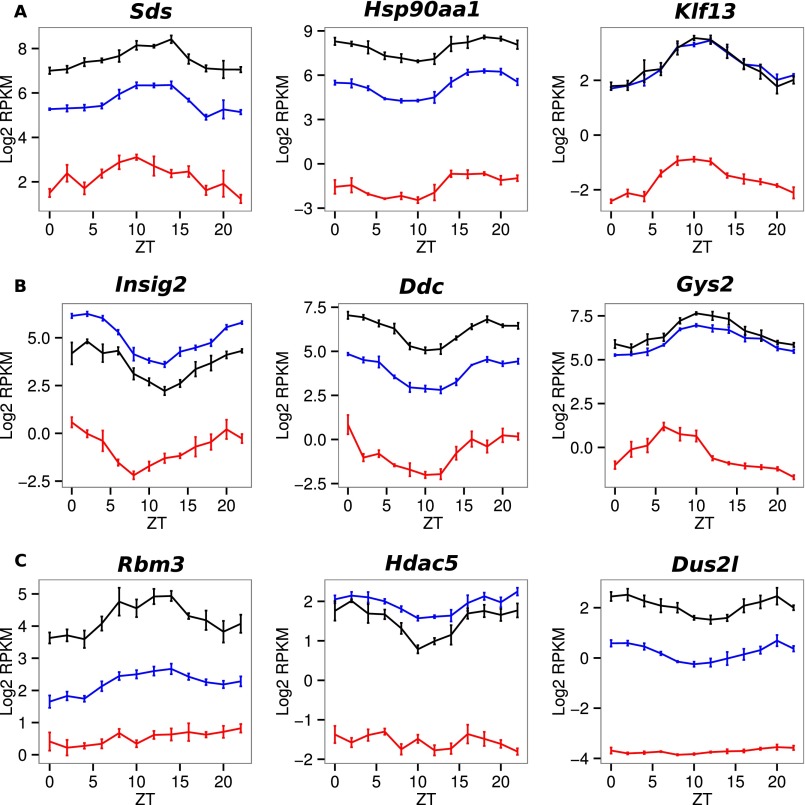
Among genes with rhythmic RFP levels that follow their corresponding mRNA accumulation, we observed three major groups with distinct transcriptional behaviors (Fig. 2D) and phase specificity (Fig. 2E). Namely, Group A is composed of genes with the same rhythmic parameters for introns and exons, introns and exons of group B genes present different rhythmic parameters, and group C genes are rhythmic only at the exonic level. Expression data for a few examples of each model are presented in Fig. S4. Surprisingly, each group shows specific gene ontology enrichment, suggesting a close relationship between biological functions, time of expression, and mode of rhythmic accumulation (Dataset S2). As described previously (25), the main difference between mRNA accumulation in groups A and B resulted from the different half-lives of these mRNAs. We used RNA-Seq exon/intron ratios as proxies for relative mRNA half-lives and found that group A genes exhibited a lower ratio than group B genes, suggesting shorter half-life (Fig. 2F). As a consequence of the increased stability of group B genes, mRNA amplitudes were reduced compared with amplitudes of pre-mRNAs (Fig. 2G). To summarize, group A genes reached their maximum expression at the light–dark transition and coded for short-lived mRNA that were mainly related to transcription and cell signaling, in particular insulin signaling. Short half-lives for translation- or signaling-related mRNA are expected to generate the required rapid responses to signals. In parallel, group B showed a bimodal distribution of mRNA phases and was mainly composed of genes with stable mRNAs that encode proteins involved in lipid and carbohydrate metabolism. Finally, rhythmic regulation for group C mRNA occurred only posttranscriptionally and concerns genes related to chromatin organization and mRNA metabolism, with sharp phase specificity at the end of the light phase.
Fig. S4.
A–C represent examples of individual profiles from genes classified in groups A, B, and C by the model selection method, respectively (Fig. 2). Individual profiles from genes classified in groups A, B, and C by model selection method (Fig. 2D). Log2 RPKM of intronic (red), exonic (blue), and RFP (black) signals are represented for genes involved in enriched biological processes identified by GO analysis (Dataset S2).
Bmal1 Deletion Affects Rhythmic mRNA Accumulation Transcriptionally and Posttranscriptionally.
To decipher the specific roles of the circadian clock and feeding rhythms on transcriptional regulation, we performed the same experiment on Bmal1 KO mice under RF (Dataset S1). To compare rhythmic pattern of genes in the two different animal models at the pre-mRNA and mRNA levels, we grouped genes into six groups according to the respective rhythmic pattern at the transcriptional and posttranscriptional levels (Fig. 3). These respective groups are composed of genes with constant pre-mRNA and mRNA (light blue), constant pre-mRNA and rhythmic mRNA (dark blue), rhythmic pre-mRNA and constant mRNA (orange), rhythmic with the same rhythmic parameters pre-mRNA and mRNA (red), rhythmic pre-mRNA and mRNA with different rhythmic parameters (brown), and finally genes with very low expression levels or noisy transcription and accumulation (gray) that cannot be included in any other category (Fig. 3A). It seems that rhythmicity is globally preserved with only moderate increase of temporally constant genes at the mRNA level in KO (light blue and orange groups, Fig. 3A). However, we observed a decreased number of genes in KO for which rhythmicity in WT is caused by posttranscriptional regulations (dark blue and brown groups, Fig. 3A), suggesting that Bmal1 deletion also affects posttranscriptional regulation. To compare how the rhythmic pattern of each gene changed between WT and KO animals, we studied their distribution among the five different groups reporting interesting patterns (Fig. 3B). Although a majority of genes remained in the same group as in WT, there was a clear spreading of the distribution of genes among the groups, indicating that Bmal1 deletion affected the general rhythmic pattern of genes. In addition, the expression phase of both pre-mRNA and mRNA was shifted in the KO compared with WT. However, this occurred in both directions, without clear further specificity (Fig. 3 C and D). Interestingly, for genes showing rhythmic patterns of expression in both WT and KO, the amplitudes of such rhythms at the mRNA level were, on average, clearly decreased in KO, whereas the amplitude of pre-mRNA was slightly increased (Fig. 3 E and F).
Concerning the change of pattern of rhythmic gene expression between WT and KO animals, one group was of particular interest, namely genes that were nonrhythmic at the pre-mRNA and rhythmic at the mRNA levels in WT (dark blue group) and that became rhythmic at both levels in the KO (red group). This group of genes was the only group whose pattern was severely perturbed in the KO (Fig. 3B). Interestingly, these genes tended to show transcription (pre-mRNA) rhythms with a period of at 12 h in WT (Fig. 3G) that morphed onto 24 h rhythms in the KO. It was previously described that 12-h-period genes changed to 24-h period in circadian clock-deficient mice (26, 27). It is likely that the two peaks of the 12-h rhythm are regulated by the circadian clock and feeding rhythms, respectively, and that the circadian component disappeared in the KO (27). Thus, the change of rhythmic pattern in this group reflects such a change of periodicity.
Translation Efficiency Is Regulated During the Diurnal Cycle for Genes with 5′-Terminal Oligo Pyrimidine Tract and Translation Initiator of Short 5′-UTR Motifs.
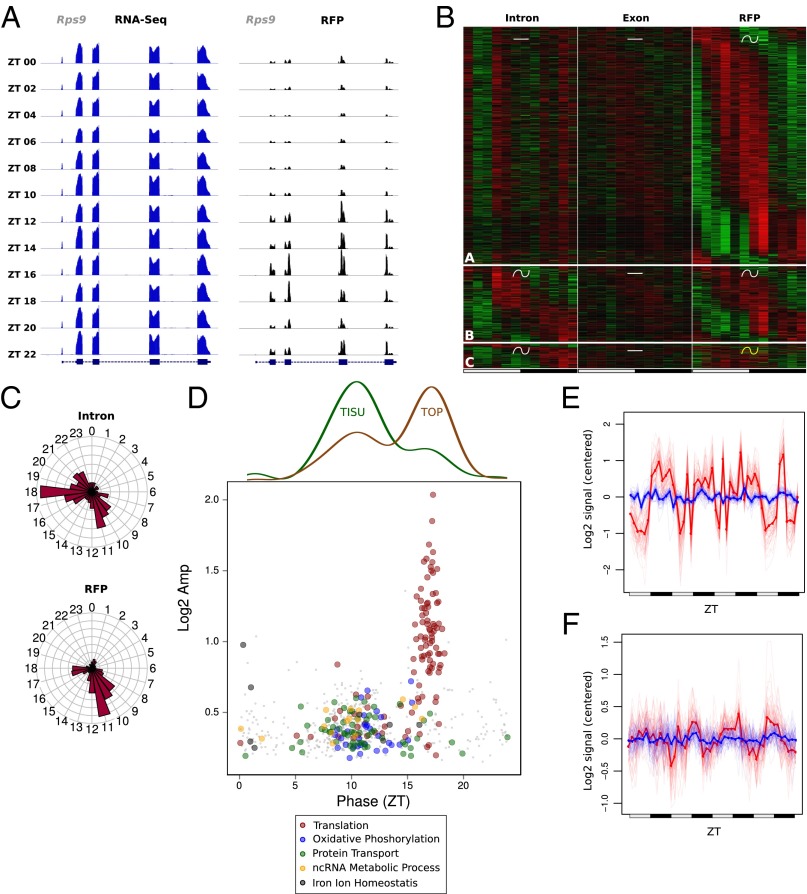
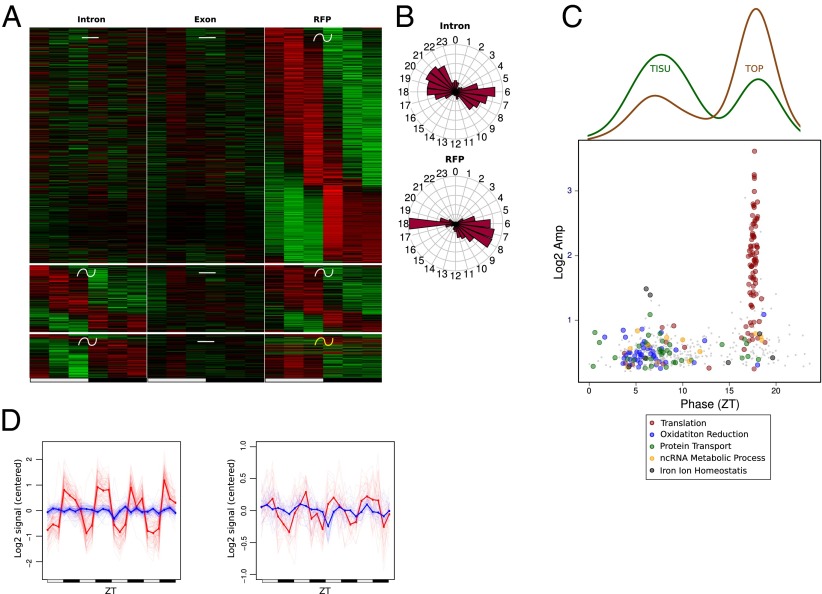
Although translation of most genes followed mRNA accumulation (Fig. 2), we identified a subset of genes exhibiting rhythmic translation efficiency in WT mice under the ALF condition. Indeed, these genes showed rhythmic translation, but their mRNA accumulated at constant levels (Fig. 2C). As we recently reported, ribosomal proteins (RPs) are translated in a rhythmic manner from constantly accumulated mRNA (12). We confirmed here that translation of RPs was regulated diurnally, as shown for Rps9 (Fig. 4A). Although some of the genes with rhythmic translation efficiency exhibited rhythmic transcription, the vast majority showed constant transcription (Fig. 4B and Dataset S1). The genes with rhythmic translation efficiency showed a bimodal phase of translation and encompassed two different main functions: The genes translated during the day at around ZT 10 are related to mitochondrial functions, whereas those translated at night around ZT 17 are related to the translation machinery, including RPs (Fig. 4C and Dataset S2). These functional clusters were enriched in genes harboring a TISU or 5′-TOP elements, respectively (Fig. 4D and Fig. S5). TISU elements have a conserved translation initiation sequence preceded by an extremely short 5′-UTR, and direct efficient cap-dependent translation initiation without scanning, thus conferring an advantageous mechanism under an inhibited translation state, as induced by nutrient starvation (28–30). However, target of rapamycin complex 1 (TORC1) has been shown to be the main regulator of TOP mRNA translation through the phosphorylation of 4E-BP and S6K proteins under nutrient availability conditions (31). Consistent with these observations, translation of TOP mRNAs occurs at the same phase as TORC1 activation (Fig. 4D and Fig. S6), as described previously (12). In addition, we compared translationally regulated genes with a study reporting on the effect of Torin 1-induced inhibition of TORC1-dependent translation (32). This showed that the translation of TOP genes was dramatically decreased, whereas the translation of TISU genes was poorly affected (Fig. S5C). Notably, rhythmic RFP accumulation exhibited variability across individual mice (Fig. 4 E and F), reflecting individual feeding behavior in this condition (33). These findings strengthened that idea that nutrient availability may affect rhythmic translation of TOP and TISU mRNAs.
Fig. 4.
Translation efficiency is regulated during the diurnal cycle for genes with 5′-TOP and TISU motifs. We used the same rhythmic analysis as in Fig. 2. (A) Exon RNA-Seq (Left) and RFP (Right) signals for the ribosomal protein gene Rps9 show increased translation during the dark phase. (B) Expression heat map for the three genes groups showing rhythmic RFP but constant mRNA levels. (Top) Constant transcription and mRNA abundance followed by rhythmic translation. (Middle) Constant mRNA abundance with similar rhythmic parameters for transcription and translation. (Bottom) Constant mRNA abundance with differential rhythmic transcription and translation. (C) Phase distribution of intronic and RFP signals for genes in B. (D, Bottom) Log2 amplitudes and phases of RFP signal for genes in B, colored by enriched functions. (Top) Densities indicate respectively fraction of TISU and TOP motifs at each phase. (E and F) Mean of relative expression profiles across genes with TOP (E) or TISU (F) motifs in D with phase respectively between ZT15–20 and ZT7–13. Exonic signals are in blue and RFPs signals in red.
Fig. S5.
Enrichment for TOP and TISU genes in model presenting rhythmic translation efficiency. (A) Schematic representation of the 29 possible rhythmic patterns. Models 2–15 and 16–29 classify genes following a 24- or 12-h period, respectively. Depending on the pattern, red-colored waves represent different rhythmic parameters compared with the black ones. (B) Genes harboring TISU (red bars) and TOP (blue bars) motif are predominantly found in models describing rhythmic translation from constantly expressed mRNAs (models 4, 7, and 8). P values were calculated using a hypergeometric test. (C) Torin 1-induced changes in translation efficiency (32) is observed for genes with TOP (blue) and TISU (red) motifs in Fig. 4. Black line represents all genes. Genes with TOP motifs, but not TISU, are responsive to mTOR inhibition.
Fig. S6.
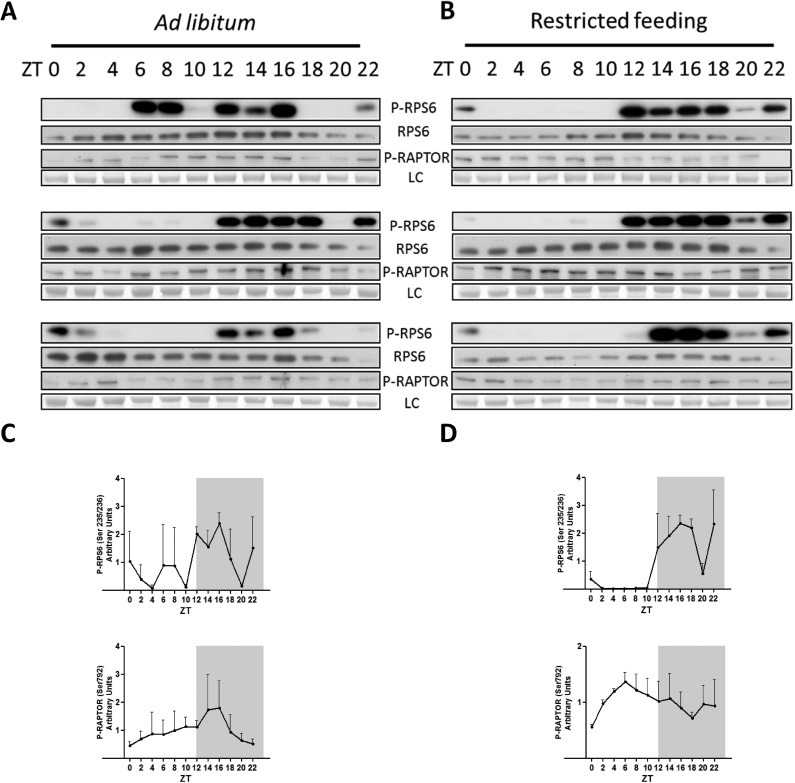
Feeding conditions differentially affect TORC1 and AMPK activities in WT mouse liver. (A) Diurnal phosphorylation profile of RPS6 (S235/236) and RAPTOR (Ser792) analyzed by Western blotting in ALF conditions. (B) Same as A in RF conditions. (C and D) Densitometry measurements of P-RPS6 (Top) and P-Raptor (Bottom) signals of A and B, respectively. Western blot analyses were normalized to the temporal means.
Rhythms in Translation Efficiency Are Sharpened Under RF.
Because nutrient availability is a key regulator of translation for both TISU and TOP genes, we wondered whether RF would affect their rhythmic translation efficiencies. Similar to the ALF condition, we observed that rhythmically translated genes were enriched in TOP and TISU genes involved in similar biological functions (Fig. 5A and Datasets S1 and S2). Interestingly, rhythmic translation of TOP and TISU genes exhibited a narrower phase with a higher amplitude compared with ALF. Rhythmic translation of mitochondrial-related TISU genes still occurred during the light phase in RF but presented an advanced phase (Fig. 5 B and C). In addition, the translational rhythms of TOP and TISU genes seemed to be reinforced by RF in the individual mice, synchronously with the availability or absence of food, respectively (Fig. 5D). TORC1 and AMP-activated protein kinase (AMPK) activities were previously shown to be synchronized to TOP and TISU translation, respectively, in response to nutrient availability (30, 31). We monitored these activities through the rhythmic phosphorylation of RPS6 and RAPTOR, showing increased coordination and amplitude under RF (Fig. S6). Taken together, these results argue in favor of a pivotal role of feeding rhythms on translation regulation of both TISU and TOP mRNAs.
Fig. 5.
Rhythms in translation efficiency are sharpened under time RF. Model selection to assess rhythmicity is applied on both WT RF datasets pooled together, and uses intronic, exonic, and RFP signals. Harmonic regression is used with a period of 24 h. Genes are assigned to one of the 15 models generated by the three conditions. An arbitrary threshold of 0.4 is set on the BIC weight. Genes with Log2 RPKM >0 at the exon and RFP levels are selected. (A) Heat map for the three genes groups showing rhythmic RFP signals and constant mRNA level under RF. (Top) Constant transcription and mRNA abundance followed by rhythmic translation. (Middle) Constant mRNA abundance with similar rhythmic parameters for transcription and translation. (Bottom) Constant mRNA abundance with differential rhythmic transcription and translation. (B) Phase distribution of intronic and RFP signals for genes in A. (C, Bottom) Log2 amplitudes and phases of RFP signal for genes in A, colored by enriched functions. (Top) Densities indicate respectively fraction of TISU and TOP motifs at each phase. (D) Mean of relative expression profile across genes with TOP (Left) or TISU (Right) motifs in B with phase respectively between ZT15–20 and ZT4–10. Exonic signals are in blue and RFPs signals in red.
Bmal1 Deletion Affects Rhythmic mRNA Translation Only for a Subset of Genes.
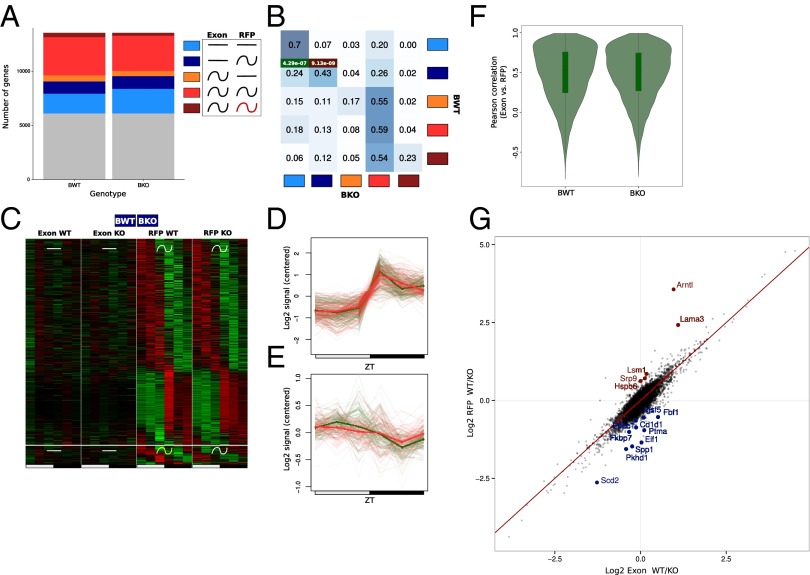
We used the same model selection method described for transcription to decipher differential translational regulation in WT and KO mice (Dataset S1). Rhythmic mRNA translation was globally conserved, although a small decrease was observed for the proportion of rhythmically translated mRNAs in the KO (Fig. 6A). Overall, genes had a tendency to follow the same pattern of rhythmic translation. We noticed, however, genes that exhibited rhythmic mRNA accumulation but constant or delayed RFP signal in WT (orange and brown groups) but synchronized mRNA and RFP signal in KO (red group) (Fig. 6B). This behavior could be hypothetically the consequence of an absence of translational regulation in KO, potentially caused by perturbed expression of RNA-binding proteins that regulate this translation.
Fig. 6.
Bmal1 deletion affects translation efficiency in a constrained group of genes. The rhythmic analysis used is the same as for Fig. 3. (A) Gray, genes not assigned in any group; light blue, constant mRNA and RFP levels; dark blue, constant mRNA and rhythmic RFP; orange, rhythmic mRNA and constant RFP; red, rhythmic mRNA and RFP; brown, rhythmic mRNA and RFP with different rhythmic parameters. (B) Fraction of genes belonging in one of the five clusters in KO compared with WT mice. Intensity of the blue color in each box represents the conservation degree of behavior of the genes between WT and KO, with darkest blue corresponding to higher conservation. Hypergeometric test for TOP and TISU enrichment in the different cluster is applied. P values for the best enrichments are displayed in green and red for TISU and TOP motifs, respectively. (C) Heat map of the exonic and RFP signal for genes harboring a constant exonic and rhythmic RFP signal in WT and KO mice. (Top) Rhythmic parameters of RFP signals are shared between WT and KO mice. (Bottom) Rhythmic parameters of RFP signals are different between WT and KO mice. (D) Mean of relative RFP profiles across genes with TOP motifs in C with phase between ZT15–20 in WT (red) and KO (green) mice. Individual genes are shown as thin lines. (E) Mean of relative RFP profiles across genes with TISU motifs in C with phase between ZT4–10 in WT (red) and KO (green) mice. Individual genes are shown as thin lines. (F) Pearson correlation between RFP and fragmented exon RNA-Seq in WT and KO test. Translation is mainly dictated by mRNA pattern in WT and KO. (G) Log2 ratio of RFP and fragmented exonic RNA-Seq signals (UMR) between WT and KO mice, taken as a measure of (relative) translation efficiency. Genes showing a significantly (false discovery rate ≤0.05) increased (logFC < −0.5) or decreased (logFC > 0.5) in translation efficiency in KO mice are colored in blue and red, respectively.
Interestingly, although the amplitudes of RFP signals of TOP genes was not affected by the Bmal1 deletion, it seemed that rhythmic translation of TISU genes showed slightly decreased amplitude (Fig. 6 D and E). Consequently, TISU genes were more enriched among the fraction of constantly translated mRNA (Fig. 6 B–E). This could be linked to the loss of rhythmic energy consumption observed in clock-disrupted animals (11). However, overall, Bmal1 deletion had only a small effect in regulating translation efficiency between WT and KO mice, with translation rhythms mostly reflecting mRNA accumulation (Fig. 6F and Dataset S1). We nevertheless identified 16 genes that showed increased or decreased translation efficiency in KO mice, with no clear evidence on the mechanism of such regulation (Fig. 6G).
Discussion
Regulation of mRNA Accumulation by Circadian and Feeding Rhythms.
We took advantage of the RNA-Seq technology to decipher the role of transcriptional and posttranscriptional regulations (17) in the establishment of the diurnally rhythmic transcriptome in mouse liver. We concluded that transcription is the main regulator of rhythmic mRNA accumulation (66% of rhythmic mRNA are rhythmically transcribed) (Fig. 2). Recent studies using different technologies to address this question yielded divergent results (Table S1). Although two studies concluded that transcriptional regulation constitutes the main origin of rhythm mRNA accumulation (ref. 34 and this study), two others concluded that this origin is mainly posttranscriptional (7, 35). Interestingly, analysis of these data through a new approach—taking into account mRNA degradation—led to the observation that ∼70% of transcripts are not subjected to posttranscriptional regulation, in accordance with our conclusion (36). At this stage, it is difficult to conclude to what extent these differences depend on the techniques used or data analysis pipelines. However, one key difference between these studies resides in the feeding conditions: night-restricted in the first case (34) and ad libitum in the second one (7, 35). Feeding rhythms play a central role in the activation of metabolically regulated pathways and transcription factors, as exemplified by the regulation of SREBP (37, 38). It is therefore possible that night-restricted feeding contributes to increased amplitude and coordination of rhythmic transcription in addition to its role in rhythmic mRNA translation for a particular subset of genes. As a consequence, RF decreased the influence of posttranscriptional regulation on mRNA accumulation compared with ALF, as suggested by the higher proportion of rhythmic mRNAs originating from rhythmic transcription. This conclusion is consistent with previous reports that night- or day-restricted feeding increases the number of cycling transcripts as well as the amplitude of their mRNA accumulation in mouse liver (10, 11).
Table S1.
Comparison of the relative importance of transcriptional and posttranscriptional regulation on rhythmic mRNA accumulation across different studies
Concerning diurnally rhythmic genes, we observed a clear relationship between the mode of rhythmic accumulation, notably related to mRNA half-life, the phase preferences, and the biological functions (Fig. 2). Correlations between mRNA stability and biological function are well-described in different animal cell models (39, 40). In particular, cells can rapidly change the levels of mRNAs coding for proteins involved in signaling and transcription, whereas longer-lived mRNAs coding for proteins involved in metabolism can contribute to damp fluctuations in the induction of such genes. In our analysis, genes showing short delay (short half-lives) between transcription and mRNA accumulation tended to code for signaling proteins linked with food response (such as insulin signaling) and were expressed at the day/night transition. Meanwhile, genes with longer delays (longer half-lives) between transcription and mRNA accumulation tended to be involved in metabolism and showed a biphasic distribution of phases with peaks at the end of the light and dark periods. These two phases could be explained by the required separation of incompatible metabolic processes (41). A last group of genes coded for proteins involved in chromatin organization and peaked at the end of the light period. Interestingly, genes involved in chromatin organization, like histones, are known to be regulated at the posttranscriptional level with a large increase in accumulation in S phase (42). Thus, it may be that the increased accumulation of these mRNA is linked to the observed rhythmic DNA synthesis that occurred in rodent liver (43, 44).
In addition, the analysis of Bmal1 KO animals allowed us to study the role of the circadian clock on rhythmic mRNA accumulation. Our results showed that Bmal1 deletion, although affecting the phases and amplitudes of many rhythmic genes, induced relatively moderate changes, which was reminiscent of previous studies showing that the ClockΔ19 mutation had a globally minor effect on rhythmic transcripts (45). More surprisingly, it seemed that Bmal1 deletion had a significant impact at the posttranscriptional regulation of mRNA levels. It was recently shown that posttranscriptional modifications involving pre-mRNA metabolism or miRNAs can influence rhythmic genes pattern (13, 46, 47). To which extent these processes are controlled by the circadian clock is presently not understood. Analysis of another genotype, the Cry1/Cry2 KO, showed that global rhythmic mRNA accumulation was strongly perturbed (11). However, these experiments were performed under ad libitum feeding and constant darkness conditions. Such conditions are very likely to induce arrhythmia in these animals (48), whereas our experiments were performed under light–dark cycles and night feeding conditions to separate feeding behavior and clock-related effects. It is thus likely that the arrhythmic feeding is responsible of the major disturbance of rhythmic mRNA accumulation observed in Cry1/Cry2 KO (11).
Mechanisms of Rhythmic Translational Regulation.
Although rhythmic coordination of transcription and mRNA accumulation has been extensively studied during the last decades, rhythmic translation has been described only recently in mammals. Indeed, it has been shown that the TORC1 pathway is rhythmically activated in mouse liver (12, 49), hippocampus (50), and SCN (51), influencing in this way rhythmic ribosome biogenesis (12) or light response and circadian behavior (52, 53). Moreover, the circadian clock, in coordination with feeding rhythms, seems to play a modulating role in this activation (12, 54, 55). Although we confirmed the rhythmic activation of the TORC1 pathway in mouse liver, it seemed that Bmal1 deletion had no effect on the rhythms of TOP genes translation; in particular, the amplitudes were indistinguishable (Fig. 6D). Even though this observation seems contradictory to our previously published results showing that rhythmic RP accumulation is perturbed in the liver of clock-disrupted mice (12), it is important to note that RP synthesis is only one part of the ribosome biogenesis. Indeed, it comprises numerous steps including rRNA synthesis and maturation and ribosome assembly that strongly influence RP stability and accumulation (56). Interestingly, recent results from our laboratory show that these steps are also rhythmic and, at least in part, under circadian clock control. It is therefore possible that these clock-regulated processes influence rhythmic RP stability and accumulation and explain this apparent discrepancy.
A recent study showed that the circadian clock can directly regulate mRNA translation through the direct interaction of BMAL1 with the translation machinery. As a consequence, total mRNA translation follows a global rhythmic pattern that is lost in the liver of Bmal1 KO mice, which also show decreased mRNA translation throughout the day (57). To date, we and others have also shown that global translation is rhythmic in mouse liver (12, 58, 59). However, the RFP experiments presented here, which measure relative translation efficiencies, highlight that diurnal relative changes in translation are predominantly regulated by transcriptional regulation and feeding rhythms. Indeed, the amplitudes associated with diurnally regulated translation efficiencies seemed to be only moderately affected by the deletion of Bmal1 (Fig. 6 D and E) and differential translation efficiency concerned only a very small number of mRNAs at the single-gene level (Fig. 6G).
One interesting finding consisted of the rhythmic regulation of TISU genes (28, 29). Recently, it was shown that these genes are protected from AMPK-induced translation inhibition (30), and the rhythmic activation of the AMPK pathway thus provides an explanation for their rhythmic translation activation (12, 60). TISU elements are predominantly found in nucleus-encoded mitochondrial protein and their translational properties affect mitochondrial activity (30). Remarkably, the TORC1 pathway was also shown to regulate translation of nucleus-encoded mitochondrial genes and mitochondrial activity (61). Incidentally, mitochondrial oxidative activity is rhythmic and under the control of the circadian clock (62–64), and this rhythmic activity is attributed in part to the clock-dependent regulation of NAD+ synthesis through the transcriptional regulation of the Nampt gene (65, 66). To what extend the rhythmic synthesis of mitochondrial proteins modulated by the circadian clock also contributes to this phenomenon remains to be demonstrated.
Consequence of Desynchronized Circadian Clock and Feeding Rhythms.
Energy balance is controlled by a complex interaction between metabolic pathways and transcriptional effectors. During the last decades, many efforts have been made to understand the transcriptional regulation orchestrated by the circadian clock and feeding cycles. For example, rhythmicity in gene expression (11) and in metabolite accumulation (33) can be partially restored in clock-deficient mice by a restricted feeding regimen. This feeding constraint consolidates rhythmicity in gene expression and metabolism and can protect against obesity in a high-fat diet regimen without reducing caloric intake (10, 67). We showed here that feeding rhythms can also sustain rhythmic posttranscriptional regulation and mRNA translation of genes coding for proteins involved in key metabolic pathways, adding a new level of regulation leading to rhythmic metabolism and physiology.
Feeding–fasting cycles seem to be the main timing cues for the liver clock, even bypassing the master clock in the brain (68, 69), which has led to the hypothesis that desynchrony of master and peripheral clocks under altered feeding conditions disrupts organisms’ homeostasis and predisposes them to chronic diseases. Recent studies linked growth and associated metabolic pathologies including metabolic syndrome and obesity to desynchronized clocks (1, 3, 6). Interestingly, many of these pathologies have also been linked to defects in ribosome biogenesis (70, 71) or mitochondrial activity (72, 73). Control of ribosome biogenesis and mitochondrial function by the circadian clock and feeding rhythms could therefore provide key factors in pathologies associated with circadian disruption. In this way, the concept of chrononutrition, in the sense of synchronizing feeding rhythms to the circadian clock-imposed rhythmic metabolism, acquires a mechanistic basis and could lead to nonpharmacological strategies to prevent metabolic diseases (74).
Materials and Methods
Animal Experiments.
All animal studies were conducted in accordance with Nestlé Animal Study Committee for animal experimentation and the regulations of the veterinary office of the Canton of Vaud. C57BL/6J mice were purchased from Charles River Laboratory. Bmal1 KO mice have been previously described (12). In all experiments, male mice between 10 and 14 wk of age were used. Mice were maintained under standard animal housing conditions, with free access to food and water, and in 12-h light/12-h dark cycles. However, for KO mice and their respective control littermates, animals were fed only at night 4 d before the organ collection to reduce effects of feeding rhythm. Parts of the collected livers were subsequently homogenized for the ribosome footprinting, and the rest of the organs were frozen in liquid nitrogen and kept at −80 °C until used for total RNA and protein extractions.
Ribosome Profiling Method.
Ribosome footprinting.
Immediately after the organ collection, livers were homogenized in lysis buffer containing 20 mM Hepes (pH 7.6), 250 mM NaCl, 10 mM MgCl2, 10 mM DTT, 20 µg/mL cycloheximide, 10 U/µL RNase inhibitor (New England Biolabs), and a protease inhibitor mixture containing 0.5 mM PMSF, 10 µg/mL aprotinin, 0.7 µg/mL pepstatin A, and 0.7 µg/mL leupeptin. The homogenates were centrifuged 10 min at 9,500 × g at 4 °C and supernatants were supplemented with 1 mg/mL heparin, 0.5% sodium deoxycholate, and 0.5% Triton ×10. Lysates were kept at −80 °C until use. Lysates were supplemented with RNase I (350 U/mg of protein; Invitrogen) and then incubated 1 h at room temperature to generate RNA footprints. Treated lysates were loaded onto sucrose gradients (7–47% wt/vol) prepared in polysome buffer containing 20 mM Hepes (pH 7.6), 100 mM KCl, 5 mM MgCl2, and 1 mM DTT. After 4.5 h of centrifugation at 130,000 × g at 4 °C, gradients were fractionated at ∼0.85 mL/min using a peristaltic pump and fractions corresponding to 40-s intervals were collected. Optic density at 260 nm of each fraction was measured to establish the monosomal profile in the gradient. Finally, fractions representing the monosomal peak were pooled and RNAs were extracted as described before (12). Briefly, fractions were precipitated by the addition of three volumes of ethanol and kept overnight at −80 °C. After 30 min of centrifugation at 5,200 × g, RNAs were extracted from the nonsoluble fraction by classic methods (75).
Size selection of RFPs.
Footprints were resolved on a 15% TBE Urea Page (Life Technologies) and gels were stained with SyberGold (Invitrogen). Using dedicated mRNA markers (76), 26- to 34-nt-long mRNAs fragments were extracted from the gels through overnight incubation into RNA gel extraction buffer [300 mM sodium acetate (pH 5.5), 1 mM EDTA, and 0.25% (wt/vol) SDS, supplemented with 2 µl of glycogen (Ambion) and 1.5 µL of GlycoBlue (Ambion)]. Footprints were recovered by overnight precipitation in isopropanol at −80 °C and dissolved in 100 μL of nuclease-free water. Purified RNAs were concentrated using the RNA Clean and Concentrator 5 kit (Zymo Research) following the recommended procedure to purify small RNA. The quality and the RNA concentration were assessed on a Bioanalyzer small RNA chip (Agilent Technologies).
RFP end modifications.
Eluted RNA (50–250 ng) was heat-denatured 3 min at 65 °C and chilled on ice. The 3′-terminal cyclophosphate resulting from the RNase I digestion was removed in a 50-μL reaction with 10 U of T4 PNK (New England Biolabs) in the absence of ATP. After 60 min of incubation at 37 °C, ATP was supplemented to a final concentration of 1 mM together with an addition of 10 U T4 PNK and the reaction was further incubated 60 min at 37 °C. RNAs were purified with the RNA Clean and Concentrator 5 kit (Zymo Research) and eluted once with 7 μL H2O.
Library generation.
Libraries were generated strictly following the Truseq Small RNA kit recommendations (Illumina). For the purpose of validation, we also tested the NEXTflex Small RNA Sequencing Kit v2 (Bioo Life Science), which uses adaptors carrying four random nucleotides at their ligated extremities, following the manufacturer’s protocol. Note that in both cases we did not directly perform the PCR amplification after the reverse transcription step (discussed below). The procedure starts with the whole RNA purified from the T4 polynucleotide kinase reaction (5 μL starting material, 25 ng to 250 ng small RNA). Only half of the ligated products was used for the reverse transcription reaction. The second half was stored at −80 °C as a backup.
Determining the optimal number of PCR cycles.
The optimal number of PCR cycles to be used was determined by a quantitative PCR (qPCR) performed on nonamplified DNA. Briefly, the nonamplified library (before the PCR step) is analyzed by qPCR using the KAPA Library Quant Kit (Kappa Biosystems), which is normally used for quantifying libraries following the final PCR amplification. No standard curve is required but the Cp (crossing point, equivalent to the threshold cycle, or Ct) obtained is corrected by a cycle correction factor (CCF) that we empirically determined after fluorimetric quantification of a final test library amplified for a known number of PCR cycles. As a result, we were able to anticipate the number of cycle that would yield the desired final concentration of each individual library. Importantly, the CCF is dependent on the qPCR cycler, qPCR mix, thermocycler in which the actual PCR amplification will be run. Its value has to be initially determined in the course of a test run but, after that, it can be considered as a constant for a given library generation protocol. Practically, the reverse transcription reaction is diluted 1/100 and the qPCR reaction assembled with 2 oligos that we specifically designed for the TruSeq Small RNA kit from Illumina (CAGAGTTCTACAGTCCGACGAT and TTGGCACCCGAGAATTCCA). The 10-μL reaction was run on a Light Cycler 480 II (Roche) according to the manufacturer’s recommendations, with 0.3 μM primers. Cycling conditions consisted of an initial denaturation step of 5 min at 95 °C followed by 40 cycles of 30 s at 95 °C and 45 s at 45 °C. The obtained Cp was first corrected for the dilution compared with the real PCR to be performed and then with the abovementioned CCF. In this experiment, we found a CCF value of +3 cycles.
Library amplification.
The PCR was designed following Illumina’s recommendation taking advantage of the indexed oligos. The calculated optimal PCR cycle number was generally homogenous among samples. In series of 48 samples, we typically found around 5 samples that had to be amplified with a different cycle numbers (± 2 cycles from the average, which is generally 10 cycles). These samples were amplified in a separate reaction.
Library postprocessing.
Libraries were purified with Ampure XP beads at a 2× ratio (cutoff below 100 bp for libraries around 150 bp) and the DNA eluted with 30 μL resuspension buffer (10 mM Tris, pH 8.0). Libraries were quantified with Picogreen (Life Technology), which usually yields 75– 200 ng DNA, and size patterns were controlled using a DNA High Sensitivity Reagent kit on a Caliper GX device (Perkin-Elmer).
Library sequencing.
Samples were pooled based on the Illumina indices used (typically 12–24 samples are pooled together). This 10 nM pool was made equimolar with individual library concentrations calculated from Picogreen data and an average peak size of 155 bp. Pools were denatured with an equivolume amount of 100 mM NaOH and neutralized with HT1 buffer (Illumina) to reach a final concentration of 20 pM. Denatured pools are spiked with 10% PhiX and loaded onto a single-read flow cell v3 at a concentration of 8 pM strictly following Illumina’s recommendations. Sequencing was performed on a HiSEq. 2500 (Illumina) for 51 cycles. Optimal sequencing depth typically required loading an equivalent of 2–10 samples per lane depending on the scope of the experiment.
Total RNA Sequencing.
Liver RNAs were extracted as previously described (25) and libraries were prepared using the TruSeq Stranded Total RNA Sample Prep Kit with the Ribo-Zero Gold depletion set (Illumina) following the manufacturer’s protocol. We used 250 ng total RNA as starting material and performed 11 PCR cycles from library amplification (optimal cycle determined with the same method as the small RNA, discussed above). Samples were pooled by 24, denatured, spiked with 3% PhiX, and loaded onto a paired-end read flow cell v3 at a final concentration of 11 pM for a paired-end 100 cycles run, strictly following Illumina’s recommendations. Sequencing depth was equivalent to three samples per lane.
All of the data are deposited in the Gene Expression Omnibus database under accession no. GSE73554.
Data Analysis.
Alignment of ribosome footprints.
We used the Ensembl Mus Musculus genome annotation (assembly GRCm38/mm10) (77). Footprint sequences were first aligned against rRNA and tRNA sequences (obtained using genome.ucsc.edu/, filtered on rRNA and tRNA) using STAR 2.3.0 (78) with options -seedSearchStartLmax 15 -clip3pAdapterSeq TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -outReadsUnmappedFastx. In this way, the STAR internal clipping function is used and reads are split into pieces no longer than 15 bp to avoid ambiguous mapping. Unmapped reads were then aligned with the same options to the reference genome. To improve accuracy on splice junctions, the index of the reference genome was built by considering the Ensembl annotation (using STAR -runMode genomeGenerate -sjdbGTFfile).
Triplet periodicity analysis in ribosome footprints.
Using a custom Perl script, 5′ ends of uniquely mapping reads (up to one mismatch) were analyzed in a window of 50 bp around the annotated translation initiation site and translation end site. The fraction of reads at every position is computed for each transcript and averaged over all annotated transcripts. This analysis was performed for the 84 samples independently.
Quantification of ribosome footprints.
For each Ensembl gene, all of the exons of the respective annotated protein-coding transcripts were considered. Reads up to one mismatch, with sizes between 29 and 35 nt, were counted considering only reads in the right orientation. Both uniquely mapping and multimapping reads were reported. Read counts were normalized by the respective library size, and by the average size of transcripts coding region. The resulting ribosome signals (in log2) are provided in Dataset S1.
NEXTFlex analysis.
Ribosome profiling reads were trimmed using fastx_clipper from fastx_toolkit with parameters -Q33 -a TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -c -l 11 -M 5. Trimmed reads were filtered for unique sequences. The output was written in separated fasta files. Both groups of files containing unique sequences or all sequences were aligned independently using the same procedure: footprint sequences were first aligned against rRNA and tRNA with the options–seedSearchStartLmax 15–clip5pNbases 4–clip3pNbases 4–outReadsUnmappedFastx. In this way, the internal trimming function in STAR is used to skip the first and last four nucleotides (representing the random barcode). Unmapped reads were then aligned with the same options to the reference genome. For the quantification of ribosome footprints, our standard pipeline was used.
Mapping of total RNA-Seq.
Paired end reads were aligned to the same genome index than for the RFP using STAR.
Quantification of exonic and intronic levels for each gene.
Considering all Ensembl transcripts annotated to a particular gene, we first defined exonic and intronic regions. A region was defined as exonic if it occurs in at least one of the transcripts. Meanwhile, intronic regions had to be shared between all of the transcripts, to maximize specificity of the intronic read counts. Using a custom Perl script, we assigned uniquely mapping paired-end reads to exonic regions (in both cases of exon–exon junctions or read pairs entirely contained in one exon), or intronic regions (in both cases of intron–exon reads pairs, and ones entirely contained in one intron), always considering reads orientation. To obtain proxies for the relative concentrations of exonic and intronic gene products, read counts were thereafter normalized by the total library sizes, as well as by the total sizes of the respective intronic or exonic regions. Log2 expression levels were calculated.
5′-TOP analysis.
A transcriptional start site sequencing (TSS-Seq) dataset from embryonic (17 d) mouse was downloaded from DBTSS (version 10) (79). TSS clusters were filtered for “confident” tag pattern, “NM_correspond” tag type, and a minimum of 10 tags. The field “Represent TSS position” was used to define the genomic position of each TSS. For each TSS, we searched in the region [−5,+20] for a pyrimidine repeat of at least 5 nt starting before the 8th nt from the TSS and with a ratio of [T] and [C] less than 0.8. Enrichment was performed using a hypergeometric test.
TISU analysis.
TSSs are defined as for the 5′-TOP analysis. We scanned, using fuzznuc (80), the region [−5,+45] around each TSS for the consensus sequence “[CG]AA[CG]ATGGCGG,” allowing one or two mismatches. Enrichment was performed using a hypergeometric test.
Gene ontology analysis.
Gene ontology (GO) analysis was performed using RDAVIDWebService (81). Enrichment test was performed for Biological Process (BP_all) and KEGG pathway annotation. Categories with adjusted P values (Benjamini–Hochberg) smaller than 0.05 and a total population count between 5 and 700 were reported.
Translation efficiency.
The ratio between RFP and exonic RNA-Seq expression levels can be used as a proxy for translation efficiency. To avoid possible bias generated by our experimental design (single-end RFP reads versus paired-ends RNA-Seq reads), RNA-Seq sequences were fragmented in silico using a custom Perl script: For each first read in the pair, a substring with a size sampled from a normal distribution (mean = 33, SD = 2) was extracted starting at a random position. The resulting fragmented RNA-Seq (FRS) reads were then aligned and quantified using the same pipeline as for the ribosome footprints (except for the adapter trimming option). Changes in translation efficiency between WT and Bmal1 KO conditions were computed using the edgeR (82) generalized linear model framework. RFP and FRS counts (unique mapping reads) were normalized using edgeR. To estimate differential translation efficiency in edgeR, conditional (WT, KO) and experimental (RFP, FRS) factors were introduced together with an interaction term, whose significance is assessed with a likelihood ratio test. Genes with FDR smaller than 0.05 and absolute Log2 fold change larger than 0.5 were analyzed.
Rhythmicity analysis.
For ALF (2-h resolution) and RF (4-h resolution) conditions, we focused on 24 h/12 h and 24 h periodicity, respectively, using harmonic regression on the log2 transformed signals . We used the relation + noise, where µ is the mean and α and β are the coefficients of cosine and sine functions with period T. To facilitate the comparison of rhythmicity between different types of signals (RFP or RNA-Seq) and conditions (e.g., genotypes or feeding), we used a model selection approach. In the case of N conditions, we allowed α and β to be either zero (no rhythm) or nonzero (rhythms), and, moreover, α and β could be shared between any subset of the N conditions. In short, a gene could thus be flat or rhythmic and share its phase and amplitude with any other conditions. The number of resulting models for N conditions is given by the Bell number BN+1 (83). Each model is solved using standard linear regression and the Bayesian information criterion (BIC) is used to control for model complexity (84). , with RSS the sum of residuals square of the multilinear regression, n the number of time points, and k the numbers of parameters. Confidence in the models can be addressed using Schwarz weights: where , with m* the minimum BIC value in the set of BN+1 models. can be interpreted as the probability that j is the optimal model (in the BIC sense) given the data and the set of candidate models.
Total Protein Extraction and Analysis.
Frozen livers were homogenized in lysis buffer containing 20 mM Hepes (pH 7.6), 100 mM KCl, 0.1 mM EDTA, 1 mM NaF, 1 mM sodium orthovanadate, 1% Triton X-100, 0.5% Nonidet P-40, 0.15 mM spermin, 0.5 mM spermidine, 1 mM DTT, and a protease inhibitor mixture. After incubation 30 min on ice, extracts were centrifuged 10 min at 21,000 × g and the supernatants were harvested to obtain total extracts. Twenty micrograms of extract were used for Western blotting. After migration, proteins were transferred to PVDF membranes and Western blotting was realized according to standard procedures using antibodies (Cell Signaling) raised against P-RPS6 (2211), RPS6 (2117), and P-RAPTOR (2083) proteins.
Supplementary Material
Acknowledgments
This research was supported by Swiss National Science Foundation Grant 31-130714 (to F.N.), the Ecole Polytechnique Fédérale de Lausanne, European Research Council Starting Grants ERC-2010-StG-260988 (to F.G.) and ERC-2010-StG-260667 (to F.N.), and the Leenaards Foundation (F.G. and F.N.).
Footnotes
Conflict of interest statement: F.A., C.G., J.M., E.M., B.W., G.L., P.D., and F.G. are employees of Nestlé Institute of Health Sciences SA.
This article is a PNAS Direct Submission. P.E. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE73554).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515308112/-/DCSupplemental.
References
- 1.Dibner C, Schibler U. Circadian timing of metabolism in animal models and humans. J Intern Med. 2015;277(5):513–527. doi: 10.1111/joim.12347. [DOI] [PubMed] [Google Scholar]
- 2.Gerhart-Hines Z, Lazar MA. Circadian metabolism in the light of evolution. Endocr Rev. 2015;36(3):289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron KG, Reid KJ. Circadian misalignment and health. Int Rev Psychiatry. 2014;26(2):139–154. doi: 10.3109/09540261.2014.911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vyas MV, et al. Shift work and vascular events: Systematic review and meta-analysis. BMJ. 2012;345:e4800. doi: 10.1136/bmj.e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane BR, Young MW. Interactive features of proteins composing eukaryotic circadian clocks. Annu Rev Biochem. 2014;83(1):191–219. doi: 10.1146/annurev-biochem-060713-035644. [DOI] [PubMed] [Google Scholar]
- 6.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161(1):84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16(6):833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jouffe C, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11(1):e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26(24):2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauvoisin D, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci USA. 2014;111(1):167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingolia NT. Ribosome profiling: New views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15(3):205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 16.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33(7):722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- 18.Howard MT, Carlson BA, Anderson CB, Hatfield DL. Translational redefinition of UGA codons is regulated by selenium availability. J Biol Chem. 2013;288(27):19401–19413. doi: 10.1074/jbc.M113.481051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connor P, Andreev D, Baranov P. 2015. Surveying the relative impact of mRNA features on local ribosome profiling read density in 28 datasets, dx.doi.org/10.1101/018762.
- 20.Gao X, et al. Quantitative profiling of initiating ribosomes in vivo. Nat Methods. 2015;12(2):147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aksenova MV, Aksenov MY, Carney JM, Butterfield DA. Protein oxidation and enzyme activity decline in old brown Norway rats are reduced by dietary restriction. Mech Ageing Dev. 1998;100(2):157–168. doi: 10.1016/s0047-6374(97)00133-4. [DOI] [PubMed] [Google Scholar]
- 23.Cui ZJ, Han ZQ, Li ZY. Modulating protein activity and cellular function by methionine residue oxidation. Amino Acids. 2012;43(2):505–517. doi: 10.1007/s00726-011-1175-9. [DOI] [PubMed] [Google Scholar]
- 24.Floyd RA. Role of oxygen free radicals in carcinogenesis and brain ischemia. FASEB J. 1990;4(9):2587–2597. [PubMed] [Google Scholar]
- 25.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4(1):25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11(1):47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Hughes ME, et al. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 2012;8(7):e1002835. doi: 10.1371/journal.pgen.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elfakess R, Dikstein R. A translation initiation element specific to mRNAs with very short 5'UTR that also regulates transcription. PLoS One. 2008;3(8):e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elfakess R, et al. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011;39(17):7598–7609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinvani H, et al. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015;21(3):479–492. doi: 10.1016/j.cmet.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849(7):801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 32.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adamovich Y, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19(2):319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Martelot G, et al. CycliX Consortium Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10(11):e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lück S, Thurley K, Thaben PF, Westermark PO. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Reports. 2014;9(2):741–751. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 37.Brewer M, Lange D, Baler R, Anzulovich A. SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. J Biol Rhythms. 2005;20(3):195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- 38.Gilardi F, et al. CycliX Consortium Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10(3):e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharova LV, et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16(1):45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang E, et al. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003;13(8):1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma. 2004;113(3):103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 42.Marzluff WF, Duronio RJ. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14(6):692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 43.Barbiroli B, Potter VR. DNA synthesis and interaction between controlled feeding schedules and partial hepatectomy in rats. Science. 1971;172(3984):738–741. doi: 10.1126/science.172.3984.738. [DOI] [PubMed] [Google Scholar]
- 44.Barnum CP, Jardetzky CD, Halberg F. Time relations among metabolic and morphologic 24-hour changes in mouse liver. Am J Physiol. 1958;195(2):301–310. doi: 10.1152/ajplegacy.1958.195.2.301. [DOI] [PubMed] [Google Scholar]
- 45.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du N-H, Arpat AB, De Matos M, Gatfield D. MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. eLife. 2014;3:e02510. doi: 10.7554/eLife.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fustin J-M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 48.van der Horst GTJ, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 49.Cornu M, et al. Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci USA. 2014;111(32):11592–11599. doi: 10.1073/pnas.1412047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saraf A, Luo J, Morris DR, Storm DR. Phosphorylation of eukaryotic translation initiation factor 4E and eukaryotic translation initiation factor 4E-binding protein (4EBP) and their upstream signaling components undergo diurnal oscillation in the mouse hippocampus: Implications for memory persistence. J Biol Chem. 2014;289(29):20129–20138. doi: 10.1074/jbc.M114.552638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181(0):79–88. doi: 10.1016/j.neuroscience.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao R, et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79(4):712–724. doi: 10.1016/j.neuron.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J Neurosci. 2010;30(18):6302–6314. doi: 10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khapre RV, et al. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany, NY) 2014;6(1):48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khapre RV, et al. Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany, NY) 2014;6(8):675–689. doi: 10.18632/aging.100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woolford JL, Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipton JO, et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161(5):1138–1151. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fishman B, Wurtman RJ, Munro HN. Daily rhythms in hepatic polysome profiles and tyrosine transaminase activity: Role of dietary protein. Proc Natl Acad Sci USA. 1969;64(2):677–682. doi: 10.1073/pnas.64.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uchiyama Y, Asari A. A morphometric study of the variations in subcellular structures of rat hepatocytes during 24 hours. Cell Tissue Res. 1984;236(2):305–315. doi: 10.1007/BF00214231. [DOI] [PubMed] [Google Scholar]
- 60.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita M, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18(5):698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Masri S, et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA. 2013;110(9):3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peek CB, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342(6158):1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohsaka A, et al. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One. 2014;9(11):e112811. doi: 10.1371/journal.pone.0112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaix A, Zarrinpar A, Miu P, Panda S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20(6):991–1005. doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stokkan K-A, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 70.Chaillou T, Kirby TJ, McCarthy JJ. Ribosome biogenesis: Emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol. 2014;229(11):1584–1594. doi: 10.1002/jcp.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23(1):63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Nasrallah CM, Horvath TL. Mitochondrial dynamics in the central regulation of metabolism. Nat Rev Endocrinol. 2014;10(11):650–658. doi: 10.1038/nrendo.2014.160. [DOI] [PubMed] [Google Scholar]
- 73.Roy M, Reddy PH, Iijima M, Sesaki H. Mitochondrial division and fusion in metabolism. Curr Opin Cell Biol. 2015;33:111–118. doi: 10.1016/j.ceb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oike H, Oishi K, Kobori M. Nutrients, clock genes, and chrononutrition. Curr Nutr Rep. 2014;3(3):204–212. doi: 10.1007/s13668-014-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 76.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue) D1:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamashita R, Sugano S, Suzuki Y, Nakai K. DBTSS: DataBase of Transcriptional Start Sites progress report in 2012. Nucleic Acids Res. 2012;40(Database issue) D1:D150–D154. doi: 10.1093/nar/gkr1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 81.Jiao X, et al. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rota G-C. The number of partitions of a set. Am Math Mon. 1964;71(5):498–504. [Google Scholar]
- 84.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90(430):773–795. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.