Significance
We screened rheumatoid arthritis (RA)-associated copy number variations (CNVs) across the whole genome and identified significant deletion variants encompassing leukocyte-specific protein 1 (LSP1) gene. Functional assays revealed that LSP1, induced by T-cell receptor activation, negatively regulates T-cell migration. Loss of Lsp1 promotes T-cell migration into antigen-instilled tissues and draining lymph nodes in mice with T-cell–dependent chronic inflammation. Moreover, patients with RA show diminished expression of LSP1 in peripheral T cells with increased migratory capacity. To our knowledge, our work is the first to demonstrate how CNVs result in immune dysfunction and a disease phenotype, highlighting the importance of LSP1 CNVs and LSP1 insufficiency in the pathogenesis of RA.
Keywords: leukocyte-specific protein 1, copy number variation, T-cell function, cell migration, rheumatoid arthritis
Abstract
Copy number variations (CNVs) have been implicated in human diseases. However, it remains unclear how they affect immune dysfunction and autoimmune diseases, including rheumatoid arthritis (RA). Here, we identified a novel leukocyte-specific protein 1 (LSP1) deletion variant for RA susceptibility located in 11p15.5. We replicated that the copy number of LSP1 gene is significantly lower in patients with RA, which correlates positively with LSP1 protein expression levels. Differentially expressed genes in Lsp1-deficient primary T cells represent cell motility and immune and cytokine responses. Functional assays demonstrated that LSP1, induced by T-cell receptor activation, negatively regulates T-cell migration by reducing ERK activation in vitro. In mice with T-cell–dependent chronic inflammation, loss of Lsp1 promotes migration of T cells into the target tissues as well as draining lymph nodes, exacerbating disease severity. Moreover, patients with RA show diminished expression of LSP1 in peripheral T cells with increased migratory capacity, suggesting that the defect in LSP1 signaling lowers the threshold for T-cell activation. To our knowledge, our work is the first to demonstrate how CNVs result in immune dysfunction and a disease phenotype. Particularly, our data highlight the importance of LSP1 CNVs and LSP1 insufficiency in the pathogenesis of RA and provide previously unidentified insights into the mechanisms underlying T-cell migration toward the inflamed synovium in RA.
Cell migration plays a central role in maintaining homeostasis and coping with a wide spectrum of perturbing stimuli for multicellular organisms. Wound healing involves the migration of several cell types, and the migration of leukocytes into lymph nodes and inflamed tissue is required for the development of immune responses (1). Moreover, excessive and uncontrolled infiltration of distinct effector leukocytes into particular organs or tissue components is a characteristic pathology found in various chronic inflammatory diseases including psoriasis, Crohn’s disease, ulcerative colitis, multiple sclerosis, asthma, atherosclerosis, and rheumatoid arthritis (RA) (1, 2).
RA is an autoimmune disorder that engages an uncontrolled influx of inflammatory cells to the joints, leading to persistent synovitis and tissue destruction (3). T cells, as one of the most abundant cell population in the RA synovium, are aberrantly activated in RA to drive chronic inflammation and joint destruction (4). RA T cells interact with other immune and resident cells, including B cells, macrophages, synoviocytes, and osteoclasts by secreting a variety of cytokines and chemokines and/or by direct cell-to-cell contact, and ultimately boost their proinflammatory action (5). The role that diverse T-cell populations play in the induction, amplification, and maintenance of inflammatory arthritis has been elucidated in various animal models of RA (6). Abnormal activation of RA T cells is associated with abnormal T-cell receptor (TCR) activation and the Ca2+ signaling pathway (7, 8). Successful outcomes for patients with RA treated with T-cell regulators, including abatacept (CTLA4-Ig) (9), highlight the importance of activated T cells in the progression of RA.
The pathologic phenotype of cellular components of a certain disease depends on the quantitative and/or qualitative abnormalities of disease-associated proteins, which might be caused by a perturbation of fundamental regulatory mechanisms, including transcription, RNA processing, and mRNA degradation and translation, in addition to genetic alterations (10). An important causal link between genomic variation and phenotypic difference includes SNPs and DNA copy number variations (CNVs). Through genome-wide association studies (GWASs), a number of non-MHC genes that potentially contribute to RA susceptibility have been identified (11). However, the majority of SNPs have modest effects and do not represent the full spectrum of genetic variations. Recently, it has been suggested that CNVs are an important source of human genetic variation—in some analyses potentially as important as SNPs (12). CNV of individual genes can result in cellular and organismal abnormalities, and cumulative effects of CNVs underlie many human diseases, including autoimmune diseases (12). A few candidate CNVs for RA susceptibility, such as CCL3L1 and FCGR3B, have been suggested (13, 14), but they have not been successfully replicated or functionally validated, suggesting that other CNVs may be yet found that significantly contribute to the overall risk model.
T-cell infiltration into the synovial compartment is an essential step for the progression of RA. T-cell accumulation primarily reflects migration rather than local proliferation (3). Therefore, regulatory mechanism of T-cell trafficking into the synovium has been focused mainly on endothelial activation in synovial microvessels, which increases the expression of adhesion molecules and chemokines. However, the intrinsic migratory mechanism of T cells and its alteration in patients with RA has garnered less attention (15). Given the importance of genetic elements in determining pathologic phenotype (16), it is necessary to explore the impact of genetic variations, such as CNV, on immune dysfunction (e.g., T-cell activation) to better understand the susceptibility and pathogenesis of RA. For this goal, we screened RA-associated CNVs across the whole genome in 500 subjects and validated them in 1,565 Korean subjects and 423 white subjects.
Results
Loss of LSP1 Gene Is Significantly Associated with RA Susceptibility.
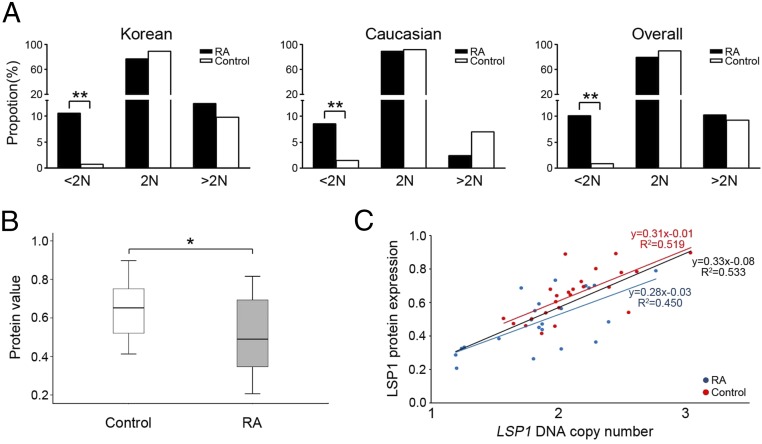
We performed a GWAS to identify RA-associated CNVs in the Korean population. Ultimately, a total of 31,373 CNVs were identified from 500 samples (100 patients with RA and 400 healthy individuals; SI Appendix, Table S1). The mean and median numbers of CNVs identified per individual genome were 62.7 and 45, respectively (range, 16-1,021), and the median size of CNVs was 18.6 kb (range, 248 bp to 8.6 Mb). Based on the CNVs, we defined 3,936 CNV regions (CNVRs) as described elsewhere (17). Using the 3,936 CNVRs, we performed logistic regression analysis after adjusting for the effects of age and sex, and seven CNVRs were found to be significantly associated with the risk of RA (false discovery rate < 0.01; SI Appendix, Table S2). There are three protein-coding genes (LSP1, TNNT3, and UGT2B28) in the seven significant CNVRs. Among them, leukocyte-specific protein 1 (LSP1) gene, located in the deletion CNVR in 11p15.5, is a specific leukocyte marker (18) with a demonstrated role in acute inflammation (19, 20). Therefore, we selected the deletion CNVR in 11p15.5, where the LSP1 gene is located, as a novel target gene for RA susceptibility, and performed independent replication and functional analyses. To this end, target-specific genomic quantitative PCR (qPCR) for the LSP1 gene was performed in a larger Korean cohort group (n = 1,565) for independent replication: 599 patients with RA and 966 healthy control individuals (SI Appendix, Table S1). We also performed the same replication in a white cohort group (n = 423, 165 patients with RA and 258 healthy individuals). Details of the study subjects, defining CNVRs, and qPCR for genomic DNA are described in Materials and Methods and in SI Appendix, Materials and Methods. As expected, the proportion of individuals with fewer than two copies of the LSP1 gene was significantly higher in patients with RA (10.5%, 63 of 599) than in controls [0.7%, 7 of 966; odds ratio (OR) = 16.1, 95% CI = 7.3–35.4; P = 3.68 × 10−20) in the Korean cohort (Fig. 1A). The proportion of the LSP1 deletion variants in the white cohort was consistent with the profile in the Korean cohort: 8.5% (14 of 165) in patients with RA vs. 1.6% (4 of 258) in controls (OR = 5.9, 95% CI = 1.9–18.2; P = 8.59 × 10−4). When we merged the two replication sets together, the significance became higher: 10.1% (77 of 764) in patients with RA vs. 0.9% (11 of 1,224) in controls (OR = 12.4, 95% CI = 6.5–23.4; P = 2.25 × 10−22). After adjusting for the effects of age and sex by logistic regression, the individuals with fewer than two copies had a significantly higher risk of RA than the individuals with two or more copies (OR = 18.9, 95% CI = 8.4–42.5, P = 1.10 × 10−12).
Fig. 1.
LSP1 copy number and LSP1 expression profiles in patients with RA. (A) Frequency distribution of LSP1 genomic copy number. (Left) Distribution patterns of LSP1 copy numbers in patients with RA (solid bar; n = 599) and normal controls (open bar; n = 966) from Korea. (Middle) Distribution patterns of LSP1 copy numbers in white patients with RA (solid bar; n = 165) and normal controls (open bar; n = 258) from the United States. (Right) Overall distribution patterns of LSP1 copy numbers from Korea and the United States (*P < 0.05 and **P < 0.001). (B) LSP1 expression in PBMCs of patients with RA and healthy controls. A ratio of LSP1 protein expression relative to GAPDH protein (LSP1/GAPDH) was measured by Western blot analysis. (C) Correlation between LSP1 protein expression level and the copy number status of LSP1 gene in PBMCs of RA and control groups.
We also compared the LSP1 protein expression level in peripheral blood mononuclear cells (PBMCs) between patients with RA and controls. For this analysis, 22 patients with RA and 24 healthy controls, whose PBMCs were available for Western blotting, were examined. As in the LSP1 CNVR association, the LSP1 protein level was significantly lower in the RA group than in the control group (0.52 ± 0.19 vs. 0.65 ± 0.14; P = 0.01; Fig. 1B). Moreover, a positive correlation between LSP1 protein expression level and the copy number status of LSP1 was observed in patients with RA and controls (R2 = 0.450, P = 0.004 in RA; R2 = 0.519, P = 6.0 × 10−6 in controls; and R2 = 0.533, P = 3.9 × 10−8 in total by Spearman rank test; Fig. 1C).
LSP1 Is Increased in Primary Human T Cells by TCR Triggering.
LSP1 is an intracellular Ca2+ and F-actin binding protein (21) that regulates murine neutrophil migration and chemotaxis (22). However, its expression and function in autoreactive T cells, including RA T cells, remain unaddressed. Based on our finding that LSP1 copy number negatively correlated with RA risk but positively correlated with LSP1 expression, we postulated that LSP1 is a negative regulator of RA development and sought to ascertain the clinical and functional relevance of altered LSP1 expression to development and progression of RA. As a preliminary experiment, we assayed the expression of LSP1 in normal PBMCs and Jurkat T cells by flow cytometry. As seen in SI Appendix, Fig. S1A, LSP1 was expressed in CD4+ T cells, CD8+ T cells, and monocytes. LSP1 also was expressed in Jurkat T cells. Western blot analysis revealed LSP1 protein with a molecular mass of 52 kDa at high levels in primary human T cells, confirming the presence of full-length LSP1 (SI Appendix, Fig. S1A, Inset).
Activated T cells play central roles in the initiation and perpetuation of RA (3, 4). We next determined which types of T-cell activators induce LSP1 expression. It is well known that TCR signaling is triggered by phytohemagglutinin (PHA) or anti-CD3/CD28 Ab (23). We investigated if these stimuli affect LSP1 expression in T cells. As shown in SI Appendix, Fig. S1B, stimulation of T cells with anti-CD3/CD28 Ab or PHA increased LSP1 expression in CD4+ and CD8+ T cells of healthy subjects, indicating that LSP1 can be induced by TCR stimulation. Additionally, phorbol myristate acetate (PMA) plus ionomycin, a Ca2+ activator, up-regulated LSP1 expression in normal human T cells as well as Jurkat T cells (SI Appendix, Fig. S1 B, D, and E), suggesting that Ca2+ signaling is required for LSP1 up-regulation in primary T cells, which is in accord with previous reports (24, 25). Another Ca2+ activator, TNF-α, also time-dependently increased LSP1 expression in Jurkat cells (SI Appendix, Fig. S1E).
To test if TCR triggering by a specific antigen increases LSP1 expression in vivo, we determined the LSP1 protein expression level in a mouse model of collagen-induced arthritis (CIA), a classic model of experimental arthritis mediated by antigen-activated T cells (26). As expected, LSP1 expression was higher in CD4+ and CD8+ T cells in draining lymph node of mice with CIA than in those of nonarthritic normal mice (SI Appendix, Fig. S1F). Taken together, LSP1 is expressed in primary T cells and Jurkat T cells, and its expression is up-regulated by TCR ligation or intracellular Ca2+ activation.
LSP1 Negatively Regulates T-Cell Migration.
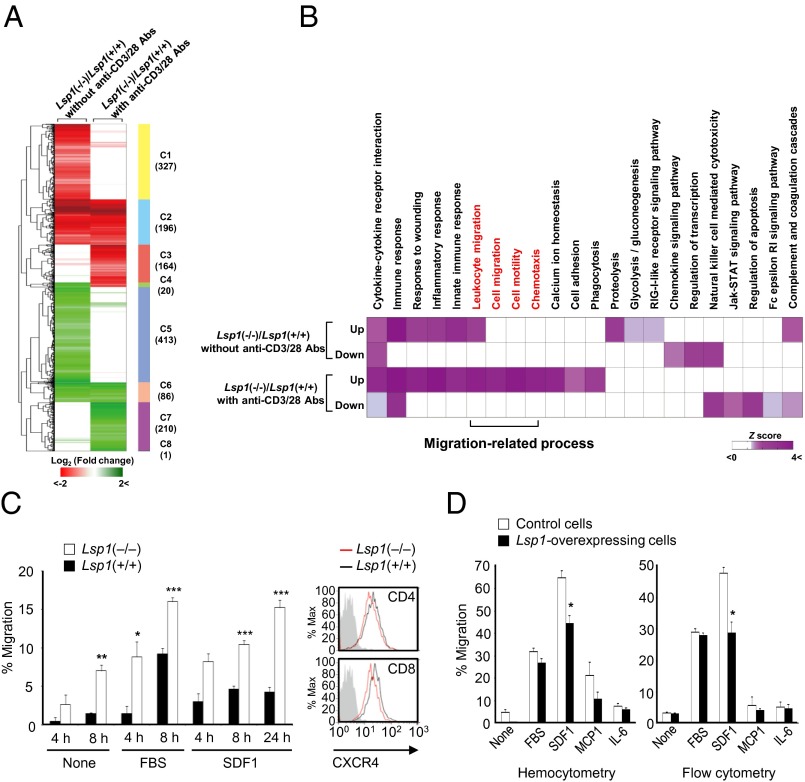
We next investigated the function of LSP1 in T-cell biology. For an unbiased and systematic analysis of LSP1 effect on T cells, we performed global transcriptome profiling of T cells obtained from Lsp1-deficient [i.e., Lsp1(−/−)] and WT mice in the presence or absence of anti-CD3/CD28 Abs. By using an integrative statistical method reported previously (27), we identified 1,043 and 677 differentially expressed genes (DEGs) in the two comparisons: Lsp1(−/−) vs. Lsp1(+/+) T cells without and with anti-CD3/CD28 Abs (Fig. 2A and SI Appendix, Table S3 and Dataset S1). A substantial proportion of the DEGs (303 genes; 21.4%) were shared in the two sets of DEGs (SI Appendix, Fig. S2A). To investigate the cellular processes represented by the DEGs, we performed functional enrichment analysis for the up- and down-regulated DEGs using Database for Annotation, Visualization, and Integrated Discovery (DAVID) software (28). As a result, we found that the up-regulated genes under TCR-activated conditions were predominantly associated with migration-related processes, including leukocyte migration, cell migration/motility, and chemotaxis (Fig. 2B and SI Appendix, Table S4). Other processes governed by LSP1 include cytokine–cytokine receptor interaction, response to wounds, innate immune response, and Ca2+ ion homeostasis (SI Appendix, Table S4).
Fig. 2.
LSP1 inhibition of T-cell migration. (A) Hierarchical clustering of DEGs in the two comparisons: Lsp1(−/−)/Lsp1(+/+) without anti-CD3/28 Abs (first column) or Lsp1(−/−)/Lsp1(+/+) with anti-CD3/28 Abs (second column). After hierarchical clustering using Euclidean distance as a dissimilarity measure and complete linkage method, we identified eight clusters (C1-C8) that reflect all possible combinations of up-regulation (red) and down-regulation (green) patterns of the DEGs in the two comparisons (SI Appendix, Table S3). The numbers in parentheses show the number of DEGs in each cluster. Color bar indicates the gradient of log2 fold change. (B) Heat map shows Gene Ontology biological processes (GOBPs) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways represented by up- or down-regulated genes in Lsp1(−/−) T cells, compared with Lsp1(+/+) T cells, in the absence and presence of anti-CD3/28 Abs. Color bar indicates gradient of Z score N−1(1 − P) where P is P value computed by DAVID and N−1(⋅) is the inverse standard normal distribution. Migration-related GOBPs or KEGG pathways are labeled in red. (C) In vitro migration of CD4+ T cells (5 × 105 cells) obtained from the spleens of Lsp1-deficient (n = 5) and WT mice (n = 5). Migration assays were performed in transwell chambers in the presence or absence of SDF1α (100 ng/mL) or 10% (vol/vol) FBS in triplicate. The results are the mean ± SD. Expression of CXCR4 (a specific SDF1 receptor) determined by flow cytometry is presented on the right. Gray-colored histogram indicates isotype control. (D) Suppression of SDF1-induced T-cell migration by LSP1 overexpression. Human recombinant SDF1α (100 ng/mL), MCP-1 (100 ng/mL), IL-6 (100 ng/mL), or 10% FBS was added to the lower chamber of the transwell inserts. The Jurkat T cells (1 × 106 cells) were loaded to the upper chamber and allowed to migrate for 4 h. The number of migrated cells were counted manually (Left) or determined by flow cytometry analysis (Right).
As migration-related processes were predominantly enriched by the DEGs, the in vitro functional assay for T-cell migration was performed in transwell chambers. As shown in Fig. 2C, the migration of CD4+ T cells isolated from the splenocytes of Lsp1-deficient mice was significantly enhanced irrespective of the presence of FBS compared with that of WT littermates. T-cell migration through the bloodstream to the target tissue is driven by chemokines. For example, stromal cell-derived factor-1 (SDF1)/CXCR4 axis induces Ca2+ flux in T cells (29) and promotes T-cell migration toward the inflamed joints, thereby contributing to RA pathology (30). In this study, we found that Lsp1(−/−) T cells exhibited increased migration in response to SDF1 compared with WT T cells (Fig. 2C). This is not attributable to the receptor level because CXCR4 expression on T cells was not different between Lsp1-deficient and WT mice as determined by flow cytometry (Fig. 2C). Additionally, there was no significant difference in the production of cytokines, including IL-10, TNF-α, IFN-γ, and IL-2 by T cells upon anti-CD3/CD28 stimulation (SI Appendix, Fig. S3 A and B), indicating that these cytokines do not contribute to increased T-cell migration as a result of Lsp1 deficiency.
The CNV in patients with RA yields a less dramatic change in LSP1 expression than complete gene loss and may be more relevant to RA pathogenesis. Thus, we investigated if Lsp1 haploinsufficiency in mice also increases T-cell migration. As expected, Lsp1(+/−) T cells still showed an increased migration in response to media, 10% (vol/vol) FBS, and SDF1 compared with Lsp1(+/+) T cells (SI Appendix, Fig. S4), but its extent was attenuated compared with Lsp1(−/−) T cells. For example, compared with WT mice, T-cell migration in response to media, 10% FBS, and SDF1 was increased by 1.9, 1.6, and 2.2 fold for Lsp1(+/−) mice, respectively, but was increased by 4.4, 2.4, 3.6 fold for Lsp1(−/−) mice, respectively (Fig. 2C and SI Appendix, Fig. S4). These results indicate a gene dose effect of Lsp1 on T-cell migration, suggesting that even a 50% reduction of Lsp1 gene is sufficient to affect T-cell function.
Based on the data in murine T cells, we wanted to determine whether LSP1 overexpression regulates human T-cell migration. To this end, we transfected the LSP1 cDNA tagged with GFP into Jurkat T cells by electroporation. After stable transfection of LSP1-GFP fusion gene, LSP1 protein as well as GFP was highly detected in Jurkat cells (SI Appendix, Fig. S1C). In contrast to LSP1-deficient cells, LSP1-overexpressing Jurkat T cells showed a lesser degree of migration in response to SDF1 in transwell chambers than the control cells (Fig. 2D). However, T-cell migration stimulated with 10% FBS, MCP-1, or IL-6 was not different between LSP1-overexpressing cells and control cells. The LSP1-dependent decrease in T-cell migration was independent of cell proliferation because BrdU incorporation revealed no difference between the two types of cells. Moreover, the number of cells counted manually was not different between the two cell lines over the 4 d of culture, indicating that LSP1 is not directly involved in T-cell proliferation (SI Appendix, Fig. S3C). In parallel, IL-2 production was not different between the two cell lines stimulated with PHA or anti-CD3/CD28 Ab (SI Appendix, Fig. S3D).
LSP1 Directly Interacts with pERK to Regulate T-Cell Migration.
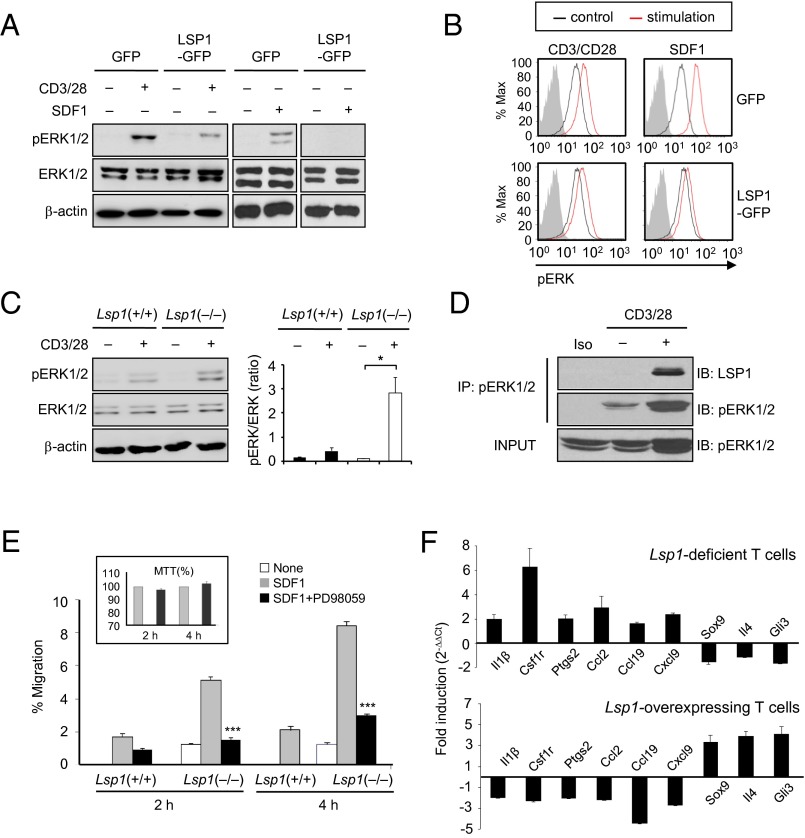
SDF1 stimulates ERK (29), which promotes T-cell migration (31). Therefore, we investigated to determine if ERK is a downstream target of LSP1 for T-cell migration. As shown in Fig. 3A, the expression level of phosphorylated ERK (pERK) was reduced in LSP1-overexpressing Jurkat T cells upon TCR and SDF1 stimulation, as determined by Western blot analysis. Flow cytometry analysis also showed that TCR or SDF1-induced increases in pERK activity in Jurkat cells were reduced by LSP1 overexpression (Fig. 3B). In contrast, TCR triggered-pERK expression was significantly higher in CD4+ T cells of Lsp1(−/−) mice than in those of Lsp1(+/+) mice (Fig. 3C), indicating that LSP1 is a negative regulator of ERK activation. Moreover, an immunoprecipitation assay revealed that LSP1 coimmunoprecipitated with pERK, (Fig. 3D), demonstrating that LSP1 directly interacts with pERK, and thereby interferes with TCR-dependent ERK phosphorylation. Additionally, the SDF1-induced increase in T-cell migration, noted predominantly in Lsp1-deficient CD4+ T cells, was almost completely abrogated by the ERK inhibitor PD98059 (Fig. 3E). This effect was not a result of the nonspecific cytotoxicity of PD98059 as indicated by an 3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 3E, Inset). Overall, these results suggest that LSP1 inhibits T-cell migration by regulating the extent of ERK phosphorylation.
Fig. 3.
LSP1 control of pERK activity for T-cell migration. (A) pERK expression in LSP1-overexpressing T cells. Jurkat T cells, stably overexpressed with LSP1 gene (LSP1-GFP), were stimulated with anti-CD3 Ab (2 μg/mL) plus anti-CD28 Ab (2 μg/mL) or SDF1α (100 ng/mL) for 30 min. The cells with GFP only were used as a control. A representative of three independent experiments is shown. (B) Decrease in pERK expression in LSP1-overexpressing T cells. The cells were stimulated with anti-CD3/CD28 or SDF1 for 15 min. Data are the representative histogram of the three independent experiments with a similar result; red curve indicates stimulated T cells, black curve represents unstimulated T cells stained with Alexa Fluor 647-labeled pERK Ab, and gray curve shows the isotype control. (C) Increase in pERK expression in Lsp1-deficient T cells. T cells were isolated from spleen of Lsp1-deficient [i.e., (−/−)] and WT mice and stimulated with anti-CD3/CD28 for 20 min. The pERK expression was determined by Western blot analysis. A ratio of pERK1/2 relative to total Erk1/2 expression is presented on the right (*P < 0.05). (D) Immunoprecipitation assay to detect LSP1 binding to pERK. LSP1-overexpressing or control Jurkat T cells were stimulated with anti-CD3/CD28 for 10 min. After cells were incubated with pERK Ab, the LSP1-pERK complexes were precipitated by centrifugation and detected with anti-LSP1 Ab. (E) PD98059 inhibition of SDF1-induced T-cell migration. Splenic T cells were isolated from Lsp1-deficient (−/−) and WT mice and stimulated with SDF1α (100 ng/mL) in the presence or absence of PD98059 (2 μM), an ERK inhibitor. Cell migration was assessed in transwell chambers. Data are the mean ± SD of five independent experiments. Cell viability (Insets) was determined by MTT assay. (F) qPCR assays for representative DEGs regulated by ERK-downstream TFs in Lsp1-deficient [i.e., (−/−)] T cells (Upper) and in Jurkat T cells (Lower), which were stimulated with anti-CD3/28 Abs for 6 h and 12 h, respectively.
To investigate LSP1-pERK axis-dependent target genes that are involved in T-cell migration, we compared 677 Lsp1(−/−)/Lsp1(+/+) DEGs in the presence of anti-CD3/CD28 Abs with 1359 migration-related genes (e.g., leukocyte migration, cell migration/motility and chemotaxis) obtained from the AmiGO database (32). We identified 67 shared genes (P < 1 × 10−6) and then examined whether the shared genes are regulated by ERK-downstream transcription factors (TFs; SI Appendix, Materials and Methods). As a result, nine ERK-downstream TFs had significant numbers of target genes (P < 0.05) in the 67 shared genes, suggesting that the shared migration-related genes are regulated by the ERK pathway (SI Appendix, Fig. S2B). Subsequently, we selected the nine representative genes from LSP1-controlled and ERK-regulated DEGs (SI Appendix, Tables S5 and S6), and validated differential expression by using quantitative real-time PCR. As shown in Fig. 3F, IL-1β, Csf1r, Ptgs2, Ccl2, Ccl19, and Cxcl9 mRNA expressions were increased in anti-CD3–activated T cells of Lsp1(−/−) mice compared with those of Lsp1(+/+) mice, but Sox9, IL-4, and Gli3 expressions were decreased in the same cells. Conversely, LSP1-overexpressing Jurkat T cells showed lower levels of IL-1β, Csf1r, Ptgs2, Ccl2, Ccl19, and Cxcl9 mRNA expression, but higher levels of LSP1, Sox9, IL-4, and Gli3 mRNA expression, than control cells (Fig. 3F). Collectively, these data support the view that LSP1 controls T-cell migration via its interaction with pERK.
T-Cell–Dependent Chronic Inflammation Is Enhanced in Lsp1-Deficient Mice.
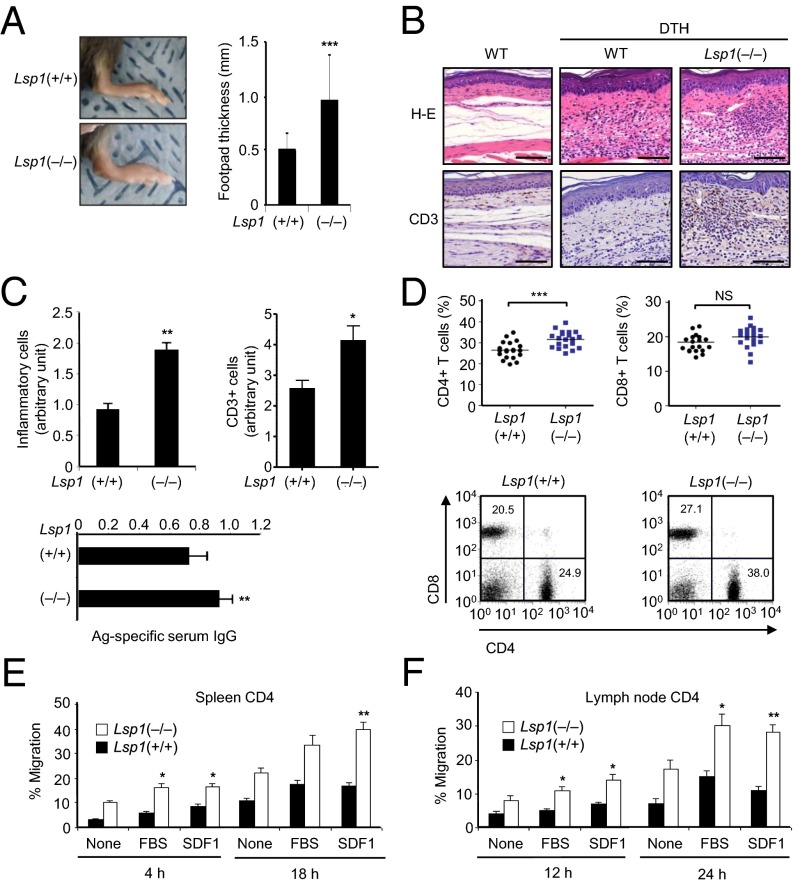
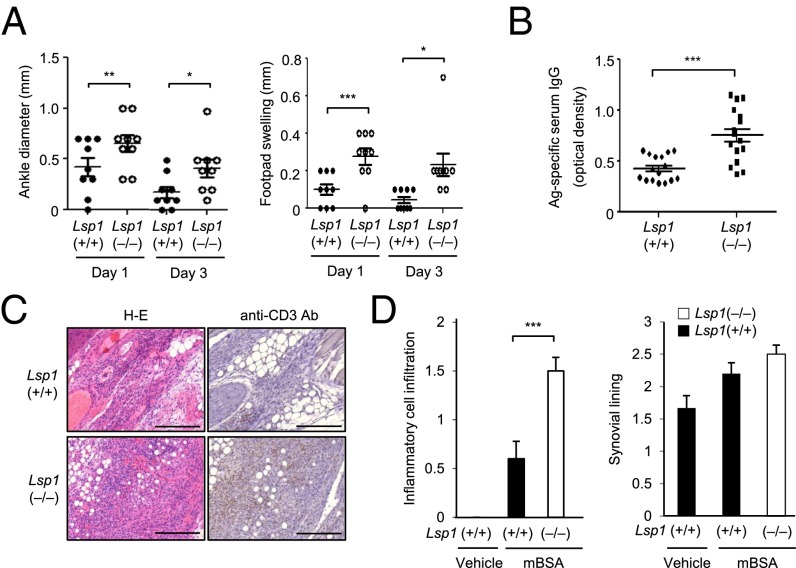
Delayed-type hypersensitivity (DTH) is a useful approach to evaluating T-cell–mediated immune responses (33). To investigate the pathology associated with LSP1 expression in vivo, DTH reactions were elicited in Lsp1-deficient and WT mice by injecting methylated BSA (mBSA) intradermally as a T-cell antigen. The result showed that footpad swelling was significantly greater in Lsp1(−/−) mice than in Lsp1(+/+) mice, as assessed 24 h after booster immunization (Fig. 4A). Histological analysis revealed that edema and total leukocyte infiltration were increased in the inflamed dermis of Lsp1-deficient mice (Fig. 4 B and C). Infiltration of T cells into the dermis, as determined by immunohistochemistry using anti-CD3 Ab, was also more pronounced in Lsp1-deficient mice than in WT mice (Fig. 4 B and C). In parallel, mBSA-specific serum IgG concentrations were significantly higher in Lsp1-deficient mice than in WT mice (Fig. 4C).
Fig. 4.
Enhanced inflammation and T-cell migration in Lsp1-deficient mice with DTH. (A) Increased foot thickness in Lsp1-deficient mice. Lsp1-deficient (n = 15) and WT mice (n = 15) were sensitized s.c. with mBSA (200 μg) plus complete Freund's adjuvant (CFA). One week after sensitization, mBSA (100 μg) was injected again into the footpads of each mouse. Footpad thickness was measured 24 h after the second challenge (***P < 0.005 vs. WT mice). (B and C) Histology of footpads of mice with DTH reaction. Sections of footpads challenged with solvent only (Left) or mBSA (Middle and Right) were stained with H&E (Upper) or anti-CD3 Ab (Lower). (B) Representative with significantly increased T-cell infiltration in the epidermis and dermis identified in Lsp1-deficient mice (C). (Scale bar: 100 μm.) Simultaneously, mBSA-specific IgG levels in the sera were determined by ELISA (Bottom). (D) Increase in the number of CD4+ T cells in lymph nodes of Lsp1-deficient mice. The proportion of CD4+ cells in the spleen and lymph node was determined by flow cytometry 7 d after the second antigen challenge. Representative dot plots are shown (Bottom). (E and F) Migration of antigen-activated CD4+ T cells (5 × 105 cells), which were obtained from the spleen and lymph node of Lsp1-deficient and WT mice with DTH. The number of migrated cells was counted manually. Data are the mean ± SEM of three independent experiments (*P < 0.05 and **P < 0.01 vs. WT mice).
Seven days after booster immunization, we assayed changes in T-cell populations in the draining lymph nodes by using flow cytometry. The frequency of CD4+ T cells, but not CD8+ T cells, was significantly higher in the lymph nodes of Lsp1-deficient mice than in those of WT mice (Fig. 4D), suggesting that antigen-activated T cells of Lsp1-deficient mice also have greater migratory and homing capacity. To confirm this observation, we isolated CD4+ T cells from the spleen and draining lymph nodes of mice with DTH, and then determined their migration in vitro (Fig. 4 E and F). As seen in Fig. 4E, splenic CD4+ T-cell migration in transwell chambers was increased by stimulation with 10% FBS and SDF1 in Lsp1-deficient and WT mice, and the increase was more pronounced in Lsp1-deficient CD4+ T cells. An experiment conducted with CD4+ T cells isolated from the lymph nodes of Lsp1-deficient mice showed similar results, suggesting that migration of antigen-activated T cells is also controlled by LSP1. Of note, the loss of Lsp1 did not affect the overall ratio (percentage gated) of naive, central memory, and effector memory populations of CD4+ and CD8+ T cells (SI Appendix, Fig. S5), eliminating the possibility that increased T-cell migration in Lsp1(−/−) mice might be caused by a difference in T-cell subsets between Lsp1(−/−) and Lsp1(+/+) mice.
Lsp1 Deficiency Increases T-Cell–Dependent Arthritis.
Antigen-induced arthritis (AIA) is another T-cell–driven disease model (34). Thus, we investigated the effect of LSP1 on the severity of AIA. After administration of mBSA into the ankle joint of preimmunized mice, arthritic signs, indicated by joint swelling and redness, rapidly developed and peaked at day 1 in Lsp1-deficient and WT mice, but they were more prominent in the former. At day 3, the foot swelling remained persistent in Lsp1-deficient mice while regressing in WT mice (Fig. 5A). Concomitantly, Lsp1-deficient mice had higher antigen-specific IgG levels in their sera than the WT mice (Fig. 5B). Histological analysis revealed increased soft tissue edema and inflammatory cell infiltration around the ankle joints, which were significantly higher in Lsp1-deficient mice. Moreover, a more dense infiltration of T cells, demonstrated by anti-CD3 immunostaining, was observed in Lsp1-deficient mice (Fig. 5 C and D).
Fig. 5.
Aggravation of AIA in Lsp1-deficient mice. (A) Increase in ankle swelling in Lsp1-deficient mice. AIA was induced in Lsp1-deficient (n = 10) and WT mice (n = 10), which had been already sensitized twice with mBSA plus CFA, by injection of mBSA into the ankle joints. Footpad and ankle diameters were measured by using microcalipers at days 1 and 3 after the intraarticular injection. (B) Levels of serum anti-mBSA Ab (IgG) determined by ELISA 7 d after AIA induction. Data are expressed in OD units. (C and D) Increase in T-cell infiltration in Lsp1-deficient mice. Seven days after AIA induction, ankle joints were removed and stained with H&E (Left) and anti-CD3 Ab (Right). Lsp1-deficient mice showed increased infiltration of mononuclear leukocytes, including T cells, in the synovial membrane and joint space, compared with WT mice. Data are the mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.005 vs. WT mice). (Scale bar: 100 μm.)
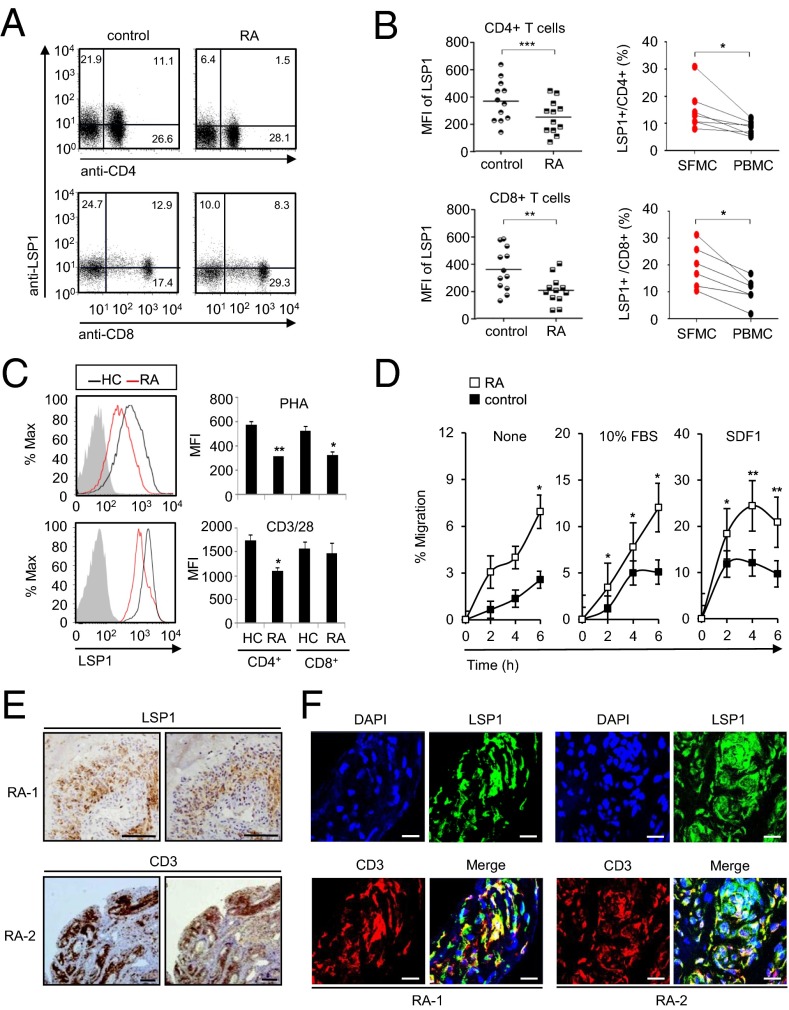
Antigen-activated T cells are crucial for the initiation and perpetuation of chronic inflammatory reactions in RA (3, 4). However, the expression and function of LSP1 in RA have never been reported to our awareness. Thus, we examined whether patients with RA have altered LSP1 protein expression in their T cells by flow cytometry. As expected, CD4+ T cells of patients with RA had lower LSP1 expression than those of healthy individuals (Fig. 6 A and B). CD8+ T cells also showed similar results (Fig. 6 A and B); these differences were not observed in monocytes (SI Appendix, Fig. S6). Interestingly, the frequency of LSP1+ cells was significantly higher in matched synovial T cells than in simultaneously obtained peripheral T cells (Fig. 6B), which concurs with our data on the increased LSP1 expression in TCR- or TNF-α–stimulated T cells as well as in arthritic mice (SI Appendix, Fig. S1). Notwithstanding, RA T cells stimulated with PHA or anti-CD3/CD28 Abs still showed a lesser induction of LSP1 expression than T cells of healthy controls (Fig. 6C). Moreover, in contrast to their LSP1 levels, RA T cells exhibited higher migration in transwell chambers with 10% FBS or SDF1 than T cells of healthy controls (Fig. 6D), which is consistent with previous studies showing that RA peripheral T cells have a greater migratory capacity toward inflamed joints containing a variety of chemokines, including SDF1 (30). In support of this notion, LSP1+ cells were frequently noted in the CD3+ T-cell zone of RA synovial tissues (Fig. 6E). Double immunofluorescence staining of RA synovium revealed that LSP1-expressing cells were also positive for CD3 (Fig. 6F), indicating that CD3+ T cells express LSP1 in RA synovia.
Fig. 6.
Decreased LSP1 expression in RA T cells. (A) Representative dot plots show LSP1 expression in the CD4+ and CD8+ T cells of healthy controls and patients with RA. PBMCs were stained with PE-labeled anti-CD4 Ab or APC-labeled anti-CD8 Ab, and then stained again with FITC-conjugated anti-LSP1 Ab. (B) LSP1 expression in T cells of patients with RA and healthy controls as determined by flow cytometry. (Left) Comparison of LSP1 expression in peripheral T cells between healthy controls (n = 12) and patients with RA (n = 12). (Right) Comparison of frequency of LSP1-expressing T cells between synovial fluid mononuclear cells (SFMCs; n = 7) and PBMCs (n = 7), which were obtained simultaneously from each patient with RA (*P < 0.05, **P < 0.01, and ***P < 0.001). (C) LSP1 induction in T cells of patients with RA (n = 4) vs. healthy controls (HC; n = 4) when stimulated with PHA (5 μg/mL) for 12 h or stimulated with anti-CD3 Ab plus anti-CD28 Ab for 48 h. Gray-colored histogram indicates isotype control (*P < 0.05 and **P < 0.01). (D) Increased T-cell migration in patients with RA. T cells were isolated from PBMCs of patients with RA (n = 6) and healthy controls (n = 6) by using anti-CD3 magnetic beads and incubated in transwell chambers in the presence or absence of 10% FBS or SDF1 (100 ng/mL). The number of migrated cells was counted manually. Data are the mean ± SEM (*P < 0.05 and **P < 0.01). (E) Immunohistochemical staining of RA synovia (RA-1 and RA-2) using anti-LSP1 Ab or anti-CD3 Ab. (Scale bar: 100 μm.) (F) Double immunofluorescence staining of RA synovia (RA-1 and RA-2) was performed by using Abs against LSP1 or CD3. Sections were stained subsequently with Cy3-conjugated anti-IgG (red) for anti-CD3 Ab and Alexa 488-conjugated anti-IgG (green) for anti-LSP1 Ab, respectively. In merged images, LSP1+ cells were costained with anti-CD3 Ab in yellow. In the same images, the nucleus was stained with DAPI in blue. (Scale bars: 20 μm.)
Discussion
Changes in DNA copy number, whether confined to specific genes or affecting whole chromosomes, can have an impact on biological homeostasis and influence interindividual differences in the susceptibility to human disorders, especially to autoimmune diseases (12–14). Even though a number of risk loci have been identified as genetic factors of RA through GWASs (11, 35), it is still unclear how CNV is related to the immune dysfunction, increasing the susceptibility of autoimmune diseases. Presently, we explored the RA-associated CNVs by a GWAS approach using SNP array analysis and identified a significant association of LSP1 deletion variants with RA. Consistent with earlier studies of other genes (36, 37), LSP1 protein expression levels in the present study correlated positively with the copy number status of the LSP1 gene. Importantly, LSP1 is critically involved in chronic T-cell–dependent inflammatory arthritis, functioning as a negative regulator of T-cell migration in mice and humans. To our knowledge, our work is the first to demonstrate how CNV contributes to immune dysfunction and a disease susceptibility phenotype.
The copy-loss CNV encompassing the LSP1 gene was reported in a previous CNV GWAS of hepatocellular carcinoma (38). The same group has demonstrated that LSP1 plays as a negative regulator of proliferation and migration of hepatoma cells (39), which supports our findings. The existence of the CNV encompassing the LSP1 gene was also identified in a study by The Wellcome Trust Case Control Consortium; however, it was not associated with RA (40). In that study, all LSP1 CNVs were gain variants, whereas we detected gain and deletion variants, and only the deletion variants showed significant association with RA. CNV calling can be dependent on the types of array platforms and analytic tools (41). Indeed, a study has reported that different analytic tools applied to the same raw data yield CNV calls with <50% concordance (41), which indicates the importance of proper candidate CNV validation.
With this in mind, we performed a strict qPCR validation with a larger cohort (764 patients with RA and 1,224 control subjects) to compensate for the potential limitation of the size of the discovery set. Additionally, to exclude the possibility that an LSP1 deletion variant might be the case for only Korean patients with RA, we also studied a cohort of white patients. As a result, a significant association of LSP1 CNV with RA susceptibility was replicated in both cohorts, which supports the reliability of our data and suggests that such association is not specific to Asian populations. Interestingly, in a subgroup analysis performed in 427 Korean patients with RA whose serum rheumatoid factor (RF) titers were available, we found that RF(−) patients (n = 139) had a greater frequency of LSP1 deletion variants than RF(+) patients (n = 288; 18.7% vs. 6.9%, respectively; P < 0.001). A similar result was observed among patients without anti-cyclic citrullinated peptide autoantibody (16.5% vs. 5.8%, P = 0.002, n = 373). These data suggest that LSP1 insufficiency may contribute more significantly to the pathogenesis of seronegative RA, and warrant further investigation to determine if LSP1 low copy reflects certain phenotypes of patients with RA, such as early onset, rapidly progressive, and HLA-susceptible allele [i.e., (+)] subgroups.
It has been reported that LSP1 is a Ca2+-activated, intracellular filamentous actin-binding protein that interacts with the cytoskeleton (21). As Ca2+ is one of the key signals for T-cell activation and the infiltration of Ca2+-activated T cells into joint tissue is an essential component of the pathogenesis of RA (3, 4, 30), we set out to elucidate the pathological role and clinical relevance of altered LSP1 expression in RA in terms of T-cell biology. We confirmed the intracellular expression of LSP1 in normal CD4+ and CD8+ T cells and Jurkat T cells (18). LSP1 expression was up-regulated upon TCR ligation as well as mitogenic stimulation promoting Ca2+ flux, such as PMA plus ionomycin or PHA, which is in line with a previous report that Ca2+ signaling is required for LSP1 up-regulation (24). Notably, the proinflammatory cytokine TNF-α also increased LSP1 expression. Given the lower LSP1 expression in patients with RA despite the higher levels of proinflammatory cytokines than in healthy subjects (Fig. 6 A and B), these results suggest that a genetic component of CNV, rather than proinflammatory cytokines, dominantly affects LSP1 expression at least in peripheral RA T cells.
Previous studies have suggested the negative regulatory effects of LSP1 on neutrophil adhesion, polarization, and migration (19, 20), but the role of LSP1 on T-cell biology remains unclear. To address this issue, we performed gene expression profiling of Lsp1-sufficient and deficient T cells, conditioned with TCR stimulation. A comparative analysis of gene expression profiles revealed that migration-related cellular processes were predominantly enriched by the DEGs in Lsp1-deficient T cells, which was subsequently confirmed by qPCR for the selected target genes. Moreover, we first found that cytokine–cytokine receptor interaction, response to wounds, innate immune response, and Ca2+ ion homeostasis also were governed by the LSP1 gene.
The mechanistic basis of regulated T-cell migration involves a chemokine gradient and the differential expression of chemokine receptors on naive and activated T cells (42). SDF1, MCP-1, and IL-6 are the main chemokines responsible for T-cell migration and are present at high levels in the synovial tissue or fluid of patients with RA (43–47). We found that LSP1-overexpressing Jurkat T cells exhibited a reduced chemotactic response to SDF1. Conversely, Lsp1-deficient primary T cells showed an enhanced migratory capacity upon SDF1 stimulation, which was not caused by differential levels of expression of CXCR4, a specific SDF1 receptor. These observations are in accordance with our results regarding gene expression profiling of Lsp1-deficient T cells. Thus, we concluded that LSP1 is a negative regulator of T-cell migration.
What is the downstream target of LSP1 responsible for T-cell migration? Evidence is emerging that, unlike MCP-1 and IL-6, SDF1-induced T-cell migration is mediated by ERK activation (43–45). In fact, the ERK pathway has been implicated in the migration of numerous cell types (48). The ERK pathway inhibitors PD98059 and U0126 inhibit cell migration in response to cell matrix proteins (e.g., fibronectin, vitronectin), growth factors (e.g., VEGF, EGF), and other stimuli, such as FCS (48). In the present study, ERK phosphorylation was blocked in LSP1-overexpressing T cells. Additionally, the enhanced migratory propensity of Lsp1-deficient T cells was almost completely abrogated by treatment with an ERK inhibitor. Moreover, as shown in Fig. 3D, LSP1 directly bound to pERK. Collectively, these results, together with previous reports (29, 45, 49), suggest that LSP1 regulation of T-cell migration is attributable to deactivation of pERK through a direct molecular contact. The central role of ERK activation in LSP1-mediated T-cell migration was corroborated by our data demonstrating nine ERK-dependent target genes and nine ERK-related TFs by comparative analysis of gene profiles.
To investigate the regulatory role of LSP1 in vivo, we used two disease models of DTH and AIA that are mediated primarily by activated T cells (33, 34). Lsp1-deficient mice exhibited marked T-cell infiltration into the inflamed tissue compared with WT mice, which correlated well with clinical severity and the levels of antigen-specific Ab response. This is in accordance with previous studies demonstrating increased neutrophil migration in a mouse model of zymosan-induced arthritis and wound healing (19, 20). It is well established that, after antigen stimulation, CD4+ T cells enter the draining lymph nodes and participate in the primary response, becoming central memory T cells (50). Given that more CD4+ T cells settled in the draining lymph node of Lsp1-deficient mice in comparison with WT mice but the overall frequency of memory or naive T-cell subsets did not differ, it is likely that a loss of Lsp1 has an intrinsic effect on CD4+ T-cell migration with no impact on expansion and differentiation.
Of note, T cells from peripheral blood of patients with RA expressed less LSP1 protein than those from healthy individuals. Considering that LSP1 negatively regulates T-cell migration, this result offers an intriguing explanation of how RA T cells accumulate into inflamed joints. RA T cells with defective LSP1 expression have a strong migratory propensity and can readily move along the SDF1 gradient into the affected joints. However, when T cells have entered the inflamed joints, they may acquire increased LSP1 expression following antigenic stimulation and the presence of proinflammatory cytokines, such as TNF-α, and then settle in the inflamed synovium. This notion is supported by our findings of higher LSP1 expression in paired synovial T cells than in peripheral T cells of patients with RA (Fig. 6B). If this is the case, the intrinsic tropism of RA T cells to the inflamed synovium, which could be predetermined genetically by CNV, may favor the stable interaction of T cells with other immune cells, including B cells to produce antigen-specific Ab, macrophages, and synoviocytes, to drive the self-perpetuation of chronic inflammation.
To our knowledge, our work is the first to demonstrate how CNV results in immune dysfunction and a disease phenotype, highlighting the importance of such genetic variants in the pathogenesis of autoimmune diseases. However, this study has some limitations. First, the LSP1 copy number loss was found in only 10.1% of RA cases, which indicates that the majority of patients with RA do not have an LSP1 low copy and that other genes or mechanisms may be involved in the increased T-cell migration in RA. Moreover, epigenetic dysregulation of LSP1 gene, which was not assessed in this study, may contribute to RA susceptibility together with LSP1 CNV to promote T-cell migration into the joints. Second, the animal models used in this study are for transient and eventually self-limiting inflammatory arthritis, and thus may not be accurate for chronic persistent arthritis like RA. Furthermore, the major cellular infiltrate in RA synovia are cells of the monocytic/macrophage lineage with appreciable numbers of neutrophils (3). As LSP1 also has been reported to have a role in neutrophil migration (19, 20), the immunopathology of RA may originate from the promigratory effects of LSP1 insufficiency on additional cell types besides T cells.
In summary, we identified a novel LSP1 deletion variant for RA susceptibility through a CNV GWAS. The copy number of LSP1 is significantly lower in patients with RA, which correlates positively with LSP1 protein expression levels. We also showed that LSP1 negatively regulates SDF1-induced T-cell migration by reducing ERK activation. Loss of Lsp1 promotes T-cell migration into antigen-instilled tissues and draining lymph nodes in mice with T-cell–dependent chronic inflammation. Moreover, patients with RA showed diminished expression of LSP1 in peripheral T cells with increased migratory capacity, suggesting that the defect in LSP1 signaling lowers the threshold for T-cell activation (e.g., cell migration) in patients with RA. Our data provide previously unidentified insight into the mechanisms of increased T-cell migration toward the inflamed synovium in RA and may open an opportunity for the development of new drugs targeting LSP1 in chronic inflammatory diseases. Additionally, the concept of an association between increased T-cell migration with LSP1 CNVs and LSP1 insufficiency might extend to the pathogenesis of other T-cell–dependent autoimmune diseases, including autoimmune thyroiditis, multiple sclerosis, autoimmune diabetes, and lupus nephritis, in which abnormal lymphocyte migration is important for infiltration and retention of T cells within pathologic sites (51).
Materials and Methods
Study Design.
For the CNV GWAS analysis, 100 Korean patients with RA and 400 healthy individuals were recruited. For the independent replication of the candidate RA-associated CNV, 764 patients with RA (599 Korean and 165 white) and 1,224 healthy individuals (966 Korean and 258 white) were recruited (SI Appendix, Table S1). LSP1 expression was confirmed from PBMCs, including T-cell subsets, by flow cytometry and real-time qPCR. Biological processes and associated molecules governed by LSP1 were explored through global transcriptome profiling and systematic analysis comparing DEGs between LSP1-deficient T cells and WT. Functional activity of LSP1 regulating T-cell migration was investigated by transwell migration assay using LSP1-deficient or -overexpressing T cells. The in vivo effect of LSP1 regulating T-cell migration on chronic inflammation was verified in two T-cell–dependent chronic inflammation models, DTH and AIA, using Lsp1-deficient mice. This study was performed with the approval of the Catholic Medical Center Office of Human Research Protection Program institutional review board (no. CUMC09U034). All patients gave written informed consent to the study protocol. The experimental procedures were approved by the Institutional Animal Care and Use Committee of the Catholic University of Korea.
CNV Discovery.
To define CNVs, we used SNP genotyping data from the 500 discovery set. In brief, 500 ng of genomic DNA was extracted from peripheral blood leukocytes and assayed on the Affymetrix Genome-Wide Human SNP array 5.0 (Affymetrix). Identification of CNVs was performed as described elsewhere (52). We designated each CNV based on its copy number status as diploid, loss, or gain. Boundaries of each CNV were determined as the distance from the linear location of the first SNP probe to that of the last probe (NCBI36/hg18; genome.ucsc.edu/index.html). Genomic qPCR for replication was performed as described elsewhere (53). Details of the study subjects, definition of CNVRs, and qPCR for genomic DNA are available in SI Appendix, Materials and Methods.
Detailed materials and methods, including microarray experiments, cell culture, cloning of overexpression of LSP1 gene, induction of DTH reaction, induction of AIA, T-cell migration assay, flow cytometry analysis of LSP1, Western blot analysis, immunoprecipitation, immunohistochemistry, and immunofluorescence staining are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs Grant HI14C3417 (to W.-U.K. and Y.-J.C.); National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology, Grants 2012R1A5A2047939 (to Y.-J.C.), 2014R1A2A1A11049812 (to W.-U.K.), and 2015R1A3A2032927 (to W.-U.K.); National Institutes of Health Grants AR049610 (to R.B.) and N01-HHSN272201100019C (to R.R.M. and R.B.); and Deutsche Forschungsgemeinschaft Grant GZ:KI 1973/1-1 (to B.-S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514152112/-/DCSupplemental.
References
- 1.Franz CM, Jones GE, Ridley AJ. Cell migration in development and disease. Dev Cell. 2002;2(2):153–158. doi: 10.1016/s1534-5807(02)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: Present and future therapeutic targets. Nat Immunol. 2005;6(12):1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 3.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 4.Tran CN, Lundy SK, Fox DA. Synovial biology and T cells in rheumatoid arthritis. Pathophysiology. 2005;12(3):183–189. doi: 10.1016/j.pathophys.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beech JT, et al. T-cell contact-dependent regulation of CC and CXC chemokine production in monocytes through differential involvement of NFkappaB: implications for rheumatoid arthritis. Arthritis Res Ther. 2006;8(6):R168. doi: 10.1186/ar2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alzabin S, Williams RO. Effector T cells in rheumatoid arthritis: lessons from animal models. FEBS Lett. 2011;585(23):3649–3659. doi: 10.1016/j.febslet.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 7.Singh K, et al. ERK-dependent T cell receptor threshold calibration in rheumatoid arthritis. J Immunol. 2009;183(12):8258–8267. doi: 10.4049/jimmunol.0901784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carruthers DM, et al. Characterization of altered calcium signalling in T lymphocytes from patients with rheumatoid arthritis (RA) Clin Exp Immunol. 1996;105(2):291–296. doi: 10.1046/j.1365-2249.1996.d01-768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporali R, Bugatti S, Cavagna L, Antivalle M, Sarzi-Puttini P. Modulating the co-stimulatory signal for T cell activation in rheumatoid arthritis: Could it be the first step of the treatment? Autoimmun Rev. 2014;13(1):49–53. doi: 10.1016/j.autrev.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Montgomery SB, Dermitzakis ET. The resolution of the genetics of gene expression. Hum Mol Genet. 2009;18(R2):R211–R215. doi: 10.1093/hmg/ddp400. [DOI] [PubMed] [Google Scholar]
- 11.Stahl EA, et al. BIRAC Consortium; YEAR Consortium Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almal SH, Padh H. Implications of gene copy-number variation in health and diseases. J Hum Genet. 2012;57(1):6–13. doi: 10.1038/jhg.2011.108. [DOI] [PubMed] [Google Scholar]
- 13.McKinney C, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67(3):409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 14.McKinney C, et al. Association of variation in Fcgamma receptor 3B gene copy number with rheumatoid arthritis in Caucasian samples. Ann Rheum Dis. 2010;69(9):1711–1716. doi: 10.1136/ard.2009.123588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarrant TK, Patel DD. Chemokines and leukocyte trafficking in rheumatoid arthritis. Pathophysiology. 2006;13(1):1–14. doi: 10.1016/j.pathophys.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Burga A, Lehner B. Predicting phenotypic variation from genotypes, phenotypes and a combination of the two. Curr Opin Biotechnol. 2013;24(4):803–809. doi: 10.1016/j.copbio.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Kim JH, et al. CNVRuler: A copy number variation-based case-control association analysis tool. Bioinformatics. 2012;28(13):1790–1792. doi: 10.1093/bioinformatics/bts239. [DOI] [PubMed] [Google Scholar]
- 18.Pulford K, Jones M, Banham AH, Haralambieva E, Mason DY. Lymphocyte-specific protein 1: A specific marker of human leucocytes. Immunology. 1999;96(2):262–271. doi: 10.1046/j.1365-2567.1999.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, et al. Modulation of Mac-1 (CD11b/CD18)-mediated adhesion by the leukocyte-specific protein 1 is key to its role in neutrophil polarization and chemotaxis. J Immunol. 2002;169(1):415–423. doi: 10.4049/jimmunol.169.1.415. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, et al. Accelerated wound healing in leukocyte-specific, protein 1-deficient mouse is associated with increased infiltration of leukocytes and fibrocytes. J Leukoc Biol. 2007;82(6):1554–1563. doi: 10.1189/0507306. [DOI] [PubMed] [Google Scholar]
- 21.Jongstra-Bilen J, Jongstra J. Leukocyte-specific protein 1 (LSP1): A regulator of leukocyte emigration in inflammation. Immunol Res. 2006;35(1-2):65–74. doi: 10.1385/IR:35:1:65. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, et al. MAPKAPK2-mediated LSP1 phosphorylation and FMLP-induced neutrophil polarization. Biochem Biophys Res Commun. 2007;358(1):170–175. doi: 10.1016/j.bbrc.2007.04.104. [DOI] [PubMed] [Google Scholar]
- 23.Kanno T, Siebenlist U. Activation of nuclear factor-kappaB via T cell receptor requires a Raf kinase and Ca2+ influx. Functional synergy between Raf and calcineurin. J Immunol. 1996;157(12):5277–5283. [PubMed] [Google Scholar]
- 24.Matsumoto N, Toyoshima S, Osawa T. Characterization of the 50 kDa protein phosphorylated in concanavalin A-stimulated mouse T cells. J Biochem. 1993;113(5):630–636. doi: 10.1093/oxfordjournals.jbchem.a124094. [DOI] [PubMed] [Google Scholar]
- 25.Jongstra-Bilen J, Wielowieyski A, Misener V, Jongstra J. LSP1 regulates anti-IgM induced apoptosis in WEHI-231 cells and normal immature B-cells. Mol Immunol. 1999;36(6):349–359. doi: 10.1016/s0161-5890(99)00055-3. [DOI] [PubMed] [Google Scholar]
- 26.Trentham DE, Townes AS, Kang AH. Autoimmunity to type II collagen an experimental model of arthritis. J Exp Med. 1977;146(3):857–868. doi: 10.1084/jem.146.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HJ, et al. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285(12):9262–9272. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Kremer KN, et al. Stromal cell-derived factor-1 signaling via the CXCR4-TCR heterodimer requires phospholipase C-β3 and phospholipase C-γ1 for distinct cellular responses. J Immunol. 2011;187(3):1440–1447. doi: 10.4049/jimmunol.1100820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nanki T, et al. Stromal cell-derived factor-1-CXC chemokine receptor 4 interactions play a central role in CD4+ T cell accumulation in rheumatoid arthritis synovium. J Immunol. 2000;165(11):6590–6598. doi: 10.4049/jimmunol.165.11.6590. [DOI] [PubMed] [Google Scholar]
- 31.Ticchioni M, et al. Signaling through ZAP-70 is required for CXCL12-mediated T-cell transendothelial migration. Blood. 2002;99(9):3111–3118. doi: 10.1182/blood.v99.9.3111. [DOI] [PubMed] [Google Scholar]
- 32.Carbon S, et al. AmiGO Hub; Web Presence Working Group AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen IC. Delayed-type hypersensitivity models in mice. Methods Mol Biol. 2013;1031:101–107. doi: 10.1007/978-1-62703-481-4_13. [DOI] [PubMed] [Google Scholar]
- 34.van den Berg WB, Joosten LA, van Lent PL. Murine antigen-induced arthritis. Methods Mol Med. 2007;136:243–253. doi: 10.1007/978-1-59745-402-5_18. [DOI] [PubMed] [Google Scholar]
- 35.Eyre S, et al. Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate; Wellcome Trust Case Control Consortium High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi Y, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110(1):164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nalesnik MA, et al. Gene deletions and amplifications in human hepatocellular carcinomas: Correlation with hepatocyte growth regulation. Am J Pathol. 2012;180(4):1495–1508. doi: 10.1016/j.ajpath.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koral K, et al. Leukocyte-specific protein 1: A novel regulator of hepatocellular proliferation and migration deleted in human hepatocellular carcinoma. Hepatology. 2015;61(2):537–547. doi: 10.1002/hep.27444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craddock N, et al. Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464(7289):713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto D, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29(6):512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: Chemokines in control of T cell traffic. Nat Immunol. 2008;9(9):970–980. doi: 10.1038/ni.f.213. [DOI] [PubMed] [Google Scholar]
- 43.Houssiau FA, Devogelaer JP, Van Damme J, de Deuxchaisnes CN, Van Snick J. Interleukin-6 in synovial fluid and serum of patients with rheumatoid arthritis and other inflammatory arthritides. Arthritis Rheum. 1988;31(6):784–788. doi: 10.1002/art.1780310614. [DOI] [PubMed] [Google Scholar]
- 44.Cambien B, Pomeranz M, Millet MA, Rossi B, Schmid-Alliana A. Signal transduction involved in MCP-1-mediated monocytic transendothelial migration. Blood. 2001;97(2):359–366. doi: 10.1182/blood.v97.2.359. [DOI] [PubMed] [Google Scholar]
- 45.Ottoson NC, Pribila JT, Chan AS, Shimizu Y. Cutting edge: T cell migration regulated by CXCR4 chemokine receptor signaling to ZAP-70 tyrosine kinase. J Immunol. 2001;167(4):1857–1861. doi: 10.4049/jimmunol.167.4.1857. [DOI] [PubMed] [Google Scholar]
- 46.Koch AE. Chemokines and their receptors in rheumatoid arthritis: Future targets? Arthritis Rheum. 2005;52(3):710–721. doi: 10.1002/art.20932. [DOI] [PubMed] [Google Scholar]
- 47.Weissenbach M, et al. Interleukin-6 is a direct mediator of T cell migration. Eur J Immunol. 2004;34(10):2895–2906. doi: 10.1002/eji.200425237. [DOI] [PubMed] [Google Scholar]
- 48.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar A, et al. CXCR4 physically associates with the T cell receptor to signal in T cells. Immunity. 2006;25(2):213–224. doi: 10.1016/j.immuni.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 50.Catron DM, Rusch LK, Hataye J, Itano AA, Jenkins MK. CD4+ T cells that enter the draining lymph nodes after antigen injection participate in the primary response and become central-memory cells. J Exp Med. 2006;203(4):1045–1054. doi: 10.1084/jem.20051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norman MU, Hickey MJ. Mechanisms of lymphocyte migration in autoimmune disease. Tissue Antigens. 2005;66(3):163–172. doi: 10.1111/j.1399-0039.2005.00434.x. [DOI] [PubMed] [Google Scholar]
- 52.Yim SH, et al. Copy number variations in East-Asian population and their evolutionary and functional implications. Hum Mol Genet. 2010;19(6):1001–1008. doi: 10.1093/hmg/ddp564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung SH, et al. Genome-wide copy number variation analysis identifies deletion variants associated with ankylosing spondylitis. Arthritis Rheumatol. 2014;66(8):2103–2112. doi: 10.1002/art.38650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.