Significance
Aggressive cancers often possess functional and molecular traits characteristic of normal stem cells. It is unclear if aggressive phenotypes of prostate cancer molecularly resemble normal stem cells residing within the human prostate. Here, we transcriptionally profiled epithelial populations from the human prostate and show that aggressive prostate cancer is enriched for a prostate basal stem cell signature. Within prostate cancer metastases, histological subtypes had varying enrichment of the stem cell signature, with small cell neuroendocrine carcinoma being the most stem cell-like. We further found that small cell neuroendocrine carcinoma and the prostate basal stem cell share a common transcriptional program. Targeting normal stem cell transcriptional programs may provide a new strategy for treating advanced prostate cancer.
Keywords: RNA-seq, prostate cancer, stem cell signature, basal cell, neuroendocrine prostate cancer
Abstract
Evidence from numerous cancers suggests that increased aggressiveness is accompanied by up-regulation of signaling pathways and acquisition of properties common to stem cells. It is unclear if different subtypes of late-stage cancer vary in stemness properties and whether or not these subtypes are transcriptionally similar to normal tissue stem cells. We report a gene signature specific for human prostate basal cells that is differentially enriched in various phenotypes of late-stage metastatic prostate cancer. We FACS-purified and transcriptionally profiled basal and luminal epithelial populations from the benign and cancerous regions of primary human prostates. High-throughput RNA sequencing showed the basal population to be defined by genes associated with stem cell signaling programs and invasiveness. Application of a 91-gene basal signature to gene expression datasets from patients with organ-confined or hormone-refractory metastatic prostate cancer revealed that metastatic small cell neuroendocrine carcinoma was molecularly more stem-like than either metastatic adenocarcinoma or organ-confined adenocarcinoma. Bioinformatic analysis of the basal cell and two human small cell gene signatures identified a set of E2F target genes common between prostate small cell neuroendocrine carcinoma and primary prostate basal cells. Taken together, our data suggest that aggressive prostate cancer shares a conserved transcriptional program with normal adult prostate basal stem cells.
Up to 90% of patients with metastasis will succumb to the disease, yet our understanding of metastasis remains limited. Metastasis is the result of cancer cells disseminating from a primary lesion and colonizing a secondary site where they reinitiate macroscopic tumor growth (1). To initiate secondary tumor growth, disseminated cells must acquire attributes that are central to malignancy such as motility, invasiveness, self-renewal, and resistance to apoptosis (2, 3). It is unlikely that every disseminated cell will retain these traits, as some may be more differentiated or reach replicative exhaustion (1). However, cancer stem cells can possess these traits and have been identified in a number of different tissues (4–8). Moreover, signaling networks and transcription factors (TFs) central to stem cells can remain activated even once a macrometastasis has formed (9–12).
Cancer stem cells and normal stem cells often share similar molecular mechanisms and functional capabilities. In colorectal cancer, primary tumor cells that give rise to metastases display many of the same traits seen in normal stem cells including long-term self-renewal (13). Genes specific for normal intestinal stem cells were found to be up-regulated in aggressive colorectal cancer and were predictive of disease relapse (14). Isolation and characterization of human normal mammary stem cells identified a gene signature capable of distinguishing breast cancers according to tumor grade. Moreover, markers for these normal stem cells enabled isolation of cancer cells that were enriched in tumor-initiating properties upon xenotransplanation (15). Breast cancer circulating tumor cells (CTCs) expressing stem cell markers were capable of forming metastatic lesions in mice. The number of stem cell marker-expressing CTCs, but not bulk CTCs, correlated with disease progression and an overall worse prognosis (16). Stem cell signaling pathways have also been found in aggressive variants of nonepithelial cancers. Leukemic and hematopoietic stem cells share a core transcriptional profile consisting of networks that regulate stemness. Gene signatures specific for each population were able to predict survival of acute myeloid leukemia (AML) patients, suggesting that acquisition of stem cell-related genes influences clinical outcome (17).
Similarly to other cancers, it has been suggested that aggressive prostate cancer acquires properties that are common to stem cells. An 11-gene BMI-1–associated gene expression signature developed from common genes between BMI-1+/+ versus BMI-1−/− neurospheres and a transgenic mouse model of prostate cancer was enriched in metastatic samples and further associated with poor prognosis in early-stage, organ-confined prostate cancer (12). Using curated signatures specific for embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and the polycomb repressive complex-2 (PRC2), Markert et al. showed that prostate cancer patients enriched for the ESC signature had a poorer survival compared with the iPSC-like tumors and PRC2-like tumors (10). An in-depth genomic and transcriptomic analysis of 150 metastatic, castration-resistant prostate cancers (CRPCs) revealed that 18% of patients had alterations in the developmental Wnt signaling pathway (18). Murine models overexpressing key components of developmental signaling pathways alone or with other genetic alterations can drive a phenotype reminiscent of late-stage prostate cancer (19–22). Although these studies provide evidence of a relationship between stem-like qualities and an aggressive phenotype, no studies to our knowledge have shown a molecular relationship between aggressive prostate cancer and uncultured stem-like cells from the human prostate.
The vast majority of prostate cancers have a glandular, adenocarcinoma phenotype; however, a subset manifests a phenotype with neuroendocrine differentiation termed neuroendocrine prostate cancer (NEPC). These tumors display many of the same markers found on neuroendocrine cells within the normal prostate such as positivity for synaptophysin, chromogranin, neuron-specific enolase, and CD56 (23). De novo, these tumors make up less than 1% of organ-confined prostate cancers; however, 20–25% of patients with CRPC exhibit an NEPC phenotype. Many believe that is an underestimate, as it is not common practice to biopsy metastases. A morphological variant of NEPC termed small cell neuroendocrine carcinoma (SCNC) is highly aggressive, has little to no response to androgen deprivation therapy, metastasizes readily, and has limited treatment options (24). Due to the relative difficulty of obtaining human tissue containing NEPC, our molecular understanding of this disease is limited. A recent important paper identified NEPC to have alterations in genes regulating cell cycling, specifically a large number with AURKA and MYCN amplifications (25). Two morphological variants of NEPC (SCNC and prostate adenocarcinoma with neuroendocrine differentiation) were grouped together in this study for bioinformatic analyses. Thus, it is unclear how NEPC morphological subtypes are molecularly different and how this compares to CRPC with an adenocarcinoma phenotype.
We have previously identified a basal cell population within the mouse and human prostate that has stem cell characteristics (26, 27). This population can give rise to all three epithelial populations and act as a tumor-initiating cell when modified to express oncogenes commonly altered in prostate cancer. In this study, we sought to molecularly characterize the Trop2+ CD49f Hi human basal stem cell population and determine if aggressive cancer reverts back to a stem cell state seen in the human prostate. We show that the functionally identified Trop2+ CD49f Hi human basal stem cell population is enriched for stem and developmental pathways. We defined a basal stem cell gene signature and showed that metastatic prostate cancer was enriched for this signature. Using a dataset comprised of different metastatic prostate cancer phenotypes, we show that metastatic small cell carcinoma was the most enriched for this signature and shared a transcriptional program with the basal stem cell population.
Results
Tissue Acquisition and RNA Sequencing Flow-Through.
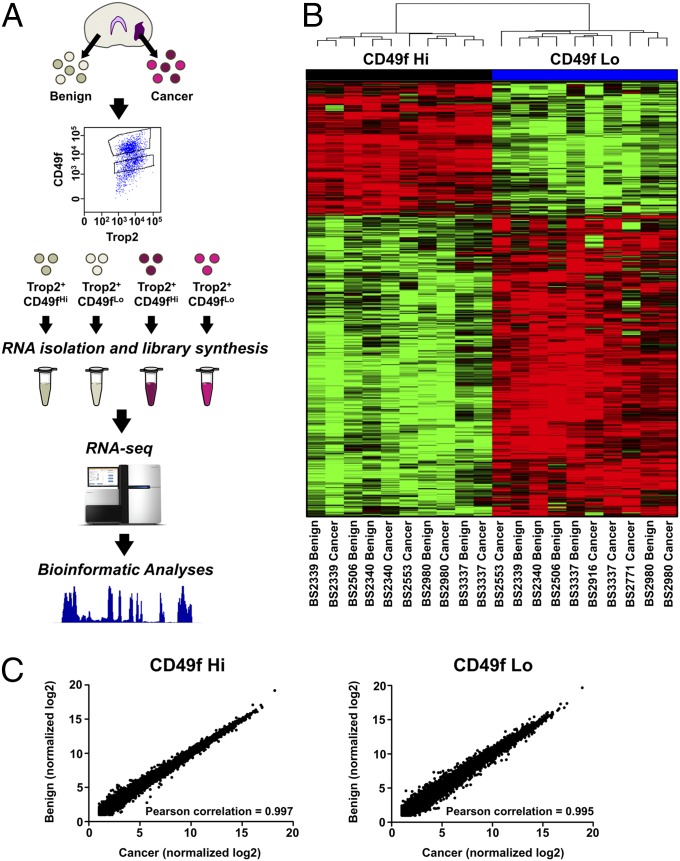
We acquired prostate tissue from eight patients that had undergone radical prostatectomy. These patients ranged in Gleason score from 6 to 9. A pathologist outlined the benign and malignant regions on an H&E slide, and a trained technician separated the benign and malignant regions of the tissue based on the outline. The tissues were digested into single cell suspensions and sorted based on Trop2 and CD49f staining as described previously (27). We aimed to collect four populations for each patient; however, due to low numbers of certain populations, we were not able to collect all four populations for each patient. We were able to collect all four populations in two patients. In total, we acquired five samples for each of the four populations. Each sample was subjected to paired-end RNA sequencing (RNA-seq) and averaged 1.0 × 108 paired reads that uniquely mapped to the human genome (Table S1 and Dataset S1).
Table S1.
RNA-seq mapping statistics for each sample
| Sample | Patient | Tissue | Population | Total RNA-seq reads | Paired mapped RNA-seq reads | % mapped |
| 1 | BS2339 | Benign | CD49f Hi | 1.81 × 108 | 1.69 × 108 | 93% |
| 2 | BS2339 | Benign | CD49f Lo | 1.64 × 108 | 1.52 × 108 | 93% |
| 3 | BS2339 | Cancer | CD49f Hi | 1.53 × 108 | 1.42 × 108 | 93% |
| 4 | BS2340 | Benign | CD49f Hi | 9.10 × 107 | 8.53 × 107 | 94% |
| 5 | BS2340 | Benign | CD49f Lo | 2.22 × 108 | 2.09 × 108 | 94% |
| 6 | BS2340 | Cancer | CD49f Hi | 1.62 × 108 | 1.52 × 108 | 94% |
| 7 | BS2506 | Benign | CD49f Hi | 2.15 × 108 | 2.02 × 108 | 94% |
| 8 | BS2506 | Benign | CD49f Lo | 2.12 × 108 | 1.99 × 108 | 94% |
| 9 | BS2553 | Cancer | CD49f Hi | 5.60 × 107 | 5.15 × 107 | 92% |
| 10 | BS2553 | Cancer | CD49f Lo | 3.35 × 108 | 3.08 × 108 | 92% |
| 11 | BS2771 | Cancer | CD49f Lo | 9.80 × 107 | 9.26 × 107 | 94% |
| 12 | BS2980 | Benign | CD49f Hi | 9.11 × 107 | 8.43 × 107 | 93% |
| 13 | BS2980 | Benign | CD49f Lo | 9.17 × 107 | 8.74 × 107 | 95% |
| 14 | BS2980 | Cancer | CD49f Hi | 9.43 × 107 | 8.97 × 107 | 95% |
| 15 | BS2980 | Cancer | CD49f Lo | 1.26 × 108 | 1.19 × 108 | 95% |
| 16 | BS3337 | Benign | CD49f Hi | 1.37 × 108 | 1.31 × 108 | 95% |
| 17 | BS3337 | Benign | CD49f Lo | 1.78 × 108 | 1.70 × 108 | 96% |
| 18 | BS3337 | Cancer | CD49f Hi | 1.60 × 108 | 1.52 × 108 | 95% |
| 19 | BS3337 | Cancer | CD49f Lo | 9.90 × 107 | 9.30 × 107 | 94% |
| 20 | BS2916 | Cancer | CD49f Lo | 4.90 × 107 | 4.57 × 107 | 93% |
Benign and Cancer Gene Expression Profiles from the Same Epithelial Population Are Very Similar.
To explore the molecular differences between the benign and cancer regions, we performed hierarchical clustering on all 20 samples. To our surprise, the samples did not cluster based on benign and cancer but rather clustered based on their epithelial population (Fig. 1B). Within the cluster, samples from the same epithelial population and same patient were more closely clustered than cancer or benign samples from the same population but different patients. Plotting the benign and cancer expression values for all 20,500 genes further confirmed that the benign and cancer samples from the same epithelial population were extremely similar (Fig. 1C). When we performed differential expression analysis on benign Trop2+ CD49f Hi and cancer Trop2+ CD49f Hi, there were only eight genes with greater than twofold change with a P value cutoff less than 0.05. Differential expression analysis on benign Trop2+ CD49f Lo and cancer Trop2+ CD49f Lo provided 62 genes with greater than twofold change, which makes up ∼0.3% of all genes. Genes up-regulated in the benign Trop2+ CD49f Lo population such as MSMB and ANPEP have been shown to have higher expression in the benign prostate (28, 29). Most of the genes up-regulated for the cancer portion have not previously been associated with prostate cancer, except for CXCL5 and APOD (30, 31). Genes typically up-regulated in prostate cancer such as AMACR and FASN were not differentially expressed between the benign and cancer regions for each epithelial population. We cannot rule out that the similarities in expression profiles may be due to contaminating normal cells within the region outlined as cancerous. The similarities in expression profiles could be also attributed to field effects. This occurs when histologically normal tissue adjacent to cancerous tissue acquires many of the same genetic alterations seen in the malignant region. Field effects have been seen in numerous epithelial cancers including head and neck, stomach, lung, and prostate (32–35).
Fig. 1.
Benign and cancer regions from the same epithelial population have similar transcriptional profiles. (A) Experimental scheme for gene expression analysis of human prostate Trop2+ CD49f Hi and Trop2+ CD49f Lo populations. (B) Hierarchical clustering of benign and cancer Trop2+ epithelial populations. (C) Scatter plots comparing the quantile-normalized log2 gene expression for each gene from the benign and cancer regions for each epithelial population.
Trop2+ CD49f Hi and Trop2+ CD49f Lo Subpopulations Are Enriched for Different Gene Sets/Pathways and Master Regulators.
Because the benign and cancer transcriptional profiles for each population were extremely similar, we combined the samples from each subpopulation to increase the statistical power for our comparison.
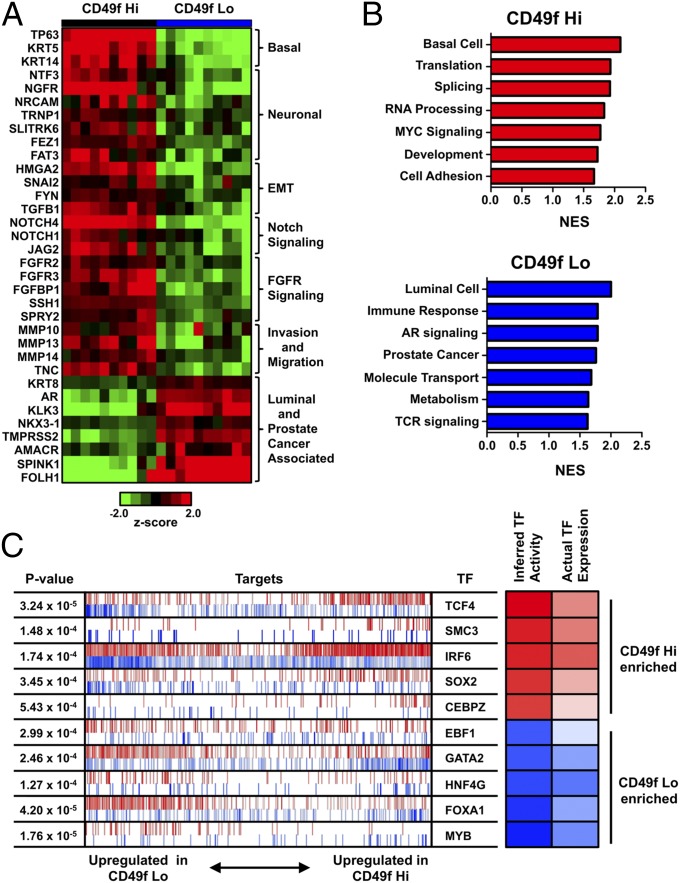
Using linear models for microarray analysis (LIMMA), we looked at differentially expressed genes between the CD49f Hi and CD49f Lo populations (36). A total of 1,501 genes were differentially expressed between the CD49f Hi and CD49f Lo populations, with 527 genes up-regulated in the Hi population and 923 genes up-regulated in the Lo population. The CD49f Hi population overexpressed a number of genes found in the NOTCH, FGFR, and WNT development pathways. Other up-regulated genes have been shown to act as epigenetic modifiers and transcriptional regulators, play important roles in neuronal processes, regulate epithelial-to-mesenchymal transitions (EMTs), and influence cell invasion and migration (Fig. 2A). The CD49f Lo population overexpressed genes commonly associated with prostate luminal cells or prostate cancer, including AR, KRT8, KLK3, NKX3-1, TMPRSS2, and AMACR (Fig. 2A).
Fig. 2.
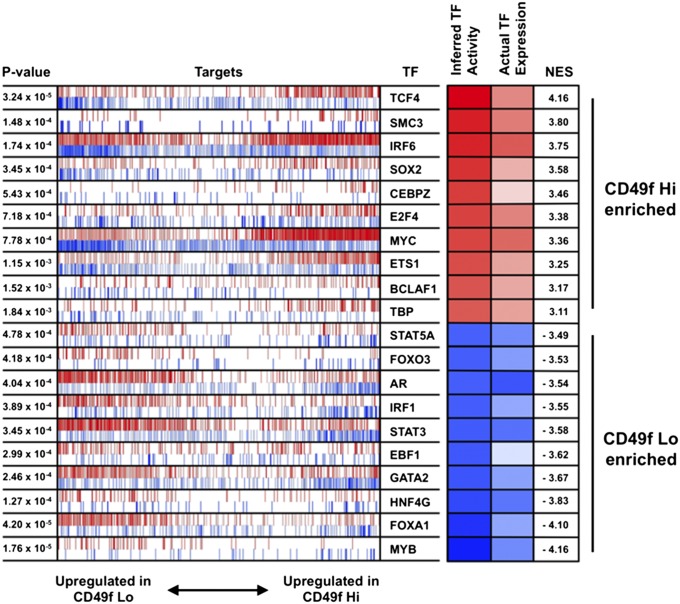
Trop2+ CD49f Hi and Lo populations are enriched for different gene sets. (A) Heat map of gene expression for selected genes. (B) Significantly enriched gene networks for CD49f Hi and CD49f Lo populations from GSEA. (C) Top 5 TFs enriched in the CD49f Hi and CD49f Lo populations using MARINa. TFs are arranged according to their P value and nominal enrichment score (NES). The shaded boxes on the right show the inferred TF activity according to the NES calculated by MARINa and the actual TF’s expression, with red indicating up-regulation in the CD49f Hi population and blue indicating up-regulation in the CD49f Lo population. The most enriched TF for the CD49f Hi population is the top TF listed in the red, and the most enriched TF for the CD49f Lo population is the last TF listed in the blue. Each row represents the MARINa results for the TF. The vertical red and blue lines represent the target genes for the TF, with positive regulated target genes in red and negative regulated target genes in blue. Increased activity of the CD49f Hi-enriched TFs is shown by enrichment of the TF’s positive targets within the CD49f Hi up-regulated genes in the CD49f MARINa signature and of its negative targets within the CD49f Lo up-regulated genes in the CD49f MARINa signature. Increased activity of the CD49f Lo-enriched TFs is shown by enrichment of the TF’s positive targets within the CD49f Lo up-regulated genes in the CD49f MARINa signature and of its negative targets within the CD49f Hi up-regulated genes in the CD49f MARINa signature.
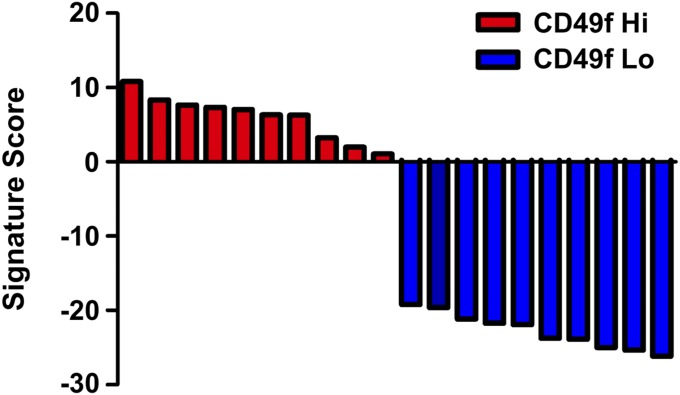
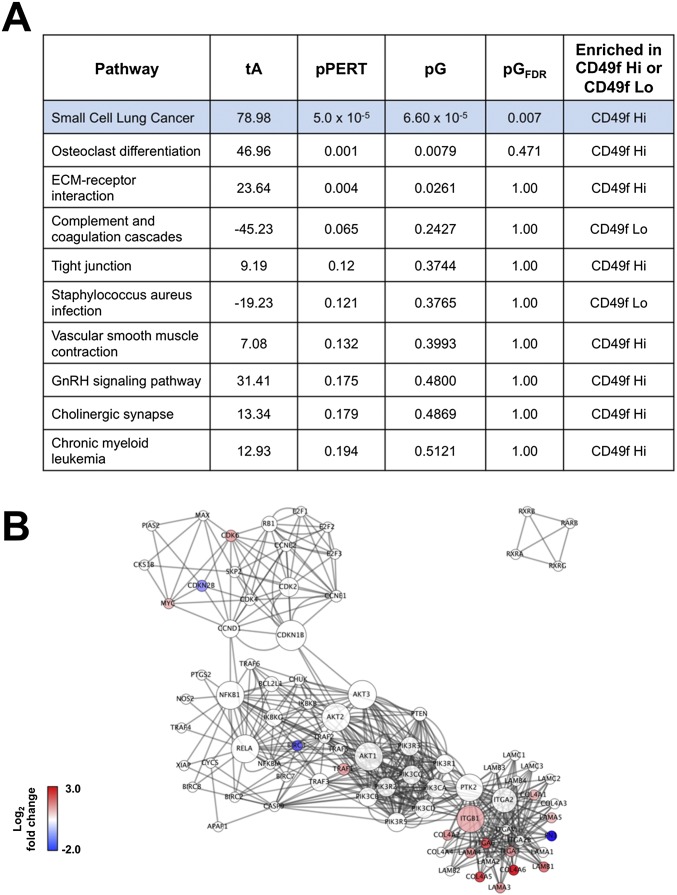
To gain more biological insight into gene networks specific for each population, we ran gene set enrichment analysis (GSEA) on a 20,500-gene-dense signature that could accurately identify CD49f Hi and CD49f Lo samples (37) (Dataset S2). In short, we first constructed a computational model to recognize CD49f Hi prostate basal cells by formulating a dichotomy between the CD49f Hi and CD49f Lo populations. Given this dichotomy, we trained a logistic regression model with elastic net regularization (38). This method produced a gene expression signature with 20,500 weights that could identify CD49f Hi and CD49f Lo samples with 100% accuracy using a leave in–leave out cross-validation scheme (Fig. S1). GSEA showed that the CD49f Hi population was enriched in gene sets associated with basal cells, translation, splicing, RNA processing, MYC signaling, stem cell and development networks, and cell adhesion (Fig. 2B). Functional studies showing that the Trop2+ CD49 Hi cell population has stem cell characteristics further supports the identified gene sets (27). The Trop2+ CD49f Lo expression profile was enriched for gene sets associated with luminal cells, prostate cancer, immune response, AR signaling, metabolism, and so forth. We also used signaling pathway impact analysis (SPIA), which is a complementary pathway analysis that takes into account the fold changes of genes along with the genes’ positions within a pathway to identify pathways that are relevant to the condition under study (39). SPIA identified a gene network associated with small cell lung cancer as the only pathway significantly activated in the CD49f Hi population (Fig. S2). No pathways were activated in the CD49f Lo population that made the false discovery rate (FDR) cutoff.
Fig. S1.
Waterfall plot of CD49f Hi basal stem cell 20,500-gene signature scores for the Trop2+ CD49f Hi (red, n = 10) and Trop2+ CD49f Lo samples (blue, n = 10).
Fig. S2.
SPIA showed enrichment of KEGG_Small Cell Lung Cancer network in the Trop2+ CD49f Hi population. (A) Table of top 10 KEGG pathways found using SPIA. The blue highlighted row indicates the only pathway that was statistically enriched for either the Trop2+ CD49f Hi or Trop2+ CD49f Lo population. (B) Network of KEGG_Small Cell Lung Cancer using gene expression values from Trop2+ CD49f Hi and Trop2+ CD49f Lo RNA-seq. Red means up-regulated in the CD49f Hi population, and blue means down-regulated in the CD49f Hi population.
To identify potential TFs that regulate each phenotype, we used the master regulator interference algorithm (MARINa), which has been used to identify master regulators for human high-grade glioma, murine prostate cancer, and normal formation of germinal centers (40–42). We created a network of TFs and their targets by combining transcriptional and genomic data from multiple databases (43–46). MARINa used this TF network to compute a score for each TF’s relative activity between the CD49f Hi and CD49f Lo populations. This activity score was derived from a combined view of the expression levels of each TF and its transcriptional targets. After filtering for the master regulators with P < 0.05 and FDR < 0.10, the top TF in the CD49f Hi population was TCF4 (Fig. 2C). TCF4 has been shown to be important for neuronal development and EMT (47, 48). Moreover, a number of TFs associated with stem cells were also enriched in the CD49f Hi population, including SOX2, MYC, and ETS1 (Fig. 2C and Fig. S3). Previous reports have shown SOX2 expression in normal prostate basal cells and in a majority of patients with castration-resistant and neuroendocrine metastatic prostate cancer (49, 50). A number of TFs were enriched in the CD49f Lo population including MYB, FOXA1, and AR, which have been previously identified in luminal cells or cancers with a luminal phenotype (51–53) (Fig. 2C and Fig. S3).
Fig. S3.
Top 10 TFs enriched in the Trop2+ CD49f Hi and Trop2+ CD49f Lo populations using MARINa. TFs are arranged according to their P value and NES. The shaded boxes on the right show the inferred TF activity according to the NES calculated by MARINa and the actual TF’s expression, with red indicating up-regulation in the CD49f Hi population and blue indicating up-regulation in the CD49f Lo population. The most enriched TF for the CD49f Hi population is the top TF listed in the red, and the most enriched TF for the CD49f Lo population is the last TF listed in the blue. Each row represents the MARINa results for the TF. The vertical red and blue lines represent the target genes for the TF, with positive regulated target genes in red and negative regulated target genes in blue. Increased activity of the CD49f Hi-enriched TFs is shown by enrichment of the TF’s positive targets among the CD49f Hi up-regulated genes in the CD49f MARINa signature and of its negative targets among the CD49f Lo up-regulated genes in the CD49f MARINa signature. Increased activity of the CD49f Lo-enriched TFs is shown by enrichment of the TF’s positive targets among the CD49f Lo up-regulated genes in the CD49f MARINa signature and of its negative targets among the CD49f Hi up-regulated genes in the CD49f MARINa signature.
The CD49f Hi Population Resembles the Normal Human Mammary Stem Cell and Uses MYC Signaling Networks.
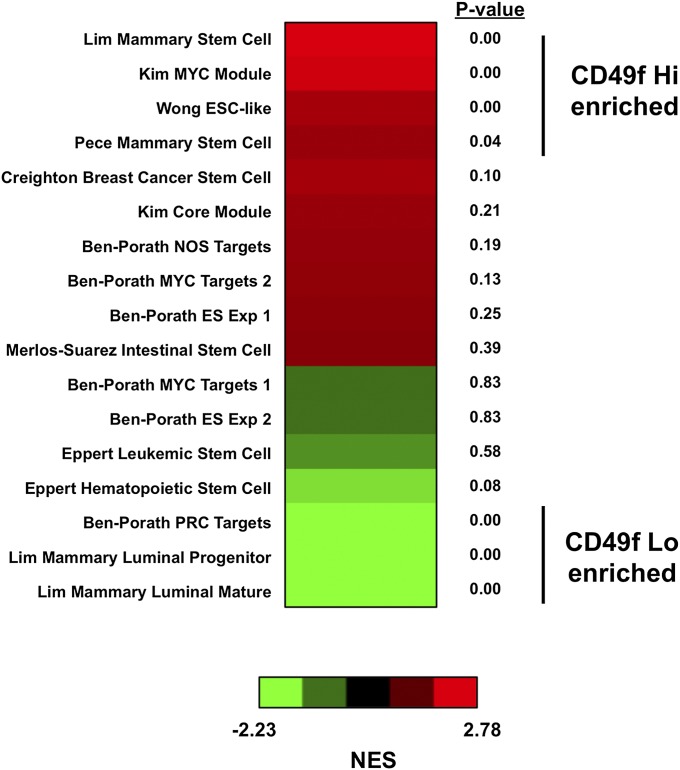
We compiled a list of published gene signatures from different human stems cells or signaling modules to determine if the CD49f Hi population resembled stem cells from other human tissues (14, 15, 54–58). We used GSEA to apply each stem cell signature against the CD49f Hi 20,500-gene-dense signature. The CD49f Hi population was most similar to normal mammary stem cell signatures from two different datasets but not stem cells from any other tissue (Fig. S4). The CD49f Hi population was also associated with a MYC signaling network and a human ESC-like signature. Integration of protein–protein and DNA–protein studies has shown that the transcription factor MYC constitutes a signaling network that is distinct from a core ESC transcriptional program, and this MYC signaling is responsible for the similarities between ESCs and cancer (57). Moreover, MYC can induce an ESC-like transcriptional profile when transduced into keratinocytes expressing known oncogenes (58). The CD49f Lo population was enriched for the normal mammary luminal mature signature and PRC2 targets, suggesting that this population is more differentiated. Interestingly, the CD49f Lo population was also enriched for the normal mammary luminal progenitor signature (Fig. S4). Using an organoid culture system, it has been shown that a small subset of human prostate luminal cells have progenitor-like capabilities (59). Gene ontology analysis of the leading-edges genes from the mammary luminal progenitor signature showed that these genes were associated with immune response, response to wounding, and defense response, but none of the terms were associated with developmental or stem cell gene networks. Although unable to form human prostate glands in the in vivo regeneration assay (27), it is possible that a subset of progenitor cells reside within the CD49f Lo population as measured by a different functional assay.
Fig. S4.
Enrichment of human stem cell signatures and gene modules in the Trop2+ CD49f Hi basal stem-like gene signature. P values were calculated from GSEA with 1,000 permutations. NES, nominal enrichment score.
Metastatic Prostate Cancer Is Enriched for the CD49f Hi Basal Stem Cell 91-Gene Signature.
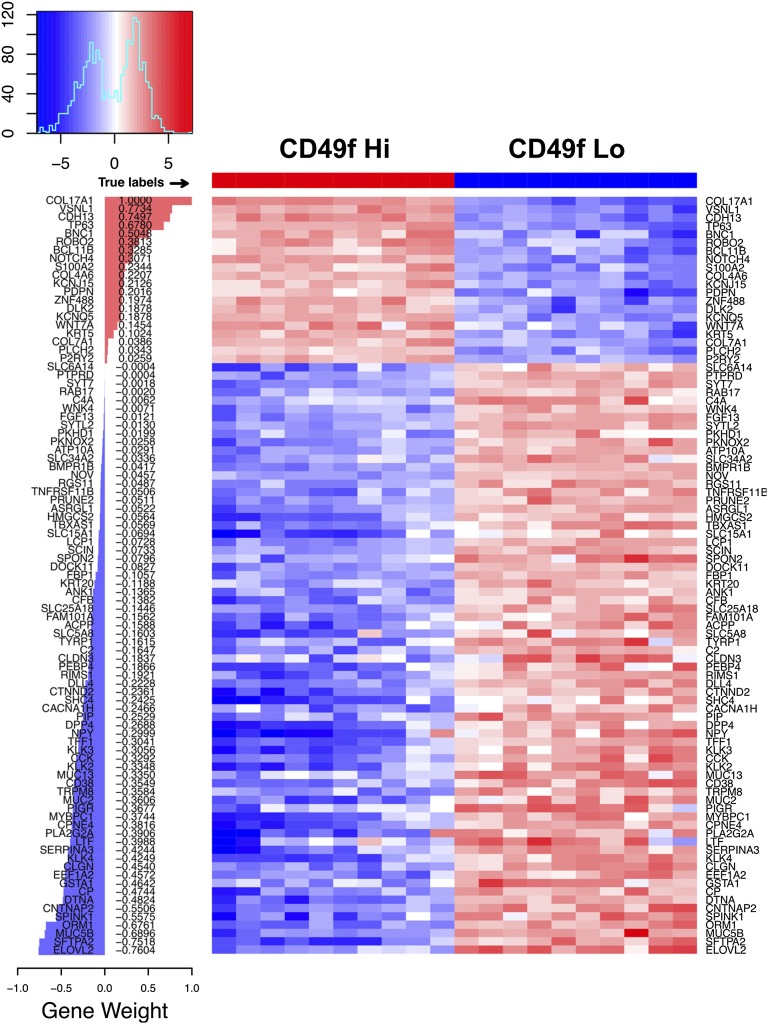
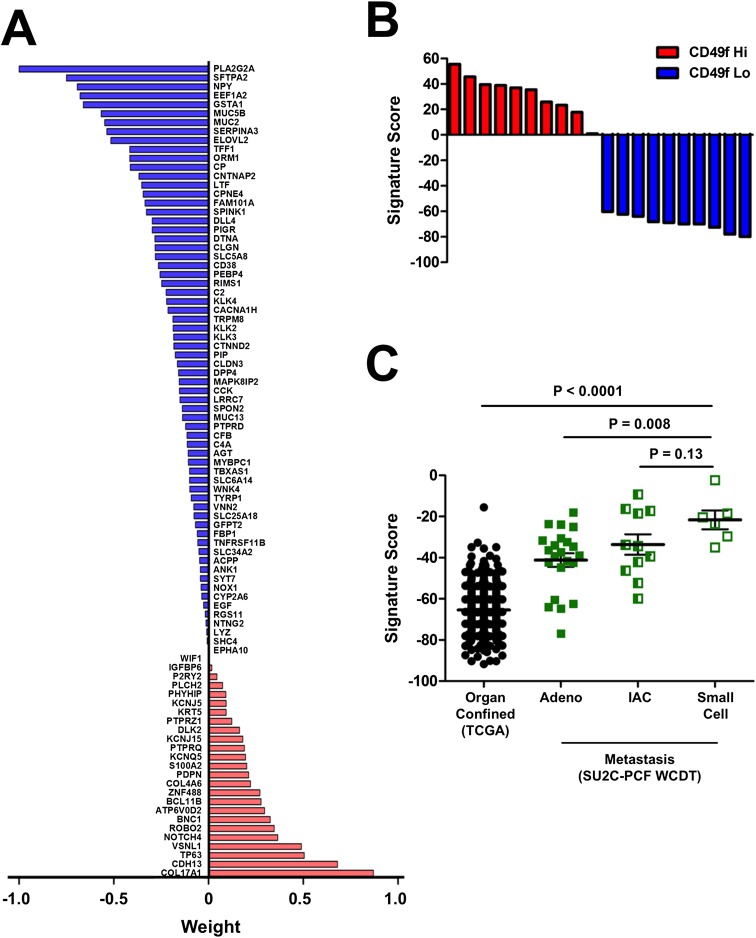
We generated a CD49f Hi basal stem cell sparse signature to investigate whether the CD49f Hi population is associated with aggressive prostate cancer. The signature was constructed using the same method as the dense signature, except we selected for the top 91 non–zero-weighted genes most predictive for the CD49f Hi and CD49f Lo dichotomy. The sparse signature contained a mixture of genes that were up-regulated in the CD49f Hi population, which carried a positive weight in the signature, and genes that were down-regulated in the CD49f Hi population, which carried a negative weight (Fig. 3). A number of genes carrying a positive weight have been associated with stem cells including NOTCH4, WNT7A, and PDPN. The majority of the genes carried a negative weight, and these genes were associated with epithelial structure maintenance, response to extracellular stimuli, and acute inflammatory responses.
Fig. 3.
Genes and associated gene weights for all 91 genes in the CD49f Hi signature.
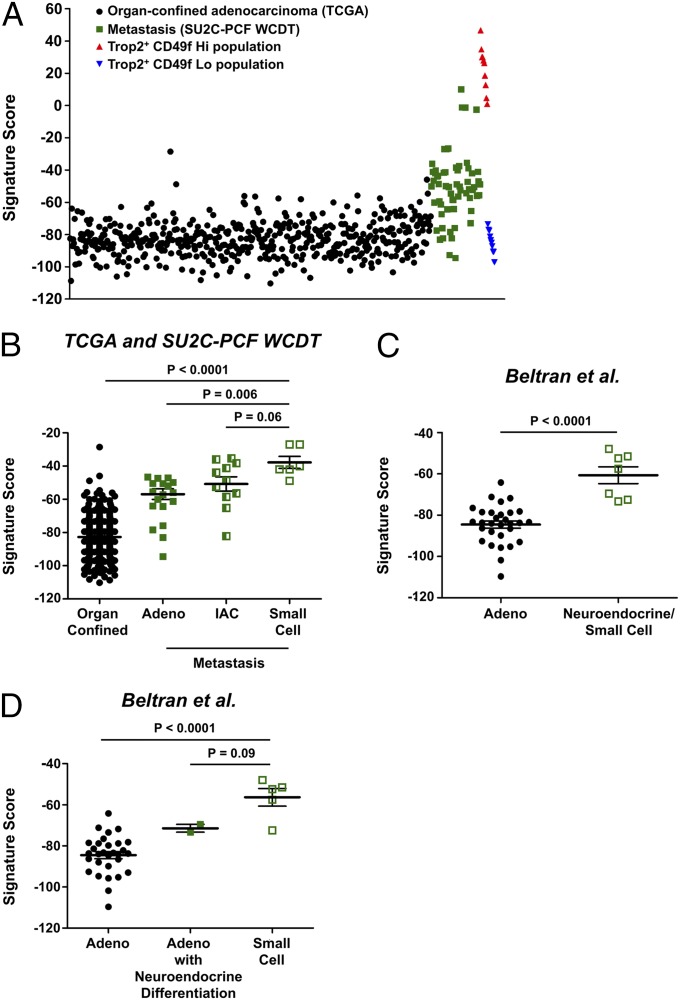
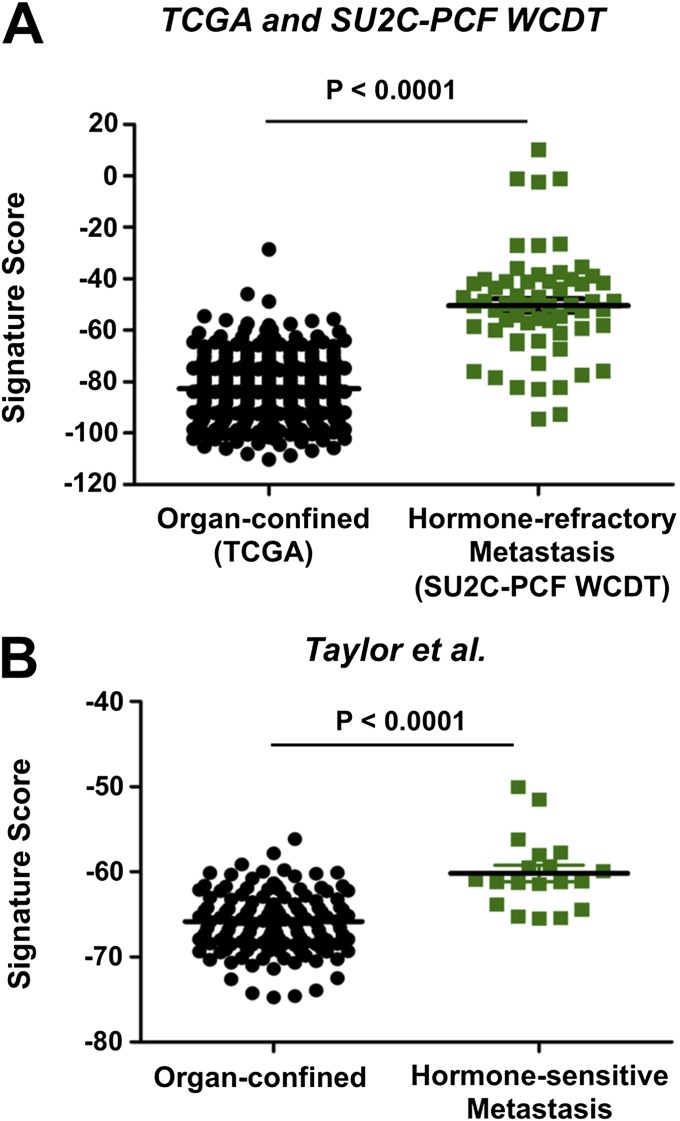
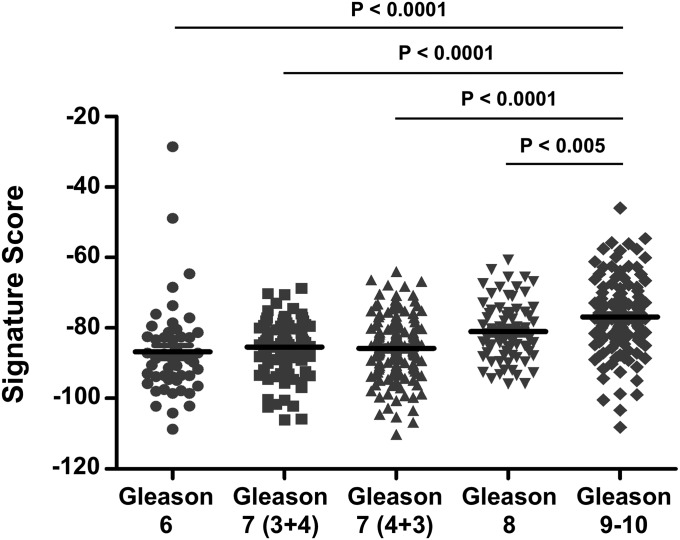
We applied the signature to organ-confined prostate adenocarcinomas from The Cancer Genome Atlas (TCGA) and to hormone-refractory metastatic prostate cancer biopsies from the Stand up to Cancer–Prostate Cancer Foundation West Coast Dream Team (SU2C-PCF WCDT) dataset to determine if aggressive prostate cancer is further enriched for the stem cell gene signature. Plotting the CD49f Hi signature scores showed that the TCGA organ-confined prostate cancer samples were similar to the sorted CD49f Lo population (Fig. 4A). This supports the GSEA findings that the CD49f Lo population is enriched for prostate cancer genes found in organ-confined prostate cancer. Moreover, as samples progressed from organ-confined to metastasis, the samples increased in the 91-gene signature toward the CD49f Hi basal stem cell population (Fig. 4A). Quantification of the signature scores showed that the aggressive SU2C-PCF WCDT samples had a significantly higher basal stem cell 91-gene signature score compared with the TCGA prostate adenocarcinomas (Fig. S5A). To determine if a possible batch effect could account for the observed differences in signature scores, we generated 30 random 91-gene signatures using an empirical phenotype-based permutation test procedure proposed in the GSEA method (37). Plotting the mean signature score for all 30 random signatures showed that the samples from all three datasets were very similar, suggesting that a batch effect was not likely responsible for the differences we saw with the CD49f Hi 91-gene signature (Fig. S6). Within organ-confined prostate adenocarcinomas, we found that samples with a Gleason score of 9 or 10 had a minor yet significantly higher CD49f Hi signature score than samples with Gleason scores of 6, 7 (3 + 4), 7 (4 + 3), or 8 (Fig. S7). We further constructed a 91-gene sparse signature comparing only the benign CD49f Hi samples (n = 5) to the CD49f Lo samples (n = 10). This benign CD49f Hi signature classified the 15 samples with 100 accuracy and showed similar results as the CD49f Hi 91-gene signature (Fig. S8). To determine if the enrichment in signature score was due to castration resistance, we applied the signature to a gene expression dataset comprised of 19 hormone-sensitive metastases and 131 organ-confined prostate cancer samples (60). The hormone-sensitive metastatic samples were significantly more enriched for the CD49f Hi gene signature compared with organ-confined prostate adenocarcinoma samples (Fig. S5B). Taken together, these results suggest that as prostate cancer progresses from an organ-confined state to metastasis, it begins to revert back to a state that resembles the normal prostate basal stem cell.
Fig. 4.
Prostate SCNC is enriched for the prostate basal stem cell signature. (A) Dot plot of CD49f Hi 91-gene signature scores for TCGA organ-confined prostate cancer (n = 498), SU2C-PCF WCDT metastatic CRPC (n = 61), Trop2+ CD49f Hi prostate basal cells (n = 10), and Trop2+ CD49f Lo prostate luminal cells (n = 10). (B) Plot of CD49f Hi signature scores of pathologist-identified pure adenocarcinoma (Adeno, n = 22), pure IAC (n = 11), and pure SCNC (Small Cell, n = 6) from the SU2C-PCF WCDT dataset and organ-confined prostate cancer samples from the TCGA dataset. (C) Plot of CD49f Hi signature scores for prostate adenocarcinoma (n = 30) and neuroendocrine/small cell (n = 7) from the Beltran et al. dataset (25). (D) Plot of CD49f Hi signature scores from the Beltran et al. dataset with the neuroendocrine/small cell samples further divided into adenocarcinoma with neuroendocrine differentiation (n = 2) and small cell (n = 5). Errors bars represent the SD. A Student t test was used to calculate the statistical significance. The distribution of scores was approximately normal (Anderson–Darling test, P > 0.05) for all categories except SU2C-PCF WCDT small cell, Beltran et al. small cell, and Beltran et al. adenocarcinoma with neuroendocrine differentiation. These phenotypes did not have enough samples to apply the Anderson–Darling test.
Fig. S5.
Hormone-refractory and hormone-sensitive prostate cancer metastases are more stem-like than organ-confined prostate adenocarcinoma. (A) Plot of Trop2+ CD49f Hi basal stem cell 91-gene signature scores for the TCGA organ-confined prostate adenocarcinoma (black circles, n = 498) and hormone-refractory metastatic (green squares, n = 61) samples. (B) Plot of CD49f Hi basal stem cell 91-gene signature scores for organ-confined (black circles, n = 131) and hormone-sensitive metastatic (green squares, n = 19) samples from the Taylor et al. dataset (60). P value was calculated using Student t test. Error bars represent the SD.
Fig. S6.
Dot plot of random 91-gene signature scores for TCGA organ-confined prostate cancer (n = 498), SU2C-PCF WCDT metastatic CRPC (n = 61), Trop2+ CD49f Hi prostate basal cells (n = 10), and Trop2+ CD49f Lo prostate luminal cells (n = 10). Signature scores for each sample were calculated for each of the 30 random 91-gene signatures, and the mean signature score for each sample was plotted. The plot is scaled the same as the similar plot for CD49f Hi 91-gene signature scores in Fig. 4A.
Fig. S7.
CD49f Hi 91-gene signature scores for Gleason 6 (n = 50), Gleason 7 (3 + 4) (n = 150), Gleason 7 (4 + 3) (n = 95), Gleason 8 (n = 72), and Gleason 9–10 (n = 125) organ-confined prostate adenocarcinomas from the TCGA prostate adenocarcinoma dataset. The P value was calculated using Student t test. Errors bars represent the SD. All distributions were approximately normal (AD test, P > 0.05), except for TCGA Gleason 6 and TCGA Gleason 7 (4 + 3).
Fig. S8.
Benign Trop2+ CD49f Hi 91-gene signature shows similar results as the CD49f Hi 91-gene signature. (A) Benign CD49f Hi 91-gene signature and associated weights. (B) Waterfall plot of benign CD49f Hi signature scores for the CD49f Hi (red) and CD49f Lo samples (blue). (C) Plot of CD49f Hi signature scores of pathologist-identified histological subtypes from the SU2C-PCF WCDT dataset and organ-confined prostate cancer samples from the TCGA dataset. P values were calculated using Student t test. Error bars represent SD.
SCNC of the Prostate Is Enriched for the CD49f Hi Signature.
The SU2C-PCF WCDT dataset contains a mixture of metastatic CRPC samples with a SCNC phenotype, an adenocarcinoma phenotype, or an intermediate phenotype termed intermediate atypical carcinoma (IAC). Because we identified a gene set associated with small cell lung cancer enriched in the CD49f Hi population, we wondered if SCNC of the prostate was also enriched for the stem cell signature. When we applied the signature to the SU2C-PCF WCDT dataset, the 91-gene signature was enriched in the SCNC samples compared with the adenocarcinoma and IAC phenotypes (Fig. 4B). We also applied the signature to a separate dataset that contains gene expression data for seven prostate neuroendocrine/small cell carcinoma samples and 30 prostate adenocarcinomas (25). The neuroendocrine/small cell samples were also significantly enriched for the CD49f Hi signature compared with the adenocarcinoma samples (Fig. 4C). Interestingly, when the neuroendocrine/small cell samples were further subdivided into pure small cell or adenocarcinoma with neuroendocrine differentiation, the pure small cell samples had a higher signature score than the adenocarcinoma with neuroendocrine differentiation (Fig. 4D). This result mimics what was seen in the SU2C-PCF WCDT dataset. Taken together, these data suggest that SCNC of the prostate is more stem-like than other histological subtypes of metastatic and organ-confined prostate cancer.
The CD49f Hi Population and Prostate SCNC Share a Gene Network Associated with E2F Targets.
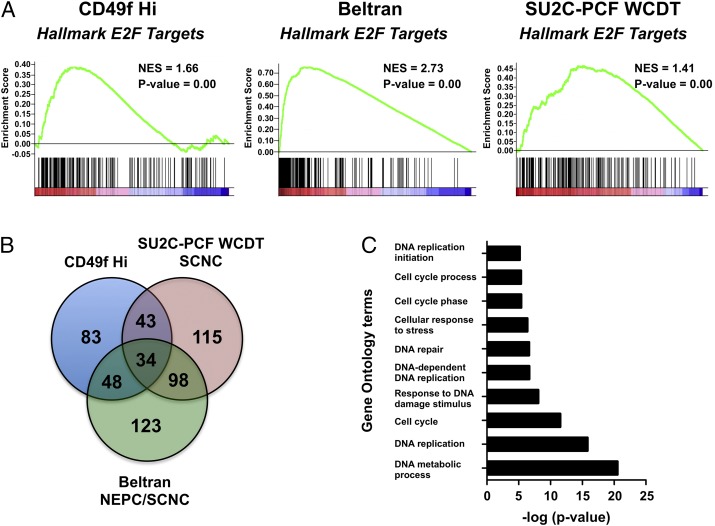
To identify common gene networks between the CD49f Hi population and prostate SCNC, we ran GSEA using the MSigDB Hallmark gene category on three separate dense gene signatures: (i) CD49f Hi versus CD49f Lo, (ii) SU2C-PCF WCDT pathologist called SCNC versus non-SCNC, and (iii) Beltran et al. NEPC/SCNC versus prostate adenocarcinoma. After filtering for Hallmark gene sets that met the P value and FDR cutoff, we found that all three signatures were enriched for a gene network associated with E2F targets (Fig. 5A). We further performed leading-edge gene analysis on the E2F targets gene set and identified 34 genes common to all three signatures (Fig. 5B and Table S2). To gain further insight into the biological processes in which these genes may be involved, we used the database for annotation, visualization, and integrated discovery (DAVID) (61). We found that these 34 genes were associated with biological processes such as DNA replication, DNA repair, and cell cycling (Fig. 5C).
Fig. 5.
The prostate basal stem cell population and prostate SCNC share a gene network associated with E2F targets. (A) GSEA plots for the E2F targets gene set significantly enriched in the CD49f Hi, SU2C-PCF WCDT SCNC, and Beltran NEPC/SCNC gene signatures. (B) Venn diagram of leading-edge genes from the E2F targets gene set found between the CD49f Hi stem cell signature, the SU2C-PCF WCDT SCNC, and the Beltran NEPC/SCNC gene signatures. (C) The 10 most statistically enriched gene ontology biological processes identified from the 34 common leading-edge genes.
Table S2.
Common hallmark E2F targets leading-edge genes found within all three gene signatures
| Gene symbol | Gene name |
| PTTG1 | Pituitary tumor-transforming 1 |
| ANP32E | Acidic (leucine-rich) nuclear phosphoprotein 32 family member E |
| GINS1 | GINS complex subunit 1 |
| UBE2T | Ubiquitin-conjugating enzyme E2T |
| CDCA3 | Cell division cycle associated 3 |
| TCF19 | TCF19 |
| BARD19 | BRCA1-associated RING domain 1 |
| CIT | Citron (rho-interacting, serine/threonine kinase 21) |
| HELLS | Helicase, lymphoid specific |
| MCM6 | Minichromosome maintenance complex component 6 |
| TIPIN | TIMELESS interacting protein |
| MXD3 | MAX dimerization protein 3 |
| DIAPH3 | Diaphanous-related formin 3 |
| MCM4 | Minichromosome maintenance complex component 4 |
| POLD1 | Polymerase (DNA directed), delta 1, catalytic subunit |
| TRIP13 | Thyroid hormone receptor interactor 13 |
| TOP2A | Topoisomerase (DNA) II alpha 170 kDa |
| TMPO | Thymopoietin |
| PCNA | Proliferating cell nuclear antigen |
| DNMT | DNA (cytosine-5-)-methyltransferase 1 |
| PLK1 | Polo-like kinase 1 |
| POLA2 | Polymerase (DNA directed), alpha 2, accessory subunit |
| CHEK2 | Checkpoint kinase 2 |
| PRKDC | Protein kinase, DNA-activated, catalytic peptide |
| CKS1B | CDC28 protein kinase regulatory subunit 1B |
| SSRP1 | Structure specific recognition protein 1 |
| MCM3 | Minichromosome maintenance complex component 3 |
| DSCC1 | DNA replication and sister chromatid cohesion 1 |
| MCM2 | Minichromosome maintenance complex component 2 |
| GINS3 | GINS complex subunit 3 |
| NASP | Nuclear autoantigen sperm protein |
| DCLRE1B | DNA cross-link repair 1B |
| NUP107 | Nucleoporin 107 kDa |
| SUV39H1 | Suppressor of variegation 3–9 homolog 1 |
Discussion
In this study, we transcriptionally profiled sorted human prostate epithelial populations using high-throughput RNA-seq to show that subtypes of metastatic CRPC vary in their stemness properties, with metastatic SCNC being the most stem-like. Although previous studies have used curated stem cell signatures to compare stemness between organ-confined and metastatic disease, this is the first study to our knowledge that (i) has developed a prostate stem cell weighted gene signature from sorted, uncultured, human prostate basal, and luminal cells; (ii) showed that an increase in neuroendocrine differentiation within late-stage, metastatic human prostate cancer leads to an increase of a stem-like transcriptional state; and (iii) has shown that SCNC and the Trop2+ CD49f Hi prostate basal stem cell share a transcriptional program associated with E2F targets.
The acquisition of stemness properties and increased activation of developmental signaling networks in aggressive cancer phenotypes has been well documented. Studies using breast and intestinal tissues have mapped the transcriptional profiles of the poorly differentiated, aggressive subtypes back to stem cell-like populations found within normal human tissue (14, 15). Our work using a CD49f Hi basal stem cell gene signature derived from freshly isolated human prostate epithelial cells supports previous reports that prostate cancer increases in a stem-like transcriptional state as it progresses from organ-confined to metastatic disease (10, 12). These previous studies used gene signatures derived from ESCs or common genes between murine metastatic prostate cancer tissue and cultured neurospheres. Interestingly, our basal stem cell gene signature had little to no overlap with either of the signatures. It is possible that all three signatures are examining the same transcriptional profile from different, narrow perspectives, enabling them to reach similar conclusions.
We found that even within CRPC metastasis, there is a difference in their degree of stemness. Metastatic samples with a SCNC phenotype were more stem-like than either metastasis with adenocarcinoma or an intermediate IAC phenotype. This is likely a general phenomenon in prostate cancer metastasis, as two different datasets containing samples that varied in their treatment regimen showed that SCNC had higher CD49f Hi signature scores than the other phenotypes within their respective studies. One question still unanswered is whether organ-confined prostate SCNC and its metastatic counterpart use different stem cell gene networks. The infrequency of organ-confined prostate SCNC (<1%) has delayed in-depth transcriptional profiling of this disease; however, these studies would be highly informative to understanding the core stem-like transcriptional component of SCNC.
Small cell carcinoma is not only found in the prostate but can present itself in a number of other anatomical sites. Little is known about the molecular underpinnings of this disease or if small cell carcinomas in different tissues share common molecular traits. Molecular profiling of the most common small cell carcinoma, small cell lung cancer, suggests that there is a stem cell component to the disease. Small cell lung cancer exhibits SOX2 amplification in 34% of patients and activation of hedgehog signaling (62, 63). Similarly, immunohistochemistry has identified SOX2 expression in a majority of patients with metastatic NEPC (50). Deregulation of the E2F-Rb pathway, which is commonly altered in small cell carcinoma, can lead to overexpression of PRC2 genes (64, 65). These genes are vital for maintaining self-renewal capacity in embryonic and adult stem cells (66). Recent evidence has also shown Rb alterations can facilitate reprogramming of fibroblasts to a pluripotent state through derepression of pluripotency factors such as SOX2 (67). In the CD49f Hi population, we found enrichment of both E2F and SOX2 targets, further supporting that these networks may be part of a stem-like component common to small cell carcinomas.
Cellular plasticity is another hallmark characteristic of stem cells that is also seen in small cell carcinomas. Studies in the lung, bladder, and prostate have shown that small cell carcinomas can share genetic alterations with a different coexisting carcinoma (68–70). These results can be explained by transdifferentiation, de-differentiation, or outgrowth of both phenotypes from a common stem-like clone. Our laboratory has shown that lentiviral introduction of NMYC and myristoylated AKT into human benign prostate CD49f Hi cells can initiate the formation of biphenotypic tumors that have an adenocarcinoma and SCNC component. This supports the idea that a tissue stem cell may be predisposed to forming biphenotypic tumors when challenged with the correct combination of oncogenic insults. In vitro, the prostate adenocarcinoma cell line LNCaP can display neuroendocrine differentiation when exposed to numerous stimuli including hormone-depleted media (71). This observation along with the increased incidence of SCNC in metastatic CRPC has led many to believe that the appearance of neuroendocrine differentiation or SCNC may be a resistance mechanism to androgen deprivation therapy and AR-targeted drugs. It is possible that multiple mechanisms may lead to the appearance of SCNC, and future work is needed to elucidate the pathways or gene networks responsible for this observed phenotypic plasticity. Moreover, further investigation is needed into the therapeutic targeting of these molecular programs that govern the stem-like component of SCNC.
Experimental Procedures
Tissue Procurement.
The acquisition of primary human prostate tissue from radical prostatectomy, dissociation into single cells, and FACS purification has previously been described (27).
Library Construction and RNA-seq of Epithelial Populations.
RNA was isolated using RNeasy Mini Kit (QIAGEN), and RNA quality was tested using an Agilent Bioanalyzer 2100 Eukaryote Total RNA Pico assay. Samples with a RNA integrity number (RIN) > 8 were used for construction of RNA-seq libraries. RNA-seq libraries were constructed using the Nugen kit. The RNA-seq libraries, after a final purification and after adapter ligation, were quantitated using both the Agilent 2100 Bioanalyzer High Sensitivity DNA assay and Qubit dsDNA HS assay (Thermo Fisher), per the manufacturer’s recommended protocols. The pooled multiplexed libraries were sequenced to generate 100-bp paired-end reads on an Illumina HiSEq 2000 platform. Raw RNA-seq files were mapped to the hg19 human genome using MapSplice, and transcripts were quantified using RNA-seq by expectation-maximization (RSEM).
Unsupervised Clustering, Differential Expression Analysis, and SPIA.
Samples were clustered based on genes that had expression values greater or equal to 1 SD from the mean expression value for all samples. Unsupervised hierarchical clustering was performed using Cluster 3.0 with Pearson correlation and complete linkage analysis and visualized using Jave TreeView. Differentially expression analysis was performed using the LIMMA R/Bioconductor package (72, 73). We kept genes with greater than or equal to twofold differential expression between the CD49f Hi and CD49f Lo populations with a P value greater than or equal to 0.05. SPIA was performed using the Graphite Web interface with an input of genes with twofold differential expression between the CD49f Hi and CD49f Lo populations and the KEGG pathway database (74). We filtered for pathways with a FDR lower than 0.05.
MARINa Analysis.
We created a compendium of TFs and their targets (TF regulons) by combining information from four databases: SuperPathway (43), Literome (44), Multinet (45), and ChEA (46). We ran MARINa master regulator analysis using the previously described TF compendium. MARINa TF scores capture each TF’s relative activity between two cohorts of interest. The activity score is derived from a combined view of the expression levels of each TF’s regulon, based on the following steps: (i) The TF regulon is split into positively and negatively regulated sets by measuring the Spearman correlation between the expression of the TF and that of each of its targets. (ii) A t statistic derived from the difference in gene expression between the two classes of interest is computed for each gene. All genes are ranked based on their t statistics to produce a CD49f MARINa gene signature. (iii) Each TF’s activation and inhibition regulons are examined for enrichment in the high or low end of the ranked gene list. The rankings of the positively and negatively regulated genes are then combined and examined simultaneously. A TF whose two target sets show consistent enrichment (i.e., the activated set is enriched for highly ranked genes and the inhibited set is enriched for lowly ranked ones, or vice versa) receives the highest/lowest activity scores, respectively. MARINa activity scores are therefore more robust measures of activity than differences in the individual expression of the TF or its targets. We compared relative TF activity between the CD49f Hi (n = 10) and CD49 Lo (n = 10) samples. We ran MARINa with its default settings, which scored TFs with a minimum of 25 targets.
Development of CD49f Hi Basal Stem Cell Gene Signatures.
We constructed a computational model to recognize CD49f Hi prostate basal stem cells by formulating a dichotomy between CD49f Hi and CD49f Lo cells. Given this dichotomy, we trained a logistic regression model with elastic net regularization (38). The elastic net regularization is characterized by two parameters: one for the ridge regression term, and one for the LASSO term. For the 20,500-gene-dense signature, we set the LASSO term penalty coefficient to 0.0 and leaving the ridge regression term coefficient at 1.0. For the 91-gene-sparse signature, we fixed the ridge regression term coefficient at 1.0 and the LASSO term parameter at 0.1. We validated our model in silico through leave-pair-out cross-validation. This cross-validation scheme iterates over all possible pairs of one CD49f Hi sample and one CD49f Lo sample, withholding each pair in turn from training. The model is then trained using all other samples and applied back to the withheld pair for evaluation. In our experiments, we found that the model was able to identify CD49f Hi and CD49f Lo samples with 100% accuracy. GSEA was performed on the 20,500-dense weighted gene signature using GSEA v2.2 with 1,000 gene set permutations. A gene set was considered to be significantly enriched in one of the two groups when the P value was lower than 0.05 and the FDR was lower than 0.25 for the corresponding gene set.
Comparing CD49f Hi Gene Signature to Other Stem Cell Signatures.
We obtained human stem cell signatures and stem cell-associate gene modules from Merlos-Suárez et al. (14), Pece et al. (15), Ben-Porath et al. (54), Creighton et al. (55), Lim et al. (56), Kim et al. (57), and Wong et al. (58). For each curated signature, we selected genes that were up-regulated for the signature indicated and had an associated Human Genome Organization (HUGO) ID. The name of the signature and the number of genes associated with each stem cell signature are as follows: Lim Mammary Stem Cell (899 genes), Lim Mammary Luminal Progenitor (342 genes), Lim Mammary Luminal Mature (534 genes), Kim Myc Module (355 genes), Kim Core Module (75 genes), Wong ESC-like (1,242 genes), Pece Mammary Stem Cell (818 genes), Creighton Breast Cancer Stem Cell (111 genes), Ben-Porath NOS Targets (179 genes), Ben-Porath Myc Targets 1 (228 genes), Ben-Porath Myc Targets 2 (774 genes), Ben-Porath ES Exp 1 (380 genes), Ben-Porath ES Exp 2 (40 genes), Ben-Porath PRC2 Targets (642 genes), Merlos-Suarez Intestinal Stem Cell (52 genes), Eppert Leukemic Stem Cell (41 genes), and Eppert Hematopoietic Stem Cell (125 genes). To compare the CD49f Hi signature to curated stem cell signatures, we ran GSEA using 1,000 permutations.
CD49f Hi Signature Scores for Prostate Cancer Phenotypes.
We downloaded the level 3 TCGA prostate adenocarcinoma RNA-seq from the TCGA Data portal (June 2015 data freeze). The gene expression data for hormone-sensitive organ-confined and metastatic prostate cancer was downloaded from GSE20134. CD49f Hi signature scores were computed for each sample within the sorted epithelial populations and prostate cancer subtypes by multiplying the weight for each gene in the signature by the normalized log2 expression value for that gene within the sample and summing the values for all 91 genes from the signature. All samples including the Trop2+ CD49f Hi samples, Trop2+ CD49f Lo samples, SU2C-PCF WCDT metastatic samples, and the TCGA prostate adenocarcinoma samples went through the same mapping and expression pipeline. A scaling value was added to the sum for all of the samples. To assess the robustness of signature scores and investigate the presence of a batch effect, we generated 30 random 91-gene signatures using an empirical phenotype-based permutation test procedure proposed in the GSEA method (37). Specifically, we randomly permuted the CD49f Hi and CD49f Lo labels and reran our method using this new permutation to produce a background weighted gene signature. The random 91-gene signature scores for each sample were computed using the same method as the CD49f Hi 91-gene signature. A Student t test was used to calculate the statistical significance when comparing two prostate cancer phenotypes.
Identification of Common Stem Cell and SCNC Gene Networks.
Dense gene signatures were constructed for pathologist-identified small cell (SCNC) versus non-small cell (non-SCNC) samples from the SU2C-PCF WCDT dataset and NEPC/SCNC versus prostate adenocarcinoma from Beltran et al. using the same method described for the CD49f Hi 20,500-gene-dense signature. This gave an 18,935-gene SU2C-PCF SCNC versus non-SCNC weighted signature and a 20,500-gene Beltran NEPC/SCNC versus prostate adenocarcinoma gene signature. GSEA was run on the 20,500-gene CD49f Hi versus CD49f Lo, 18,935-gene SU2C-PCF SCNC versus non-SCNC, and 20,500-gene Beltran NEPC/SCNC versus prostate adenocarcinoma gene signatures using the Hallmarks category in MSigDB. A cutoff of P ≤ 0.05 and FDR ≤ 0.25 was applied to identify statistically enriched gene sets. Leading-edge genes analysis was used to identify genes that drove a signature’s enrichment for each specific gene network. The common leading-edge genes found within all three signatures were uploaded to the DAVID website (david.abcc.ncifcrf.gov/). Gene Ontology terms for biological processes were then identified.
Supplementary Material
Acknowledgments
We thank members of the O.N.W. and J.M.S. laboratories for helpful comments and discussion on the manuscript. We thank the Tissue Procurement Core Laboratory at University of California, Los Angeles (UCLA) for assistance on tissue processing and H&E staining, UCLA Clinical Microarray Core for construction of the RNA-seq barcoded libraries, and the High Throughput Sequencing Core at the Eli and Edythe Broad Stem Cell Research Center for performing RNA-seq. This work was supported by UCLA Tumor Immunology Training Program T32 CA009120 (to B.A.S.). J.M.S. is supported by NIH Grant U24-CA143858. O.N.W. is an investigator of the Howard Hughes Medical Institute and partially supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research. O.N.W. and J.M.S. are supported by Stand up to Cancer/American Association for Cancer Research/Prostate Cancer Foundation Grant SU2C-AACR-DT0812 (O.N.W. co-principal investigator). This research grant is made possible by the generous support of the Movember Foundation. Stand up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 14406.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518007112/-/DCSupplemental.
References
- 1.Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell. 2014;14(3):306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24(4):410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488(7412):527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Santagata S, Ligon KL, Hornick JL. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am J Surg Pathol. 2007;31(6):836–845. doi: 10.1097/PAS.0b013e31802e708a. [DOI] [PubMed] [Google Scholar]
- 10.Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci USA. 2011;108(52):21276–21281. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shats I, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2011;71(5):1772–1780. doi: 10.1158/0008-5472.CAN-10-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115(6):1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieter SM, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9(4):357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Merlos-Suárez A, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8(5):511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Pece S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Baccelli I, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31(6):539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 17.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 18.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karhadkar SS, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431(7009):707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 20.Acevedo VD, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12(6):559–571. doi: 10.1016/j.ccr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Wang Y, DeGraff DJ, Wills ML, Matusik RJ. Wnt/β-catenin activation promotes prostate tumor progression in a mouse model. Oncogene. 2011;30(16):1868–1879. doi: 10.1038/onc.2010.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoyanova T, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci USA. 2013;110(50):20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terry S, Beltran H. The many faces of neuroendocrine differentiation in prostate cancer progression. Front Oncol. 2014;4(60):60. doi: 10.3389/fonc.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadal R, Schweizer M, Kryvenko ON, Epstein JI, Eisenberger MA. Small cell carcinoma of the prostate. Nat Rev Urol. 2014;11(4):213–219. doi: 10.1038/nrurol.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beltran H, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci USA. 2008;105(52):20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329(5991):568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitaker HC, Warren AY, Eeles R, Kote-Jarai Z, Neal DE. The potential value of microseminoprotein-beta as a prostate cancer biomarker and therapeutic target. Prostate. 2010;70(3):333–340. doi: 10.1002/pros.21059. [DOI] [PubMed] [Google Scholar]
- 29.Sørensen KD, et al. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br J Cancer. 2013;108(2):420–428. doi: 10.1038/bjc.2012.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez JC, et al. Apolipoprotein D expression in benign and malignant prostate tissues. Int J Surg Investig. 2000;2(4):319–326. [PubMed] [Google Scholar]
- 31.Begley LA, et al. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10(3):244–254. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci. 2009;39(4):331–337. [PubMed] [Google Scholar]
- 33.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 34.Cooper CS, et al. ICGC Prostate Group Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47(4):367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risk MC, et al. Differential gene expression in benign prostate epithelium of men with and without prostate cancer: Evidence for a prostate cancer field effect. Clin Cancer Res. 2010;16(22):5414–5423. doi: 10.1158/1078-0432.CCR-10-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform. 2015;16(1):59–70. doi: 10.1093/bib/bbt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 39.Tarca AL, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aytes A, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25(5):638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lefebvre C, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6(377):377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon H, Quirk C, DeZiel C, Heckerman D. Literome: PubMed-scale genomic knowledge base in the cloud. Bioinformatics. 2014;30(19):2840–2842. doi: 10.1093/bioinformatics/btu383. [DOI] [PubMed] [Google Scholar]
- 45.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLOS Comput Biol. 2013;9(3):e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lachmann A, et al. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forrest MP, Waite AJ, Martin-Rendon E, Blake DJ. Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013;8(8):e73169. doi: 10.1371/journal.pone.0073169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flora A, Garcia JJ, Thaller C, Zoghbi HY. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci USA. 2007;104(39):15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ugolkov AV, Eisengart LJ, Luan C, Yang XJ. Expression analysis of putative stem cell markers in human benign and malignant prostate. Prostate. 2011;71(1):18–25. doi: 10.1002/pros.21217. [DOI] [PubMed] [Google Scholar]
- 50.Yu X, et al. SOX2 expression in the developing, adult, as well as, diseased prostate. Prostate Cancer Prostatic Dis. 2014;17(4):301–309. doi: 10.1038/pcan.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Badve S, et al. FOXA1 expression in breast cancer--Correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13(15 Pt 1):4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 53.Bernardo GM, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32(5):554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 57.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor BS, et al. Integrative genomsic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 62.Rudin CM, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44(10):1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park KS, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med. 2011;17(11):1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coe BP, et al. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One. 2013;8(8):e71670. doi: 10.1371/journal.pone.0071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 67.Kareta MS, et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16(1):39–50. doi: 10.1016/j.stem.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng L, et al. Molecular genetic evidence for a common clonal origin of urinary bladder small cell carcinoma and coexisting urothelial carcinoma. Am J Pathol. 2005;166(5):1533–1539. doi: 10.1016/S0002-9440(10)62369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williamson SR, et al. ERG-TMPRSS2 rearrangement is shared by concurrent prostatic adenocarcinoma and prostatic small cell carcinoma and absent in small cell carcinoma of the urinary bladder: Evidence supporting monoclonal origin. Mod Pathol. 2011;24(8):1120–1127. doi: 10.1038/modpathol.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen R, et al. Transdifferentiation of cultured human prostate cancer cells to a neuroendocrine cell phenotype in a hormone-depleted medium. Urol Oncol. 1997;3(2):67–75. doi: 10.1016/s1078-1439(97)00039-2. [DOI] [PubMed] [Google Scholar]
- 72.Smith GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3(1):Article 3.
- 73.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sales G, Calura E, Martini P, Romualdi C. Graphite Web: Web tool for gene set analysis exploiting pathway topology. Nucleic Acids Res. 2013;41(Web Server Issue) W1:W89–W97. doi: 10.1093/nar/gkt386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.