Significance
A tumor comprising many cells can be compared to a natural population with many individuals. The amount of genetic diversity reflects how it has evolved and can influence its future evolution. We evaluated a single tumor by sequencing or genotyping nearly 300 regions from the tumor. When the data were analyzed by modern population genetic theory, we estimated more than 100 million coding region mutations in this unexceptional tumor. The extreme genetic diversity implies evolution under the non-Darwinian mode. In contrast, under the prevailing view of Darwinian selection, the genetic diversity would be orders of magnitude lower. Because genetic diversity accrues rapidly, a high probability of drug resistance should be heeded, even in the treatment of microscopic tumors.
Keywords: intratumor heterogeneity, genetic diversity, neutral evolution, cancer evolution, natural selection
Abstract
The prevailing view that the evolution of cells in a tumor is driven by Darwinian selection has never been rigorously tested. Because selection greatly affects the level of intratumor genetic diversity, it is important to assess whether intratumor evolution follows the Darwinian or the non-Darwinian mode of evolution. To provide the statistical power, many regions in a single tumor need to be sampled and analyzed much more extensively than has been attempted in previous intratumor studies. Here, from a hepatocellular carcinoma (HCC) tumor, we evaluated multiregional samples from the tumor, using either whole-exome sequencing (WES) (n = 23 samples) or genotyping (n = 286) under both the infinite-site and infinite-allele models of population genetics. In addition to the many single-nucleotide variations (SNVs) present in all samples, there were 35 “polymorphic” SNVs among samples. High genetic diversity was evident as the 23 WES samples defined 20 unique cell clones. With all 286 samples genotyped, clonal diversity agreed well with the non-Darwinian model with no evidence of positive Darwinian selection. Under the non-Darwinian model, MALL (the number of coding region mutations in the entire tumor) was estimated to be greater than 100 million in this tumor. DNA sequences reveal local diversities in small patches of cells and validate the estimation. In contrast, the genetic diversity under a Darwinian model would generally be orders of magnitude smaller. Because the level of genetic diversity will have implications on therapeutic resistance, non-Darwinian evolution should be heeded in cancer treatments even for microscopic tumors.
The level of genetic diversity in a natural population is determined by several evolutionary forces, including mutation, genetic drift, migration, and natural selection (1–3). Tumors can be regarded as asexual populations of cells, so they are subjected to similar forces to those of natural populations (4–7). Therefore, the genetic diversity in tumors of the same patient is informative about how various forces drive their evolution. The level of diversity may also influence how tumors respond to environmental perturbations, either natural or medical (5–7). In the prevailing view, Darwinian selection for and against new mutations is the main driving force of intratumor diversity (4, 8–18). Because selection generally reduces genetic diversity within populations (19–21), studies assuming Darwinian evolution usually described MALL (the total number of coding region mutations within the whole tumor) in the range of tens to hundreds of coding mutations (22, 23).
Despite its wide acceptance, the Darwinian view has never been subjected to hypothesis testing, by which the observed diversity is compared with quantitative predictions. This study is to our knowledge the first one that uses high-density sampling in a single tumor and compares the observations with theoretical predictions. In this test, we consider a null model of non-Darwinian evolution in which MALL is a function of N (population size), u (mutation rate per generation), and growth parameters. In tumors, N is large, generally 106, and u is the mutation rate of the entire functional portion of the genome (at the level of 10−2 per cell division) (18, 24). Hence, the expected genetic diversity of tumors by non-Darwinian evolution would be large, probably on the order of millions of mutations, most of which are present at low frequencies (25).
We ask whether the observed intratumor genetic diversity can be largely explained by non-Darwinian forces and we invoke positive selection only when the null model of non-Darwinian evolution is rejected. There was a controversy in molecular evolution generally known as the neutralism–selectionism debate (1, 26, 27). In the postdebate modern view, genetic polymorphisms in natural populations are largely consistent with the non-Darwinian model (1–3, 26–28). There are further reasons to question the efficacy of selection within populations of cells that make up tumors (Discussion). For instance, although selection against nonsynonymous mutations is nearly universal in natural species (1, 3, 27), selection against such mutations in tumors is not apparently stronger than against synonymous ones (29).
In the recent literature, there has been increasingly more attention on assessing the non-Darwinian model of tumor evolution vs. the prevailing Darwinian view (30, 31). Tao et al. (31) studied 12 cases of multitumor hepatocellular carcinomas (HCCs) and concluded that competition often occurs between tumors large enough to be visible. In contrast, the genetic diversity contained within the same tumor does not deviate from the predictions of the non-Darwinian model. A caveat is that whereas the number of population samples used in testing Darwinian selection in natural populations is often in the hundreds, the sample number rarely exceeds 10 in intratumor studies (12, 13, 15–18, 30, 31). Therefore, the power to reject the null model in tumor studies might have been too low. Clearly, there is a need to sample a large number of regions in one single tumor. In this study we sampled close to 300 regions to examine the spatial distribution of single-nucleotide variants and to estimate the amount of genetic diversity in the tumor. We used these data to give a rigorous test of the null hypothesis of non-Darwinian evolution.
Results
Sampling, Sequencing/Genotyping, and Mutation Calling.
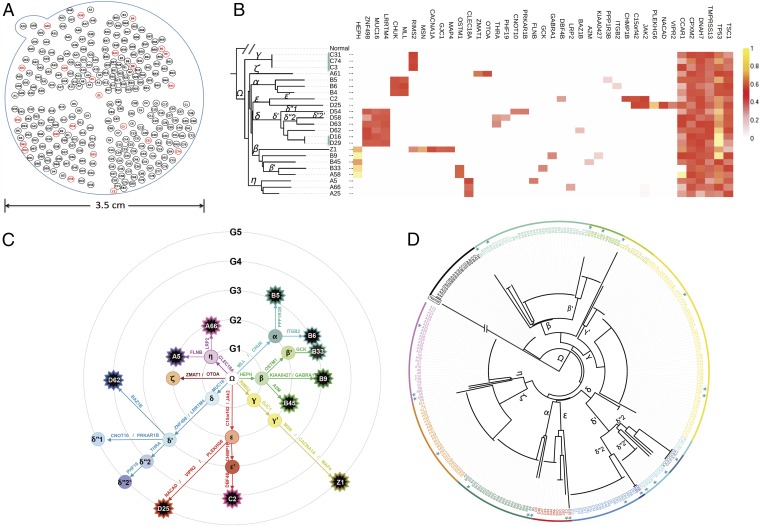
The honeycomb-like microdissections yielded 286 tumor samples on a plane of a single HCC tumor (Materials and Methods, section 1), each sample being a cylinder of 0.5 mm in diameter and 1 mm in height (Fig. 1A and Fig. S1). A sample contained, on average, 20,000 cells (Fig. S2 and Materials and Methods, section 2) and permitted precise delineation of clones. Fig. 1A displays the spatial distribution of the 286 tumor samples, which were evenly distributed among the four quadrants of the tumor slice, labeled A–D clockwise. The 23 sequenced samples (red color in Fig. 1A) were also evenly distributed, with 12 on the periphery of the tumor and 11 in the interior.
Fig. 1.
Sampling scheme and clonal genealogy of HCC-15. (A) Samples were taken from a 1-mm-thick slice cut through the middle of a HCC tumor, 3.5 cm in diameter. Of the 286 samples, 23 were subjected to whole-exome sequencing (red numbers) and the rest (black numbers) were used in genotyping for mutations discovered in sequencing (Materials and Methods, sections 1–5). The numbers correspond with those of Fig. 2. Across the sequenced samples, the average read depth was 74.4× (Dataset S1). On average, these samples contained 85% cancerous cells estimated by ABSOLUTE (52). This level of purity is consistent with previous reports regarding hepatic tumor samples (12), especially when the sample volumes are small (∼20,000 cells). Pathology reports, when available for the matched HCC samples, generally agreed with the purity estimates. (B) All 35 polymorphic nonsynonymous mutations in the sequenced samples are shown in the heat map, which depicts the observed frequencies (from 0 in white to 1 in yellow) with mutation names at the top of the map. Each row presents the mutations in a sequenced sample. Far Right shows six fixed mutations that are potential drivers. Left shows the genealogy of the 24 samples. Only two clones, indicated by blue bars, are represented by more than one sample. (C) The genealogy of clones arranged to reflect their spatial relationships. The ancestral clone, Ω, is in the middle and the descendant clones radiate outward. These clones are arranged on six rings with each outer ring having one more nonsynonymous mutation (indicated) than its interior neighbor. Each star symbol represents a singleton clone. (D) The expanded genealogy that includes all 286 samples. The blue stars designate the sequenced samples.
Fig. S1.
Sample collection with honeycomb-like microdissection. The tumor tissues were embedded in optimal cutting temperature (OCT) compound and sliced into 1-mm-thick pieces. One 1-mm-thick slice was subjected to high-density microdissection, using the Harris Micropunch. In total, the slice yielded 286 microsections, including one in the middle of the slice (Z1), and 60–80 microsections in each of the four quadrants (A–D). All sample IDs were marked, indicating their position in the tumor. The 23 sequenced samples were marked by red circles.
Fig. S2.
Estimated number of cells in microdissected tissues. The x axis represents the samples; the y axis is the estimated number of cells based on the amount of DNA extracted from each of (A) 23 whole-exome sequenced samples and (B) 286 genotyped samples.
For sequencing, the average read depth was 74.4× per sample (Dataset S1), yielding a total of >1,700× for the plane of Fig. 1A (SI Materials and Methods). With the additional genotyping over 286 samples, the coverage is to our knowledge the highest ever carried out on a single tumor. The average sample purity is 85% as described in the legend of Fig. 1A (Materials and Methods, section 3). In total, we found 269 single-nucleotide variations (SNVs) in coding regions or at splice sites (Materials and Methods, sections 4 and 5 and Dataset S2). Due to the dense sampling, SNVs found in multiple samples are unambiguous by the cross-validation among samples, using whole-exome sequencing (WES) and/or Sequenom. Singleton SNVs (i.e., occurring in only one sample) required additional validations. By Sequenom genotyping, and sometimes Sanger sequencing, all singleton SNVs presented have been confirmed to be true positives (Datasets S2 and S3 and Fig. S3). Therefore, the final SNV calls for this study are considered free of false positives. Furthermore, given the large number of samples, false negatives would likely be negligible.
Fig. S3.
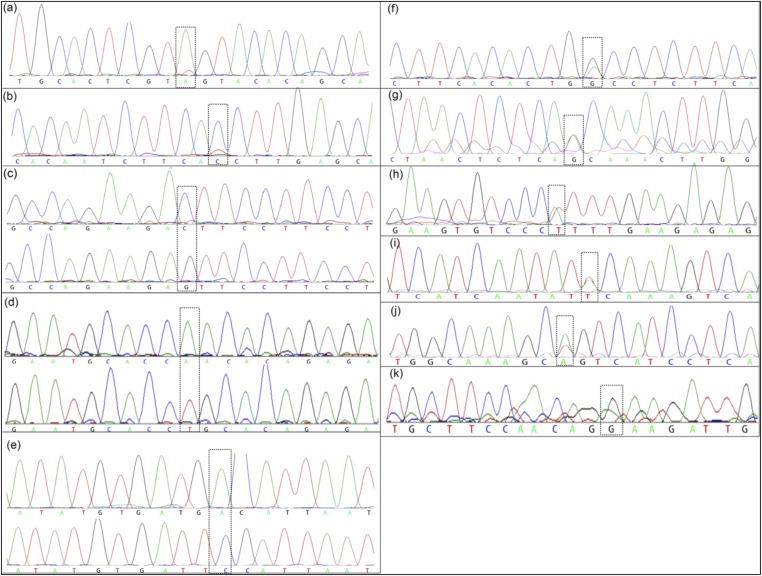
The chromatograms of Sanger sequencing for SNV validation. The boxes indicate the mutations. (A–G) Polymorphic SNVs. (H–K) Fixed SNVs that were not detected from the exome sequencing data in a few samples due to low coverage at the regions. (A) B5-specific A to T on PPP1R3B; (B) D62-specific C to T on BAZ1B; (C) A5-specific G to C on FLNB; (D) A66-specific T to A on LRP2; (E) B45-specific C to A on A2M; (F) B33-specific G to A on GSK; (G) B6-specific A to G on ITGB2; (H) C31 has the T to A mutation on DSCAM; (I) C31 has the A to T mutation on ENPP3; (J) C31 has the A to T mutation on COL21A1; and (K) B4 has the G to A mutation on HYDIN.
Copy number alterations (CNAs) are another common source of somatic genomic aberration. We used the program package CAScnv to call CNAs from our data (Materials and Methods, section 3). On average, each sample contained 23.6 CNAs, distributed among 14 chromosomes (SI Materials and Methods and Dataset S4). Because the mechanisms of CNA production are very different from those for SNVs, and because the latter also are much easier to ascertain, this study focused on SNVs (Discussion).
Fixed and Polymorphic Somatic Mutations.
Somatic mutations discovered in the sequenced samples were classified as either fixed or polymorphic. In this study, the terminology of population genetics is applied to facilitate theoretical analyses. Fixed mutations were those present in the entire cancerous cell population but absent in the noncancerous sample. These mutations must have already occurred at the onset of tumorigenesis. Polymorphic mutations, on the other hand, were present in some but not all cancerous samples (Materials and Methods, section 6).
Among the 269 SNVs observed in HCC-15, 209 and 35 mutations were confirmed to be fixed and polymorphic, respectively (Datasets S2 and S3 and Fig. S4). The remaining 25 mutations, divided into 22 possibly fixed and 3 possibly polymorphic SNVs, were not used in the analysis. The 35 validated polymorphic SNVs would define clone sizes and delineate clonal boundaries according to the genotypes of the 286 samples (Materials and Methods, section 7 and Dataset S3).
Fig. S4.
Identification of fixed and polymorphic mutations. A total of 269 putative SNVs were found from the 23 sequenced tumor sections. Diamond, 3 SNVs were randomly chosen for validation; cross-star and star, these SNVs were chosen for validation in the whole-exome sequencing (WES) samples where they were missing, usually due to low read depth. These 269 SNVs are divided into 209 fixed and 38 polymorphic mutations. In addition, 22 SNVs are possibly fixed but have been lost occasionally (i.e., LOH in CNA regions; Materials and Methods, section 4) and 3 SNVs are possibly polymorphic but could not be reliably confirmed by Sequenom across samples due to PCR difficulties.
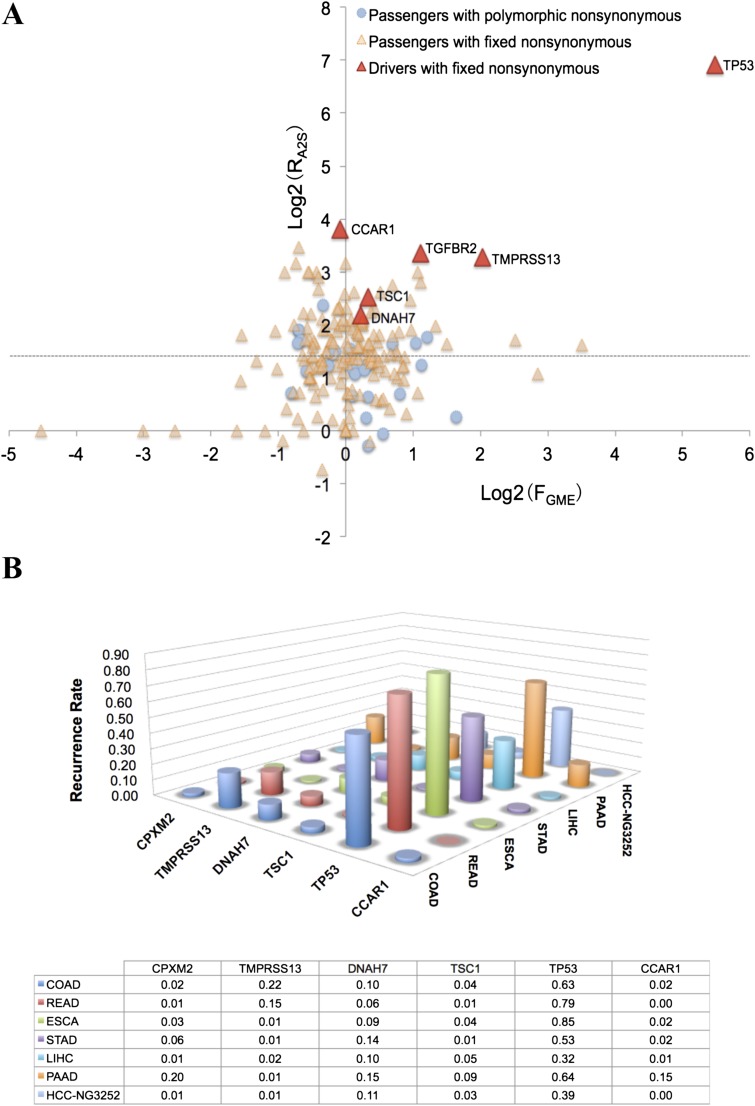
The 209 fixed mutations are divided into 166 protein-altering mutations (comprising 148 missense, 11 nonsense, and 7 splicing mutations) and 43 synonymous changes. In Materials and Methods, section 8, Fig. S5, and Dataset S5, a list of “driver” genes that are significantly more commonly mutated in cancer samples, especially in gastrointestinal and HCC tumors [Dataset S6; https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp; Schulze et al. (32)], was compiled from published data. In reference to this list, we identified 6 putative driver genes among the fixed mutations, which were CCAR1, CPXM2, DNAH7, TMPRSS13, TP53, and TSC1. In contrast, none of the 35 polymorphic mutations is in the driver group. The pathways represented by the fixed and polymorphic mutations are also somewhat dissimilar, as shown in Dataset S7.
Fig. S5.
The somatic mutation pattern of mutated genes of HCC-15 in 1,363 patients with gastrointestinal cancer. (A) The mutation pattern of fixed and polymorphic mutations of HCC-15 in 1,363 patients with gastrointestinal cancer. FGME is the fold of gene coding mutation enrichment. RA2S is the ratio of the number of nonsynonymous to the number of synonymous mutations. FGME and RA2S were calculated from all 460,967 somatic mutations in 1,363 patients with gastrointestinal cancer. (B) The occurrence rate of six putative driver genes of HCC-15 in 1,363 patients with gastrointestinal cancer, including 202 liver hepatocellular carcinomas (LIHC), 183 esophageal carcinomas (ESCA), 288 stomach adenocarcinomas (STAD), 220 colon adenocarcinomas (COAD), 81 rectum adenocarcinomas (READ), and 147 pancreatic adenocarcinomas (PAAD) from TCGA datasets and 242 hapatocellular carcinomas in Schulze et al. (32) (HCC-NG3252), respectively.
Clonal Diversity and Genealogy.
The 35 validated polymorphic SNVs delineated 20 cell clones among the 23 sequenced samples. A clone is defined as a cell population carrying a unique set of somatic mutations. We denoted Φi as the number of clones that appeared i times in n samples. The vector of [Φi, i in 1 to n − 1] is the allele frequency spectrum in population genetics (2, 3). In our data, [Φi = 18, 1, 1, 0, 0, 0 …; i = 1–22] and n = 23 = 18 × 1 + 1 × 2 + 1 × 3. In other words, 20 (= 18 + 1 + 1) clones consisted of 18 singletons, 1 doubleton, and 1 tripleton, which were, respectively, cell clones represented by one, two, or three samples. The small number of samples (3 of 23) yielding redundant information was indicative of the extensive diversity in the coding regions of the tumor. In particular, Simpson’s diversity index, H = 1 − Σ(Φi/n)2, was 0.941, indicating that two random samples would have a very high probability of being genetically different.
The genealogical relationship of the 20 clones is shown in Fig. 1B. The same genealogy with spatial information is given in Fig. 1C, in which clones were shown to emanate from the ancestral Ω clone in the center. For visual clarity, these clones were arranged on five rings, denoting the number of mutations away from Ω. The 7 direct descendants of Ω, labeled from α to η, all carried 1–2 mutations in addition to that of the Ω clone. Their descendant clones, each having additional mutations, were denoted with primes (δ′ and δ′′, for example). Some clones at the end of a branch were marked by a star symbol, which represented a singleton. On average, the number of coding mutations (U) accrued since the tumor began to grow from a single progenitor cell was 2.65 (Fig. 1C). As shown in Table 1, U is an important parameter in determining the genetic diversity of the entire tumor and, at U = 2.65, the mutation rate in HCC-15 is unexceptional among studies of intratumor diversity (12, 13, 16–18, 31). The genealogy of Fig. 1C was further expanded to include all 286 samples as portrayed in Fig. 1D (Materials and Methods, section 7).
Table 1.
Expected clonal diversity, HT, according to Eq. 3
| NT = 103 | NT = 104 | NT = 105 | |
| Exponential growth: dN/dt = r N and Nt = ert | |||
| r = ln(2) × 0.1 | 0.850 (u = 0.02, T = 100) | 0.910 (u = 0.015, T = 133) | 0.936 (u = 0.012, T = 167) |
| r = ln(2) × 0.01 | 0.586 (u = 0.002, T = 1,001) | 0.772 (u = 0.0015, T = 1,335) | 0.860 (u = 0.001, T = 1,668) |
| 3D growth: dN/dt = r N2/3 and Nt = (1 + rt/3)3 | |||
| r = (36π)1/3 × 0.1 | 0.944 (u = 0.036, T = 56) | 0.968 (u = 0.016, T = 127) | 0.976 (u = 0.007, T = 282) |
| r = (36π)1/3 × 0.01 | 0.776 (u = 0.0036, T = 558) | 0.902 (u = 0.0016, T = 1,274) | 0.952 (u = 0.0007, T = 2,817) |
| 2D growth: dN/dt = r N1/2 and Nt = (1 + rt/2)2 (u = 0.03, r = 2π1/2 for all cases below) | |||
| Simulations under a well-mixed population (calculation by Eq. 3) | 0.667 ± 0.075 (0.643) | 0.968 ± 0.01 (0.965) | 0.9997 ± 0.0005 (0.9997) |
| Simulations under spatial rigidity | 0.728 ± 0.096 | 0.978 ± 0.012 | 0.9999 ± 0.0001 |
T and u are also given. Three different growth models reaching different final cell numbers (NT) are used in the calculation. U = u × T = 2, which corresponds to the number of coding region mutations acquired during tumor growth (main text and SI Materials and Methods). T is the number of generations to reach NT and u is the mutation rate per generation. When cells double every generation with no cell death, r = ln(2). Hence, r = ln(2) × 0.1 would mean 10% of the growth rate of the pure cell-doubling populations. In the 2D “simulations under a well-mixed population,” the results are checked against the theoretical values given by Eq. 3. The simulated values match the theoretical calculations well.
Sizes of the Mutation Clones in Relation to Darwinian Selection.
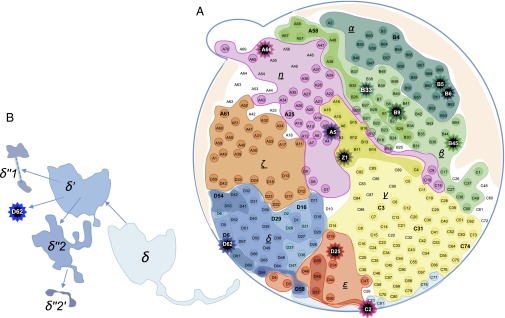
To delineate the size and spatial limit of each clone, the 286 samples were genotyped. Although a cell clone is typically defined by a suite of mutations (Fig. 1C), it may often be more informative to define a “mutation clone” by the collection of clones that share that mutation. For example, the MUC16 clone in Fig. 1C was composed of δ, δ′, δ′′1, δ′′2, δ′′2′, and D62 clones, whereas the THRA clone, which included δ′′2 and δ′′2′, was a subclone of the MUC16 clone.
Fig. 2 displays the sizes and spatial patterns of the mutation clones observed, with the subclones shown in increasingly darker shades. Genealogically, separate clones were observed to be segregated, revealing limited cell movement within solid tumors. The “sectoring” patterns of Fig. 2 suggested that clones grow outwardly, as the derived subclones were consistently observed on the outer flank of the parental clone.
Fig. 2.
Map of the mutation clones of HCC-15. A mutation clone is the aggregate of all samples carrying that mutation (main text). Hence, subclones (with increasingly darker hues) are nested within their parent clones. (A) Each star symbol indicates a singleton clone, represented by one sample. The clonal boundaries are delineated by the genotypes of all 286 samples. Many samples straddle two clones (including A3, B17, B19, B20, C78, D6, D9, and Z1). In this “sectoring” pattern of growth, δ′ grew outward from δ and, subsequently, δ′′s (−1, −2) grew outward from δ′. Note that tumors grew in three-dimensional (3D) space but the observations made were on a two-dimensional (2D) plane. This was apparent in the “northeast” direction, along which both the α and β clones were extending from the interior toward the periphery. It appears that α grew above or below β in their expansion toward the periphery. (B) The δ lineage clones are pulled out to display the overlaying pattern of mutation clones. The clonal map was also used to compute the mutation frequency spectrum, ξi, which is the number of sites where the frequency of the mutation was between (i − 1)/23 and i/23 from the 286 samples. We kept the number of frequency bins at 23 because the mutations discovered remained based on the initial 23 samples. The spectrum, as given in the text, is [ξi = 26, 7, 1, 1, 0, 0, …] for i = 1–22 (Materials and Methods, section 9 and Dataset S8).
We now evaluate whether certain clones grew faster than others. The null hypothesis of non-Darwinian evolution was that all clones have the same (or neutral) growth rate, whereas the alternate hypothesis of Darwinian selection posits faster growth of some clones. To test the null hypothesis, we compared the sizes of the observed mutation clones with the expected sizes, often referred to as the mutation frequency spectrum and denoted as [ξi, i = 1 to n − 1]. ξi is the number of sites where the mutant appears i times in n samples in the infinite-site model of population genetics (2, 3). In HCC-15, [ξi = 26, 7, 1, 1, 0, 0, …] for i = 1–22 (Fig. 2 legend and Dataset S8), where Σiξi = 35 was the number of mutations in the sequenced samples (Materials and Methods, section 9).
In a population with a constant effective size of NT, E(ξi) = θ/i, where θ = 2NTu (2, 3). In exponentially growing populations, the corresponding E(ξi) has been defined by Durrett (25) as
| [1] |
where r is the rate of population growth, the difference between cell birth and death rates (see below). In addition, u is the mutation rate per cell generation, and n is the sample size (Materials and Methods, section 10). Because Σi>1ξi = (7 + 1 + 1) = 9 = 23 × u/r × Σi>11/i(i − 1) = 23 × u/r × 0.95, we obtained u/r = 0.41 by Eq. 1. For the total of 35 sites, [E(ξi) = 26.0, 4.72, 1.57, 0.79, 0.47, 0.31, …], which was very close to the observed spectrum of [ξi = 26, 7, 1, 1, 0, 0, …] ( = 2.53 and P = 0.865 for ξi>1s). Hence, the size distribution of the mutation clones (Fig. 2) was as expected under the neutral model, and no clones were of unusually large proportion.
The next question is whether the analysis would have the power to reject the non-Darwinian model if selection was indeed in operation. A key feature of the neutral spectrum is that it has very few high-frequency mutations. In our samples, only ∼0.5 site is expected to have a frequency greater than 50%. Thus, even a very small number of mutations that have been driven to a high frequency by selection would stand out, as noted before (19). For example, if only one of the 35 mutations in our samples was driven to a high frequency of 90%, or 3 of the 35 were driven to the medium frequency of 50%, the new spectra would be rejected as neutral with P < 0.05. This can be seen in the simulations based on Eq. 1 and presented in Materials and Methods, section 11 and Fig. S6. Of course, a true comparison between the non-Darwinian and Darwinian models is possible only when the mode and strength of selection are specified in the Darwinian model. It may hence be more appropriate for investigators with a defined selection scheme to carry out such a test.
Fig. S6.
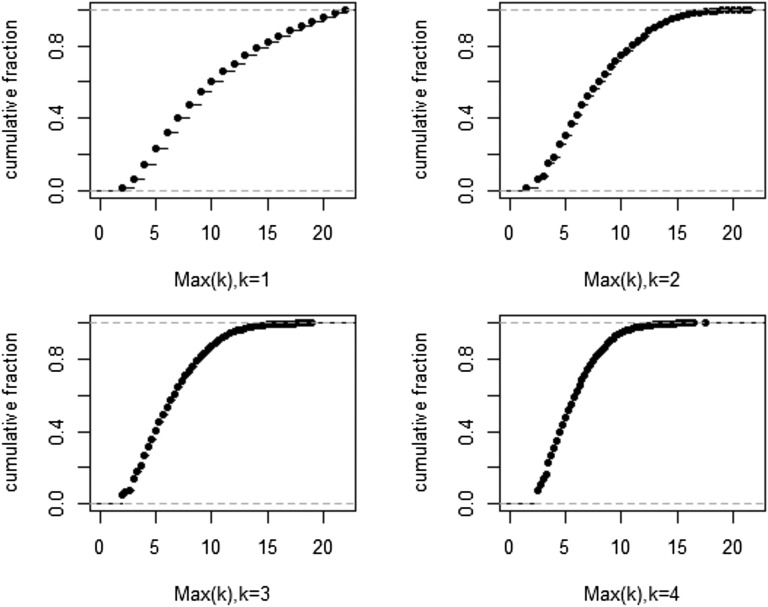
Cumulative distribution of Max(k) based on 10,000 times of simulation, for k = 1–4. Max(k) measures the average frequency of the most common k mutations (details in Materials and Methods, section 14).
The simplest form of selection does make a qualitative prediction in which larger clones, driven by selection, may have taken less time to become larger than the smaller clones. When time is measured by mutation accumulation, the larger clones may be younger, whereas in the non-Darwinian model the larger clones would be older (2, 3). In a previous study, Tao et al. (31) showed that, among physically separated HCC tumors, younger but larger tumors appeared to have been driven by Darwinian selection. The authors also detected many small and visible tumors, presumably neutrally growing, by molecular means. Within the same tumors, Tao et al. (31) found the expected non-Darwinian pattern in which the younger clones are smaller than the older (parental) ones. The trend is also observable in HCC-15. For example, γ→γ′→Z1, β→β′→B33, and ε→ε′→C2, where A→B means the B clone is derived from and is smaller than the A clone. Taken together, in this first study with the necessary empirical data that were analyzed by modern population genetics theory, the evolution within this single tumor appears largely non-Darwinian.
The Genetic Diversity of the Entire HCC-15 Tumor.
The ability of a tumor to respond/adapt to challenges may depend on MALL (the total number of coding mutations in the entire tumor). MALL has not been estimated before because under a Darwinian model it would vary greatly, depending on how selection operates. Estimation is feasible under non-Darwinian evolution as shown by the four methods used to estimate MALL in HCC-15. The most conservative estimate is Mmin. When a tumor grows from one cell to NT cells, the minimal number of cell divisions and mutations should be NT − 1 and Mmin = NT × u, respectively. The highest estimate of diversity was obtained from exponentially growing populations (Mexp) in which the number of mutations with frequency >x in the entire population is given by Durrett (25) as
| [2] |
In between these two estimates are Meq and M3D. The estimates of MALL are given in the Fig. 3 legend, which explains the four methods, with the details given in Materials and Methods, sections 12–14.
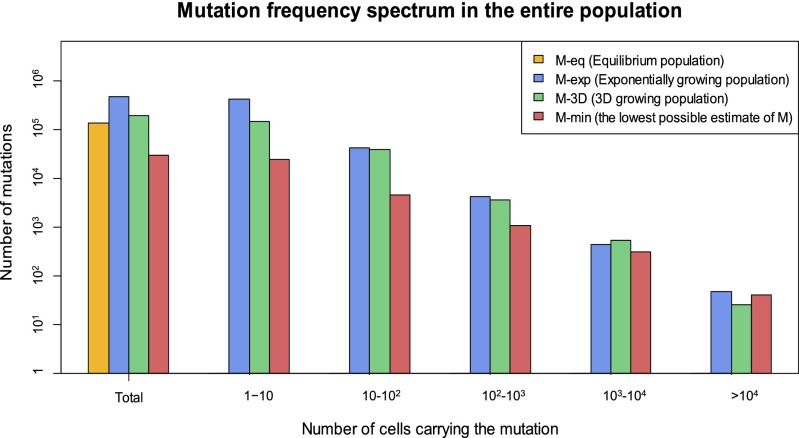
Fig. 3.
Estimated mutation frequency spectrum in the entire HCC-15 tumor. Four estimates assuming different modes of population growth to NT = 106 cells are given (Materials and Methods, sections 10 and 12–14), all within the same order of magnitude of 105 mutations. (i) Mmin, the lowest possible estimate of MALL, is (NT – 1)u (Materials and Methods, section 12). It is here simulated in populations that grow on the periphery, but the interior cells neither divide nor die. (ii) Meq is the estimate of the total diversity assuming that the population has remained at a constant size, equivalent to the long-term average of nonconstant populations. Based on the standard population genetic formulas for constant populations (2, 3), the higher-frequency bins tend to be overestimated and lower-frequency ones underestimated. Overall, Meq would be an underestimation (details in Materials and Methods, sections 12–14). (iii) Mexp is obtained for populations that have grown exponentially from a single cell with the cell birth rate being larger than the death rate (Eq. 2 and Materials and Methods, section 12). (iv) M3D is for the 3D cell population that grows on the periphery with frequent cell turnover in the interior (Materials and Methods, section 14).
When HCC-15 had only 106 cells (∼1.0 mm in diameter), less than 0.1% of its final size, all four estimates are within an order of magnitude of 105 coding mutations. If MALL is extrapolated to the final tumor size of >109 cells, it would be greater than 100 million. In comparison, under the specific model of Darwinian evolution of Tao et al. (31), MALL would be orders of magnitude smaller (Materials and Methods, section 15 and Dataset S9).
The estimated large diversity of HCC-15 consisted mostly of low-frequency mutations. Small local regions of the tumor are each expected to harbor some levels of diversity, which are the building blocks of the total diversity. In Fig. 4, using the rules of clonal growth and mutation accumulation for HCC-15, we simulated the total diversity. The clonal diversity of the plane through the middle of the tumor is illustrated in Fig. 4A (clones >50,000 cells were shown). Importantly, the observation in Fig. 2 and the simulation in Fig. 4A provided visual confirmation of the statistical test based on [ξis]. The size distribution, the growth dynamics, and the geography of the clones of HCC-15 therefore agreed well with the non-Darwinian growth model. When the simulations of Fig. 4A magnify into smaller areas, the diversity continues to increase as shown by Fig. 4B (resolution >4,000 cells) and Fig. 4C (resolution >100 cells). If we randomly sample and sequence ∼50 cells from a local area at the scale of Fig. 4C, the observed genetic diversity should match the simulations. Using the 23 WES samples, we indeed verify the simulated high local diversity in the Fig. 4D legend.
Fig. 4.
Simulated vs. observed fine-scale diversity in HCC-15. (A–C) Simulated clonal diversity at three levels of resolution. Adjacent clones are differentiated by different colors but nonneighbors often have to be depicted by the same color. Neighboring clones usually differ by one to two coding mutations. The three panels zoom in with finer resolution. The axis labels are the numbers of cells. The minimal clone sizes to be displayed are 50,000, 4,000, and 100 cells in A–C. The mutation rate is u = 0.03 in coding regions and NT = 1.15 × 109. The simulations are done in the 3D space (Materials and Methods, section 14) and samples are taken from a 2D plane cut through the middle as in the actual sampling. Note that clones sometimes go around one another in the third dimension. The simulated A and the observed Fig. 2 are roughly in the same scale. (D) Observed local diversity. From each of the 23 WES samples with an average read depth of ∼75×, the equivalents of 37–38 random cells are sequenced. The mean numbers of mutations in each size bin (ranging from 1 to 40 cells in increments of 5) as well as the SDs across the 23 samples are given. The simulated numbers when 40 cells are sampled from the equivalents of C are also shown. The agreements between the observed and simulated mutation numbers are generally good, except in the smallest-size bin of one to five reads where sequencing errors are high.
Intratumor Genetic Diversity—A General Theory.
This high-density study suggests that previous reports on intratumor diversity should be reevaluated in light of the non-Darwinian model (8, 11–18). Under this simpler model, the diversity estimates of Figs. 3 and 4 can be generalized because a tumor’s diversity depends only on how much time (measured by mutation accumulation) it has taken the tumor to grow to a given size (Materials and Methods, sections 16 and 17). The expected genetic diversity at generation T (HT, the probability that two randomly chosen cells are genetically different in the coding region) can be expressed as
| [3] |
where Ni is the population size at generation i, T is the time (measured in generations) of tumor growth from a single progenitor cell, and u is the mutation rate in the coding region (Materials and Methods, section 16). A generation is the time between cell divisions. An alternative formulation based on the birth-and-death process yields nearly identical results (Eq. 6 in Materials and Methods, section 16). Although T and u in Eq. 3 are not known, their product (U = uT) is observable. U, the number of somatic mutations accrued during tumor growth, has been well documented (12, 13, 16, 17). When a population of cells grows from a single progenitor to NT in a duration measured by U, NT and U will largely determine the level of genetic heterogeneity (Materials and Methods, section 17). Eq. 2 shows the diversity to be the product of NT and U.
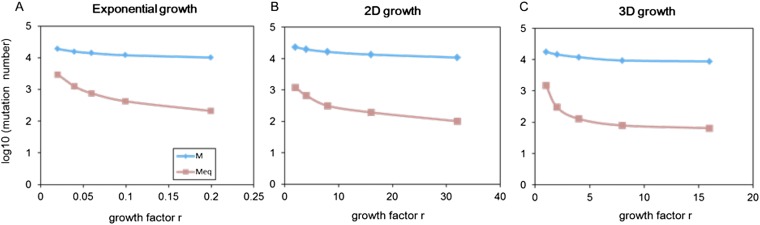
In Table 1, we computed the clonal diversity by setting low NTs, between 103 and 105 cells, under three different growth models (Materials and Methods, section 17). A tumor with fewer than 106 cells is not detectable by current imaging technologies and U = 2 corresponds to two coding region mutations during tumor growth, which is also conservative (12, 13, 16, 17). Even given these parameter values, the neutral clonal diversity is still very high, in the range of 0.6–0.99. For NT > 105 cells, two random cells should almost always be genetically different. Importantly, H is not greatly affected by the assumed model (exponential, 3D, or 2D growth) of tumor growth because T and u would vary in opposite directions to yield similar H values (Table 1). The conclusion of high diversity should therefore be generally applicable.
Discussion
Darwinian selection is undoubtedly the driving force of biological evolution but even Darwin himself was puzzled by the amount of genetic diversity within a species. As pointed out by Fisher (20), the better genotypes should have taken over the populations, leaving little room for within-population diversity (19, 21). In the modern Darwinian view (26, 27), complex forms of selection might be able to maintain high intratumor diversity but quantitative predictions, against which observations can be compared, need to be generated first (4, 6, 33, 34).
We propose that non-Darwinian evolution be considered the null model, under which one can generate testable predictions. If the non-Darwinian predictions are rejected, it will then be necessary to incorporate some forms of selection into the model (1, 3). In this study, we test the evolution of SNV. The non-Darwinian prediction is consistent with the high Ka/Ks ratios (nonsynonymous/synonymous SNVs per site) observed in 400 cancer genomes (29) and in The Cancer Genome Atlas (TCGA) data (35). The ratio is statistically indistinguishable from 1 in most studies (36), thus indicating ineffective selection against protein sequence changes in tumors. Cases of Ka/Ks ∼ 1 are rarely seen in nature; for example, Ka/Ks < 0.3 between humans and other primates (1).
The level of intratumor diversity is very different between Darwinian and non-Darwinian evolution. Under non-Darwinian evolution, HCC-15 may have 100 million coding mutations and those in the very low-frequency range account for the bulk of the diversity. Fig. 4D based on the polymorphisms within the 23 sequenced samples corroborates this estimate. Under the selection model of Tao et al. (31), the high diversity could be realized only when the selective coefficients are small, i.e., when Darwinian evolution converges with non-Darwinian evolution.
In view of our estimate of the presence of hundreds of millions of SNPs in a tumor the question then arises, “Why is there little Darwinian selection?”. One reason is that the bulk of the mutations are in very low frequencies. The frequency spectrum in a rapidly growing population approaches θ/x2, where x is the mutation frequency. In fact, ∼99% of the mutations are found in fewer than 100 cells. Given the strong random drift on low-frequency mutations, it is not surprising that the bulk of mutations appear to be subject to no selection. However, a more important reason may be that in a solid tumor cells stay together and do not migrate, so that when an advantageous mutation indeed emerges, cells carrying it are competing mostly with themselves. These mutations may confer advantages in fighting for space or extracting nutrients but they are stifled by their own advantages. In a nonsolid tumor such as leukemia, cells are not spatially constrained and a selection sweep may indeed occur.
In a physiological sense, good mutations may emerge now and then but in solid tumors the cell populations are so structured that selection may often be blunted. The physiological effect has to be very strong to overcome those constraints. That may be what a drug treatment does—it “loosens up” the population for effective competition to occur.
It is important to note that several types of genetic changes, including synonymous and nonsynonymous SNVs, CNAs, and epigenetic changes, are evolving in the same genomes. Although the constraints on selection discussed above may apply to all mutations, different types of changes, even synonymous and nonsynonymous SNVs in the same genes, may nevertheless experience different selective pressures and exhibit different evolutionary dynamics. The conclusion of this study applies to SNVs. Whether CNAs or other changes may evolve in the Darwinian mode cannot be tested at present because the underlying forces such as mutation rate are largely unknown.
Patient survival has been shown to be negatively correlated with the level of genetic diversity within tumors (5, 7–9). When mutations can be found in nearly all possible coding regions within a tumor, resistance to most drugs seems highly likely. Read et al. (37) pointed out that aggressive strategies against cancerous cells are effective only in the absence of resistance at treatment and various strategies for administering drugs in the face of resistant clones have been proposed (38–42). Finally, a key feature of the non-Darwinian model is the rapidity with which mutations accrue. Even microscopic tumors with fewer than 105 cells, which are often targets of postsurgery adjuvant therapy, would be genetically diverse (Table 1). The possibility of high intratumor diversity even in small tumors suggests a need to reevaluate treatment strategies.
Materials and Methods
The following sections present essential technical information that is referred to as Materials and Methods, sections 1–17 in the text. Additional details can be found in Supporting Information).
1) Clinical Information.
The patient was a 75-y-old man with chronic Hepatitis B Virus (HBV) infection and liver cirrhosis. The tumor, ∼35 mm in diameter, was on the left lobe of the liver and well encapsulated. It was a histopathological grade III hepatocellular carcinoma (HCC) diagnosed at Peking University Cancer Hospital. The pathology report indicated that the tumor sections contain ∼90% hepatoma cells. Two sections of 35 × 35 × 10 mm from the tumor and an adjacent nontumor sample were obtained. This study was approved by the Ethics Review Committee of Peking University Cancer Hospital. Informed consent was signed according to the regulations of the institutional ethics review boards.
2) Number, Volume, and Geographical Distribution of Samples.
The honeycomb-like sampling is further described in Fig. S1. One 1-mm-thick slice of the tumor sample was subjected to high-density microdissection, using the Harris Micropunch with 0.5 mm inner diameter. In total, 286 microsections were obtained, equally distributed in the four quadrants (labeled A–D; Fig. S1). An adjacent nontumor sample was used as the control. Genomic DNA was extracted using the TIANampMicro DNA Kit (Tiangen) and quantified using a Qubit 2.0 fluorometer according to the manufacturer’s instructions.
Special attention was paid to minimizing the sample volume (number of cells per sample) as genealogical information is better preserved in samples of smaller volume. Given that the diameter of a HCC cell is about 25 μm (20–30 μm), and the volume of a microsection is ∼0.2 mm3, the number of cells in a microsection was estimated to be ∼24,000. DNA was extracted and quantified from 10,000 tumor cells that were precisely collected by laser capture microdissection (LCM). The cell number in each of the microsections was estimated based on the reference quantity. For the 286 microsections, the median number of cells per sample was ∼20,000, which approximates the number estimated by volume (Fig. S2).
3) Detection of Copy Number Alterations and Estimation of Tumor Purity.
CAScnv (an in-house software) was used to detect the somatic copy number alterations (Supporting Information). We used ABSOLUTE (49) (www.broadinstitute.org/cancer/cga/ABSOLUTE) to infer the purity and ploidy for our samples. The copy number alterations called from whole-exome sequencing reads using CAScnv were input into the ABSOLUTE program (49). Based on precomputed models of recurrent cancer karyotypes, the ABSOLUTE algorithm examined possible mappings from relative to integer copy numbers by jointly optimizing two parameters, α(purity) and τ (ploidy). The inferred tumor purity and ploidy of all 23 tumor samples are shown in Dataset S1, which are consistent with the estimates in the pathology report.
4) Detection of Somatic SNV.
Tagmentation-based library preparation (Fig. S7), WES, and sequence alignment are described in Supporting Information. Somatic SNV calling was performed using the in-house software, CASpoint, which has been extensively tested in the public domain (Dataset S10; also see the result in the International Cancer Genome Consortium-TCGA DREAM Somatic Mutation Challenge (SMC): https://www.synapse.org/#!Synapse:syn312572/wiki/70726). We compared the false positive and negative rates, sensitivity, and accuracy of CASpoint in SNV calling with the performances of other published software installed in the Beijing Institute of Genomics (BIG) computational center and bioinformatics facility, including Mutect (43), SomaticSniper (44), JoinSNVmix (45), Varscan2 (46), and Samtools (47). Simulated sequencing reads in the SMC and a large set of whole-genome or -exome sequencing reads produced from various genomics projects in solid tumors (31) and leukemia (48) in Beijing Institute of Genomics were used to evaluate the performance of CASpoint. The overall accuracy of CASpoint is comparable to the others for the SMC simulated reads. Because CASpoint showed better performance in reducing false positive rates than other programs for real sequencing data according to validation results using Sequenom and Sanger sequencing, the in-house program was used in this study to minimize the false positive rate.
Fig. S7.
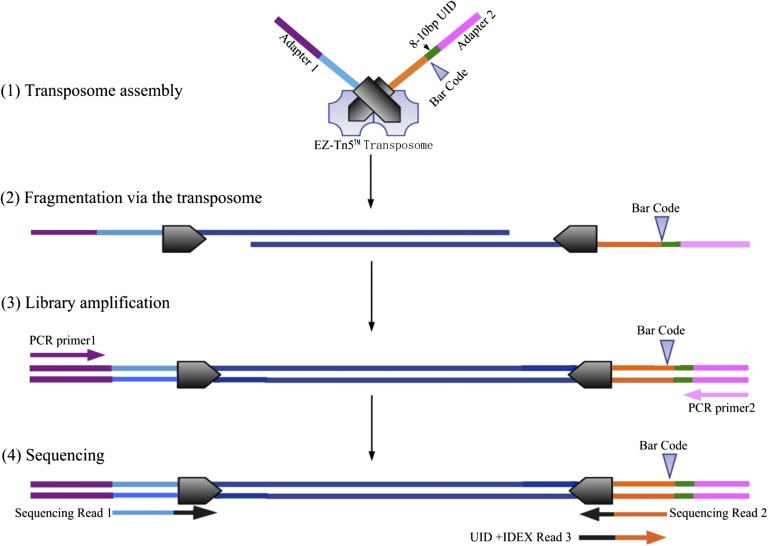
Schematic overview of the library preparation using in-vitro transposition. The EZ-Tn5 transposase mediates the fragmentation of double-stranded DNA and simultaneously ligates the DNA fragments with synthetic oligonucleotides including a unique DNA identity (UID) plus sample barcode plus sequencing adapters. The procedure includes (1) the assembly of a transposase complex (2), DNA fragmentation, and (3) library amplification. After purification, the library is ready for sequencing or whole-exome capture. Dark blue, genomic DNA; light blue/orange, partial sequencing primer; purple/pink, PCR primers and sequencing platform adapters; green, UID of 8-bp random oligonucleotides; gray hexagon, transposon; and gray triangles, 6-bp sample barcode.
As described in Zhu et al. (48), two statistical tests are introduced in the program. One-sided Fisher testing calculates the statistical significance of tumor mutant allele frequency (MAF) that is higher in the tumor population than in the normal cell population. Binomial testing calculates the significance of tumor mutant allele number observed from the aligned tumor sequencing data that meet a binomial distribution. In addition, 10 filtering criteria were applied to detect somatic SNVs as described in Supporting Information. All somatic mutations are shown in Dataset S2.
5) Validation of the Observed SNVs Across the 286 Samples.
SNVs discovered by WES were validated by Sequenom genotyping on the 286 samples. These discovered SNVs fall into three classes: (i) The ALL class has 178 SNVs that were discovered in all 23 WES samples (all red dots in Fig. 1A), (ii) the MOST class has 53 SNVs that were present in most samples and missing in only a few (usually 1–4 where read depth was low), and (iii) the SOME class has 38 SNVs that were present in some (≤6 samples; Fig. 1B) but missing in all other samples. These partitions are shown in Fig. S4 and the mutant allele frequencies are shown in Dataset S2.
The three classes of mutations (ALL, MOST, and SOME) require different levels of efforts of validation by Sequenom genotyping: (i) For the 178 mutations in the ALL class, their ubiquity is certain. We chose 3 of these mutations for validation in the 286 samples and confirmed their ubiquity. (ii) The 53 mutations in the MOST class were validated in the few WES samples where they were found missing. In samples where the mutant was missing due to low read depth, Sequenom confirmed the presence of these 31 mutations (Dataset S2). There are 22 ambiguous cases where the mutant is missing in 1–3 samples that have copy number alterations in the regions of the mutations. These are likely cases of loss of heterozygosity (LOH). Although we suspect the mutations to be “possibly fixed” with a few LOH samples, these 22 mutations are not included in subsequent analyses. (iii) The 38 mutations in the SOME class were validated in all or most of the 286 samples. The results are shown in Dataset S3. Of the 38 mutations, 3 could not be reliably detected across samples due to PCR difficulties. Hence, we analyzed only the remaining 35 mutations as true polymorphisms.
We used the Sequenom MassARRAY Assay Design 3.1 software to design the PCR and MassEXTEND primers (Dataset S11) for multiplexed assays. MassEXTEND reactions and iPLEX Gold assays were subsequently used for primer extension and allele frequency measurement. The allele-specific extension products for different allelic types were quantitatively analyzed, using the MALDI-TOF mass spectrometer. Using the mutant signal of nontumor as a negative control, mutation calling and allele frequencies for each SNV site were determined using the MassArrayTyper 4.0 Analyzer according to the manufacturer’s specifications. To estimate mutant allele frequency and degree of heterozygosity, the peak areas of the mutant and the wild-type allele were quantified and the mutant allele frequencies were determined as the average of (mutant peak)/(mutant peak + wild-type peak). We wrote scripts to extract mutation frequencies from Sequenom Typer 4.0 software. Genomic positions for all validation SNVs were retrieved using the HG19 as reference. Some SNVs found in only one sample were further validated by PCR and Sanger sequencing (Supporting Information).
6) Identification of Fixed and Polymorphic Somatic Mutations.
Based on the descriptions in Materials and Methods, section 4 and the results of Datasets S2 and S3 and Fig. S4, the 269 SNVs are classified as 209 confirmed fixed SNVs, 35 confirmed polymorphic SNVs, and 25 less certain mutations. These 25 mutations, including respectively 22 possible fixed and 3 possible polymorphic mutations, were not used in the analyses. The partition of these 269 SNVs summarized in Fig. S4 is as follows:
-
i)
The confirmed 209 fixed mutations include 178 from the ALL class and 31 from the MOST class described in Materials and Methods, section 4 above. The 178 SNVs were observed in all 23 WES samples (Dataset S2) and the limited validation among unsequenced samples indeed confirmed their ubiquitous presence. The 31 MOST class mutations were present in all but a few (1–4) WES samples, due to low depth coverage of such sites in these samples. Sequenom results validated their presence in these samples.
-
ii)
The 35 confirmed polymorphic mutations are listed in Dataset S2 among WES samples. They were further validated across the 286 samples by Sequenom (and occasionally by Sanger sequencing) as shown in Dataset S3, which is the basis of the spatial distribution of these mutations shown in Figs. 1 and 2 of the main text.
-
iii)
For the remaining 25 SNVs, 22 mutations are missing in 1–4 samples (Dataset S2, under “SNV in CNA regions”). These mutations occurred in regions of frequent CNAs, which would result in LOH. LOH could be inferred directly from these data when AB (mutations A and B occurred in 20 samples), A+ (mutation A but not B occurred in 2 samples), and +B (mutation B but not A occurred in 1 sample) were all observed. In this pattern, B is lost twice and/or A is lost once. From the pattern shown in Dataset S2, it is likely that all of the 22 mutations are fixed but it is prudent to exclude them from subsequent analyses, as was done here.
-
iv)
The 3 possibly polymorphic mutations were detected in some WES samples but could not be reliably genotyped across the 286 samples by Sequenom. They are almost certainly polymorphic mutations but could not be used in this study to delineate the spatial boundaries of clones or their sizes.
7) Clone Map Delineation and Phylogenetic Analysis.
The 35 polymorphic SNVs unaffected by CNAs were validated in the 286 tumor samples, using Sequenom and/or Sanger sequencing (Dataset S3). The neighbor-joining method of Saitou and Nei (50) was used to construct the phylogenetic tree (Fig.1 B and D). A consolidated matrix was created, containing the mutations of all samples with “1” and “0” representing the presence and absence of a mutation based on genotyping results of the 35 SNVs. We used the “APE” R package (51) and iTOL (itol.embl.de) for constructing and plotting the phylogenetic trees (Fig. 1 B and D). The positions of eight samples (A3, B17, B19, B20, C78, D6, D9, and Z1) that carried mutations of two neighboring clones were marked with blue stars in the phylogenic tree in Fig. 1D. The boundaries and space of the subclones in HCC-15 were delineated in the two-dimensional clonal map based on both the presence of the polymorphic SNVs and the phylogenic relationship (Fig. 2).
8) Identification of Putative “Driver” Genes.
We attempted to identify driver genes from among the 269 mutated genes in our study. As in common practice, driver genes are defined as those that are significantly overrepresented in the cancer databases. The data we used here comprise 460,967 somatic mutations (402,716 SNVs, 42,886 small deletions, and 12,249 small insertions) detected in whole-exome sequencing data of 1,363 patients with gastrointestinal cancer (Dataset S6), including 202 hepatocellular carcinoma (LIHC) (72,862 mutations), 183 esophageal carcinoma (ESCA) (54,042 mutations), 288 stomach adenocarcinoma (STAD) (115,357 mutations), 220 colon adenocarcinoma (COAD) (114,594 mutations), 81 rectum cancers (READ) (25,003 mutations), and 147 pancreas cancers (PAAD) (56,815 mutations) from TCGA datasets and 242 hepatocellular carcinomas (22,294 mutations) in Schulze et al. (32). We applied the program MutSigCV 1.4 (52), which corrects for variation by incorporating a patient-specific mutational spectrum and gene-specific background mutational burden, and by measuring gene expression and replication time as well, to detect significantly mutated genes.
In total, we identified 372 driver genes from the somatic mutations dataset of 1,363 gastrointestinal cancer cases (Dataset S5). Comparing the 166 fixed protein-altering mutations in our study to the driver genes identified from the databases, we identify 6 putative driver genes (q-value ≤ 0.2): CCAR1, CPXM2, DNAH7, TMPRSS13, TSC1, and TP53; the last of the 6 genes has a high frequency in all gastrointestinal cancers (Fig. S5). We note that none of the genes carrying any of the 35 polymorphic mutations in this study belongs in the driver group. Ingenuity pathway analysis (IPA) (www.ingenuity.com) and Fisher’s exact test were carried out to identify significantly enriched pathways for the genes with polymorphic and fixed protein-sequence altering SNVs (Dataset S5).
9) Observed Mutation Frequency Spectrum.
The mutation frequency spectrum is denoted as [ξi, i = 1 to n − 1] in the main text, ξi is the number of sites where the mutant appears i times in n samples in the infinite-site model of population genetics. In Fig. 1B, the heat map is equivalent to a spectrum of [ξi = 24, 2, 3, 2, 1, 3, 0, 0, …] for i = 1–22, where Σiξi = 35 is the number of mutations in the 23 WES samples. In this spectrum, the mutation in each sample is scored as either present or absent.
Because the frequency of each mutation was more accurately determined by genotyping, ξi is represented by the number of sites where the frequency of the mutation was between (i − 1)/23 and i/23 from the 286 samples. We kept the number of frequency bins at 23 because the mutations discovered were still based on the initial 23 samples. The spectrum, as given in Dataset S8, is [ξi = 26, 7, 1, 1, 0, 0, …] for i = 1–22. There are two methods to compute the frequency spectrum using the data from the 286 samples. One is to score the presence/absence of each mutation in each sample. This will tend to inflate the frequencies of the mutations as samples with only a fraction of cells carrying the mutation would be scored as a full site. To obtain the spectrum of [ξi = 26, 7, 1, 1, 0, 0, …], we used a second method by averaging the frequency of each mutation over all samples.
Finally, Fig. 4D presents local diversity by scoring mutations that have lower counts in each WES sample. In calling such mutations, stringent validation is necessary to determine the level of confidence, which would be lower as the frequency decreases. At 21 of 22 sites, SNV calls based on 6–10 mutant reads were validated by Sequenom genotyping (Dataset S2), a validation rate of 95.5%. SNV calls supported by >10 reads are accurate with a 99% validation rate. The validation rates suggest that the calls in bins >5 reads are of high confidence. We disregard calls with ≤5 reads in Fig. 4D, which gives the mean and SD of mutation number in each of the larger size bins.
10) Expected Mutation (Site) Frequency Spectrum in Exponentially Growing Populations.
| [4] |
where r is the rate of exponential growth, u is the mutation rate per cell generation, n is the sample size, and NT is the cell population size at time T (25). For HCC-15, [ξ23,i = 26, 7, 1, 1, 0, 0, …; i = 1–22]. Because Σi>1ξ23,i = (7 + 1 + 1) = 9 = 23 u/r, we obtain u/r = 0.41. The expected site frequency spectrum for 35 mutations is hence [E(ξ23,i) = 26.0, 4.72, 1.57, 0.79, 0.47, 0.31,…].
11) Max (k)s—Frequency of the Most Common k Mutations in a Sample.
We note that the observed frequency spectrum given in the main text is [ξi = 26, 7, 1, 1, 0, 0, …] for i = 1–22. Hence, the average frequency of the k most common mutations would be 4, (4 + 3)/2, and (4 + 3 + 2)/3 for k = 1–3, respectively. Because Darwinian selection would drive the advantageous mutations to a high frequency (19, 21), it would be informative to compare the observed frequencies of the most common k mutations with those expected for the detection of selection.
Under the neutral model, we can determine the average frequency of the most common k mutations in our sample, denoted Max(k). E(ξn,i) is the expected number of mutations that were found in i of the 23 samples. In exponentially growing populations, the corresponding E(ξn,i) has been defined by Durrett (25) as Eq. 1,
where r is the rate of population growth, the difference between cell birth and death (main text). For the total of 35 sites, [E(ξn,i) = 26.0, 4.72, 1.57, 0.79, 0.47, 0.31, …] which sum up to 35. We took 35 mutations from this distribution and determined the Max(k) for k = 1–4. The distributions of Max(k) in 10,000 repeats are given in Fig. S6. For example, the 95% cutoff for k = 1 (i.e., the most common mutation) would be 20 of 23 samples. Likewise, the 95% cutoff for k = 3 (the average frequency of the top 3 common mutations) would be 12 as presented in the main text.
12) Four Estimates of the Total Number of Mutations (MALL).
The minimal number of mutations accrued during tumor growth was referred to as Mmin. When cells of a tumor divide from 1 to NT cells, the minimal number of cell divisions should be NT − 1, if no cells die during tumor growth, resulting in Mmin = NT × u. We carried out computer simulations in which tumors grew outward as a 3D mass. In our model, cells were “frozen” when they become encapsulated inside the tumor; only cells on the periphery were able to divide (Materials and Methods, section 14). This mode of growth does not require many more cell divisions than the minimum of NT − 1. Fig. 3 shows that the number of mutations under such a growth mode, with NT = 106 and u = 0.03 (per cell division in the coding region, equivalent to 10−9 per cell division per base pair), was very close to the minimum of Mmin = NT × u = 3 × 104 mutations. Even at this minimum, MALL was substantial. The choice of u = 0.03 is in agreement with several previous studies (18, 24), as well as with the estimate by the approximate Bayesian computation (ABC) method (Supporting Information and Fig. S8).
Fig. S8.
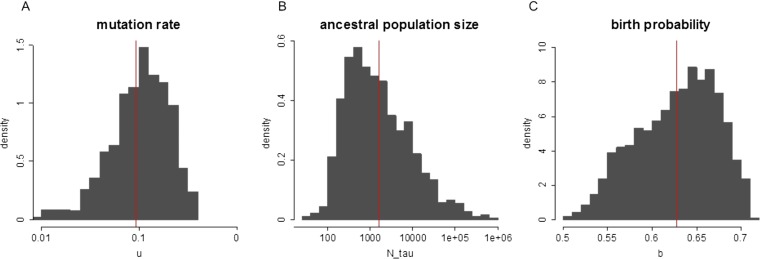
Posterior histograms of parameters by ABC inference. (A) u, mutation rate per cell division in the coding region; (B) Nτ, ancestral population size during the growth of HCC-15; (C) b, the probability that a cell has two offspring in each generation in the discrete-time branching process. The exponential growth rate r = ln(2b). The posterior means are u = 0.093, Nτ = 1,585, and b = 0.627 [thus r = ln(2b) = 0.226]. The red lines indicate the mean values.
The second method used to estimate MALL was to assume that the cell population was maintained at a constant size, close to the long-term average of changing population sizes. Standard population genetic formulaes can then be used to estimate the equilibrium genetic diversity, Meq, analytically (2, 3). Nevertheless, because the cell population is likely to have been growing, Meq would almost always underestimate the true diversity. This is because the imposition of the equilibrium conditions on the data would result in adequate estimation of diversity only in the observable portion of the spectrum. Low-frequency mutations were expected to be underestimated. In Materials and Methods, section 13, we provide the details of obtaining Meq, as well as the simulation data that corroborated the conjecture of Meq < MALL. As most tumors are growing, albeit not necessarily in any specific mode, Meq should be a reasonable lower bound of a tumor’s diversity. For HCC-15, Meq was roughly 14% of NT and substantially larger than Mmin as shown in Fig. 3.
In the third estimate, Mexp, the mode of population growth is specified. If cells divide and die at a constant rate, the population would be growing exponentially. The net growth (i.e., the difference between the birth and death rates) could be positive, negative, or net zero. Under this exponential model developed by Durrett (25), the number of mutations with frequency >x in the entire population is given by Eq. 2.
The elegant simplicity of Eq. 2 is not unexpected because the genetic diversity in a tumor is largely determined by two parameters: the number of mutations (U) each cell accumulates during tumor growth and the population size (NT = erT). The expression, U = uT = (u/r) ln(NT), thus anticipates the simplicity. The total number of mutations in the tumor, Mexp(x = 1/NT), is projected to be (u/r) × 1/(1/NT). From the observed mutation frequency spectrum [ξis] and Eq. 1, we have obtained u/r = 0.41.
Given NT = 106 cells, HCC-15 would have Mexp= 4.1 × 105 coding mutations (Fig. 3), which was more than 10-fold larger than Mmin. The mutation frequency spectrum is also given in Fig. 3. Even for such a small tumor, there would still be 5,000 mutations, each of which can be found in more than 100 cells. In a different approach to estimating u/r, we used an approximate Bayesian computation method (53) by simulating a branching process with cell birth, death, and mutation often used for modeling tumor growth (54). We obtained the posterior mean u and r that showed u/r = 0.412 (Fig. S8), which was nearly equal to 0.41 obtained from Eq. 1.
In the fourth estimate, M3D, the growth mode is also specified and the increase in cell number is assumed to occur only on the periphery of a tumor in 3D (Materials and Methods, section 14). In the interior, each cell division results in the birth of one cell, which would replace a neighboring cell. Because the births and deaths cancel out in the interior, the growth rate of the tumor (dN/dt) is proportional to N2/3, instead of N as in the exponential growth. Simulation results of Fig. 3 showed that the 3D growth mode yielded similar mutation numbers Mexp, except in the lowest-frequency bin of fewer than 10 cells.
13) Computer Simulations of Meq, a lower bound of MALL.
Meq is the number of mutations in the population by artificially imposing the mutation–drift equilibrium on the tumor. Thus, Meq = θ ln(NT), where θ = 2Neu is the scaled mutation parameter in tumor growth and Ne is the effective cell population size. We implemented computer simulations to prove that Meq is a proper lower bound of mutation number M in a growing population. Meq is expected to always be smaller than M under any mode of tumor growth. Three typical growth models were simulated, including exponential growth, 2D growth, and 3D growth. It should be noted that the cell populations with models of 2D growth (dN/dt = r N1/2) and 3D growth (dN/dt = r N2/3) are essentially well mixed and belong to the power law family of tumor growth models.
For exponential growth, we simulated a discrete-time birth–death process, in which an individual divides and gives birth to two daughter cells with probability b and dies with probability d (b + d = 1) in each generation. Hence the expected exponential growth rate r = ln(2b). In simulation of 2D growth or 3D growth, the birth probability varied with time, depending on the population size N(t). In particular, birth probability b = (1 + r/N(t)1/2)/2 and (1 + r/N(t)1/3)/2, respectively, for 2D growth and 3D growth, where r is the factor determining the growth rate.
Because Meq = θ ln(NT) and θ is unknown, we use the maximum-likelihood method based on the Ewens sampling formula to estimate θ under a particular growth model and associated parameters (2). To do this, we need to know the allele frequency spectrum, which can be obtained by randomly sampling 23 cells at a time from NT = 105 cells (i.e., similar to sampling 23 cell populations for exome sequencing in HCC-15). Therefore, we can obtain both M and Meq. Mutation rate u = 0.03 (the whole coding region) was applied in all of the simulations. In each model, 20 simulations were implemented and the average was treated as the estimate for the mutation numbers M and Meq (Fig. S9).
Fig. S9.
The number of mutations M and Meq from simulation of different growth models. M is the number of mutations in a population of size NT = 105 grown from (A) the exponential model, (B) the 2D growth model, and (C) the 3D growth model. Meq is the equilibrium number of mutations formulated as θ ln(NT). θ is the mutation parameter that was estimated by randomly sampling 23 cells in the population of NT = 105. The method is Ewens’ sampling formula, using the allele frequency spectrum of the 23 sampled cells. Mutation rate in the coding region was set as u = 0.03 per cell division in all simulations.
14) Simulation of Genetic Diversity in Growing Populations.
To simulate the clonal diversity in a tumor and to compare with the theoretical predictions, we designed cellular automata models (55, 56) to simulate tumor expansion and mutation accumulation in 3D space.
15) The Expected Frequency Spectra Under Selection.
Here, we develop a model of selection to compare the total genetic diversity under Darwinian and non-Darwinian evolution (SI Theory). The full model was developed to study the evolution of tumor size (31) with selection and migration. Because the dynamics with migration are the same as those with mutation, the model is interchangeable for mutation and migration (31).
16) Mathematical Derivation of Clonal Diversity HT.
Let Nt be the population size at generation t and u be mutation rate per generation in the coding region of the human genome. We denote by Pr(coalescence) the probability that two randomly chosen cells at generation t find their common ancestor at generation t − 1. And we denote by Jt the probability that two random cells are genetically identical at generation t (1 − Jt is equivalent to Simpson’s diversity index).
Jt can be expressed as a recursive formula:
In a Wright–Fisher growing population, Pr(coalescence) can be approximated by 1/Nt−1, and thus
| [5] |
which can be solved by substituting Ji by Ji−1 successively:
If N0 = 1, the last term can be removed. Then
When u is small, it can be approximated by
Therefore, the expected clonal diversity H, the probability that two random cells or cell clones are genetically different, at time T will be
which is Eq. 3.
The Wright–Fisher model of tumor growth assumes Poisson distribution for the number of offspring cells that a cell generates in a division, which may not be rigorous in modeling cell dynamics. To investigate the generality of Eq. 3, we also derived the exact formula of clonal diversity (HT) under a discrete-time birth–death process of tumor growth. In particular, a cell gives birth to two daughter cells with probability a and dies with probability b (a + b = 1) in a generation. The population grows exponentially with an expected population size of Nt = N0(2a)t at time t. We are primarily interested in a lineage that starts with one single cell and propagates in a total of t generations. Therefore, N0 = 1 and Nt = (2a)t. Suppose two cells are randomly selected from generation t; the probability that coalescence occurs in the previous generation between the two cells is Pr(coalescence) = 1/(Nt − 1). Solving previous recursion in the same way gives rise to
| [6] |
17) Estimating Clonal Diversity, HT, Under Different Growth Models.
Eq. 3 can be applied to arbitrary time variable-size cell populations. However, we examined three models in this study to estimate the clonal diversity HT in a growing cell population (Dataset S8), including an exponential growth model and two power-law growth models (resembling 2D and 3D growth, respectively).
In the exponential model, the population dynamics follow dN/dt = r*N in continuous time. It can be solved as Nt = ert. In discrete time, r = ln(2) corresponds to the situation that all cells duplicate with no cell death in each generation. We showed the results using two growth rate values as r = ln(2) 0.1 and r = ln(2) 0.01, respectively. The other two models, 2D growth (dN/dt = rN1/2) and 3D growth (dN/dt = rN2/3), belong to the power-law family of tumor growth models. In these two models, r = 2π1/2 and r = (36π)1/3 correspond to the condition that the population generates exactly one layer of cells in the periphery in each discrete generation for 2D growth and 3D growth, respectively. In the 3D growth model, two growth rate values were tested, where r = (36π)1/3 × 0.1 and r = (36π)1/3 × 0.01.
We set NT at three levels between 103 and 105 cells and Nis for i between 0 and T, depending on which of the three growth models were used. Once the growth model and the r value were defined, the number of cell divisions required to reach NT could be calculated. We calculated the expected clonal diversity HT from Eq. 3 (Table 1).
SI Materials and Methods
Sample Volume.
The sample volume (number of cells per sample) is a critical factor in sampling (Fig. S2). A large-volume sample with 106 cells, for example, would span many clones, each in low frequency as illustrated in Fig. 3. Such a sample would underrepresent the genetic diversity, as low-frequency mutations cannot be reliably called. On the other hand, sampling with a very small volume, consisting of a single cell, as an extreme example, would have many technical and logistical difficulties. Given the current error rate in single-cell sequencing, a somatic mutation can be called only when it is found in multiple single cells (18). Because only clonal groups of cells can yield reliable information on somatic mutations, a small local patch of cells, which are generally clonal, may be the most efficient sampling strategy at present. When a sample of n cells is sequenced, mutations that are discovered would be those present in the single cell (i.e., the most recent common ancestor of the n cells), from which the n cells descend. Mutations that occur after the ancestral cell started to proliferate would be missed. In this study, n is about 20,000 cells as shown below.
Given that the diameter of a HCC cell is about 25 μm (20–30 μm), and the volume of a microsection is ∼0.196 mm3, the number of cells in a microsection was estimated to be ∼24,000. To test DNA yield from a certain number of cells, DNA was extracted and quantified from 10,000 tumor cells that were precisely collected by laser capture microdissection (LCM). We obtained 30 ng DNA per 10,000 tumor cells on average in four replicates. The cell number in each of the microsections was estimated based on the reference quantity. For the 286 microsections, a median yield of 60 ng DNA (range 20–100 ng) per section was obtained. The median number of cells per sample was ∼20,000, which approximates the number estimated by volume (Fig. S2).
Tagmentation-Based Library Preparation Using the EZ-Tn5 Transposase.
The standard protocols of library construction for next-generation sequencing suggest 1–10 μg DNA as input material per library, which is not appropriate for the DNA yield per microdissected tissue sample in this study. We used the EZ-Tn5 transposase to mediate the fragmentation of double-stranded DNA and ligate synthetic oligonucleotides based on the procedure developed by Adey et al. (57). A catalytically active complex of transposase comprises two transposases, each combined with the specific 19-bp recognition sequences at the ends of the transposable element. The construction of fragment libraries by in vitro transposition requires ∼50 ng of input genomic DNA for whole-exome sequencing (57). In this study, the library preparation for exome sequencing was started with 20 ng genomic DNA. Because PCR amplification of limited amounts of DNA template carries an increased risk of product redundancy, we modified the procedure by ligating a unique DNA identity (UID) (8 bp random oligonucleotides, giving ∼2 × 107 possible barcodes for each encoding oligonucleotide) to the complex of transposase (transposome). The UIDs labeled each genomic DNA template with an individual sequence tag before PCR amplification and were used to remove redundancy in the alignment of sequencing reads. The patent of this method is currently pending (CN201410158327.6). The modified procedure includes the assembling of the transposome, DNA fragmentation, and then library amplification (Fig. S7).
Transposome assembly.
We synthesized two DNA oligonucleotides, with each insert oligo containing one of the sequence platform adapters at the 5′ end and the EZ-Tn5 transposon at the 3′ end (named as insert DNA1 and insert DNA2, Dataset S11). In addition, insert DNA2 includes a sample barcode and UID (Dataset S11). Each insert DNA was annealed with the complementary sequence of the EZ-Tn5 transposon to form double-stranded DNA with a final concentration of 1 μL of 2 μM. Transposome assembly was carried out in a 5-μL volume containing 1 μL double-stranded insert DNA1 and 1 μL double-stranded insert DNA2, 2 μL 100% glycerin, and 1 μL EZ-Tn5 transposase enzyme (EZ-Tn5 <KAN-2> Insertion Kit; Epicentre), by an incubation at 25 °C for 20 min.
Fragmentation via the transposome.
A total of 20 ng genomic DNA was fragmented with 5 μL prepared transposome from step 1, 2 μL 5× fragmentation reaction buffer [50 mM Tris⋅OAc, pH 8.0, 25 mM Mg(OAc)2] (58), and nuclease-free water to a total volume of 10 μL, at 55 °C for 5 min.
Library amplification by PCR.
The fragmented DNA libraries were purified using a QIAquick enzyme reaction purification kit (Qiagen). We combined 20 μL of the purified DNA library with 10 μM of each paired-end PCR primer (the sequences of primer 1 and primer 2 are in Dataset S11) and added 50 μL Phusion 2× HF PCR Master mix (Thermo Scientific) to a total volume of 100 μL. PCR reactions were heated at 72 °C for 5 min, followed by10 cycles of denaturing at 98 °C for 10 s, annealing at 53 °C for 30 s, and extension at 72 °C for 3 min; this was followed by a final extension at 72 °C for 10 min. A 350–550 bp size selection was carried out on a 2.0% agarose gel, followed by purification using the QIAquick Gel Extraction Kit (Qiagen). In total, we constructed 24 tagmentation-based libraries for 23 tumor and 1 adjacent nontumor sections.
Whole-Exome Sequencing and Sequence Alignment.
We performed whole-exome capture on every four fragmented DNA libraries. The four libraries, each ligated with a unique barcode (Dataset S11), were equally pooled. According to the manufacturer’s instructions, 800 ng of pooled DNA libraries was captured using the Human All Exon kit SureSelect Target Enrichment System (Agilent) and custom blockers based on the insert DNA1 and insert DNA2 sequences (Dataset S11). The captured libraries were amplified by 10 cycles of 98 °C for 10 s, 53 °C for 30 s, and 72 °C for 3 min, followed by extension at 72 °C for 10 min, and purified using the QIAquick Gel Extraction Kit (Qiagen). The insert size and the concentration of purified libraries were exanimated using Agilent Bioanalyzer and qRT-PCR. Paired-end (2 × 100 bp) multiplex sequencing of samples on the Illumina HiSeq2000 platform was performed to the desired median sequencing depth (>70×) at the core facility of Beijing Institute of Genomics (BIG) (Dataset S1). Paired-end reads in FastQ format were aligned to the human reference sequence (National Center for Biotechnology Information build hg19), using the Burrows–Wheeler Aligner (BWA) (59) with default parameters. PCR redundancy in sequencing reads was removed according to the 8-bp UID sequences.
Detection of Somatic Single-Nucleotide Variation.
In addition to the descriptions in Materials and Methods, section 4, the following filtering criteria were applied to detect somatic single-nucleotide variations (SNVs): (i) Only reads mapping uniquely to the genome with less than three mismatches are considered; (ii) only nucleotides with Phred quality of 20 and mapping quality of 30 or greater are considered; (iii) the number of start points in the reads covering the mutant sites is >1, and both forward and reverse reads are required; (iv) score of somatic mutation is greater than 35; (v) P-value of Fisher’s exact test (one tailed) for the mutant frequency in the tumor is <0.05; (vi) P-value of binomial test for the tumor alleles is <1 × 10−15; (vii) the number of unique UIDs in the reads that support a mutation is larger than 4; (viii) the minimum coverage of 10× and 6× is required in at least one sequenced tumor sample and in the normal sample, respectively; (ix) the minimum coverage of 150× is required in merged exome sequencing reads in 23 tumor samples; and (x) the tumor MAF must be greater than 0.1. All somatic mutations are shown in Dataset S2.
Detection of Copy Number Alterations and Estimation of Tumor Purity.
CAScnv (an in-house software) was used to detect the somatic copy number alterations (CNAs). We computed the average sequencing depth of each gene and used the circular binary segmentation algorithm (CBS) to segment CNA regions according to logarithm reads ratio (LRR),
where RC and RN are the sequencing read counts in a gene region in cancer and normal samples, respectively, and γ is the correction coefficient denoting the read counts ratio of normal and cancer DNA in a chromosome (48). Finally, we divided the genome into contiguous regions according to the CBS segmentation significance level (P < 1 × 10−5). The CAScnv called 542 CNAs from 23 whole-exome sequencing datasets. On average, each sample contained 23.6 CNAs. These CNAs are distributed among 14 chromosomes. All detected CNAs are listed in Dataset S4. We used ABSOLUTE (49) (www.broadinstitute.org/cancer/cga/ABSOLUTE) to infer the purity and ploidy for our samples as described in Materials and Methods, section 3.
SI Theory
Inference of u, r, and Nτ by Approximate Bayesian Computation.
We have demonstrated that the growth of HCC-15 is likely to be a neutral process in the main text. To gain a sense of the parameters during tumor growth, e.g., mutation rate and growth rate, we modeled the growth of HCC-15 by a neutral discrete-time birth and death process. In particular, a cell divides and gives rise to two daughter cells with probability b and dies with probability d in each generation. The population is expected to grow exponentially with growth rate r = ln(2b). Thus, EN(t) = eln(2b)t when started from a single cell. Because of the complexity of parameter space, we used the approximate Bayesian computation (ABC) (53) approach to infer relevant parameters, which include growth rate r = ln(2b), mutation rate u per cell division, and ancestral population size Nτ (defined as the average tumor size at the time that the observed clones with polymorphic mutations were first derived during the growth of HCC-15).
The ABC inference scheme is as follows:
-
i)
Sample the parameter Θ = [b, u, Nτ] from the prior distributions f(Θ).
-
ii)
Simulate the tumor growth process with Θ; sample 23 cells at the time of Nτ and calculate the summary statistics S′.
-
iii)
If ρ(S′, Sobs) < ε, accept Θ.
-
iv)
Go to step i.
We use six summary statistics, grouped as S = [H, Am, Vm, ξ1, ξ2, ξ3], where H is clonal diversity, Am is average number of polymorphic mutations acquired by the individual cells (or clone), Vm is variance of the number of mutations per cell (or clone), ξ1 is number of singletons in the site frequency spectrum, ξ2 is number of doubletons in the site frequency spectrum, and ξ3 is number of tripletons in the site frequency spectrum.
The observed summary statistics in HCC-15 are Sobs = [0.941, 2.652, 1.692, 26, 7, 1]. The prior parameters we used are uniform distributions, where b ∼ Uniform[0.5, 0.7], Log10(u) ∼ Uniform[−4, −1], and Log10(Nτ) ∼ Uniform[2, 7]. ρ is a distance measure and ε is the error threshold. The R package abc (60, 61) was applied to obtain the posterior distribution of parameters (Fig. S8), in which a nonlinear (neural network) regression algorithm and 0.1% tolerance were used.
The Expected Frequency Spectra Under Selection.
Here, we develop a model of selection to compare the total genetic diversity under Darwinian and non-Darwinian evolution. The full model was developed to study the evolution of tumor size (12) with selection and migration. Because the dynamics with migration are the same as those with mutation, the model is interchangeable for mutation and migration, which is designated u in Eqs. S1 and S2. The relevant text from Tao et al. (31) is reproduced below.
To understand the forces driving multitumor evolution, it is necessary to derive the size distribution of clones. Within the same tumor, clones are defined by new mutations but, in multitumor cases, a new tumor seeded by migrant cells can be considered a new clone as well. By substituting the rate of migration for the mutation rate, we can derive the size distribution of tumors, just as we calculated clone sizes in Fig. 4, using the infinite-allele model (2, 3). A cell clone is thus either a population of cells sharing the same mutation (a mutation clone) or a population of cells originating after the same migration event (a migrant clone or a new tumor).
In formulating the clone size distribution, the neutral null model assumes that cells divide and mutate (or migrate) with a given probability without invoking selection or other complex interactions (4). Let the migration rate (number of migrant cells per cell per generation) be u. Given any u, we ask (i) how many tumors are visible, defined as those that are larger than, for example, 1% of the total tumor mass, and (ii) how many are too small (<1%) to see (31). Various tumor migration models (14, 17) make similar predictions on visible tumors and the dynamic model of Iwasa and Michor (62) is adopted here.
The model assumes that a tumor grows exponentially with a rate of r (12). The number of emigrant cells at time t is u[N(t)]α = ueαrt, where α is a fractal dimension factor that corrects the spatial effect on cell migration. When only cells on the periphery of a 3D sphere are capable of emigration, α = 2/3. Let G(x) be the number of migrant clones that have more than x cells at time T. For neutrally growing tumors,
| [S1] |
Eq. S1 has been extended to include growth advantages. In the extension, the newly seeded tumor has a growth rate, s, which may be higher or lower than r (the growth rate of the parent tumor). We assume that s follows an exponential distribution with the mean of r. With this distribution, the growth rate of most new tumors would be lower than that of the parental tumor, but a few would have a much greater growth rate. G′(x) is the number of migrant clones with more than x cells,
| [S2] |
Both G(x) and G′(x) [see Tao et al. (31) for the derivation] will be used to estimate the total tumor mass that will include the unobserved clones.
The clonal size distributions under the selection model and under neutrality are shown in Dataset S9. It can be seen that the diversity pattern under selection is dominated by a small number of large clones at the expense of many small clones. The largest bin with >108 cells accounts for more than 90% of the cell mass under selection whereas the largest bin accounts for only 10% of the mass under neutrality.
SI Discussion
The main difference between this study and previous publications on intratumor diversity (11–17, 63) is the application of the neutral population genetic theory to estimating the total diversity (2, 3). Previous studies have not compared the observed diversity with the neutral predictions, partly because most subclonal mutants were assumed to be under Darwinian selection. To use the non-Darwinian (i.e., neutral) theory, it is necessary to show that the clonal sizes follow the neutral predictions as given by Eqs. 1 and 2 of the main text.
Sample Volume and the Delineation of Clones.
Given the large number of clones expected, the size of each of them can be determined only when the sampling scheme fulfills several criteria. First, the number of samples has to be adequate, preferably larger than 100. To our knowledge, no other study has that level of saturation sampling. Second, the samples have to be evenly distributed in space, given that clones are highly spatially clustered. This is to our knowledge the only study that carries out honeycomb-like even sampling. Third, the sample volume has to be small, preferably <20,000 cells per sample. Only Sottoriva et al. (30) met that criterion. Although single-cell sequencing would be the ultimate fulfillment of the third criterion (18, 64, 65), the technology at present is not ready for the large-scale applications required here. Given the high error rate, many cells from a local cluster have to be sequenced and mutually validated to yield usable polymorphism data. In that case, it would seem easier to sequence a local cluster of a few hundred or a few thousand cells. Of course, when the error rate is sufficiently low such that the mutual validation from two single cells would yield reliable polymorphism, single-cell sequencing will be most informative. In Fig. 4D of the main text, the analysis is akin to this approach of multiple single-cell sequencing. We estimated that each sample consisted of 40 single-cell data.
Two Different Definitions of Clones.
An important aspect of the analysis of this study is the distinction between “mutation clones” and “cell clones.” A mutation clone comprises all cell clones that harbor a particular mutation. The distinction permits the uses of two classes of population genetic models—the infinite-allele model that corresponds with cell clones and the infinite-site model that corresponds with mutation clones (2, 3). Studies that do not make such a distinction are unable to compare the observations with the theoretical predictions.
Darwin’s Puzzle in Relation to Intratumor Genetic Diversity.
Darwinian selection is expected to reduce genetic diversity when the better genotypes take over the population and the deleterious genotypes are quickly eliminated. The same forces should reduce the level of genetic diversity within tumors. Imagine a tumor with 100 cell clones among which 90, 9, and 1 clones grow at the rate of r, 1.5r, and 2r, respectively. Initially, all 100 clones are detectable (>0.1% of the tumor mass). In 10 cell generations, only 10 clones will be detectable and in 20 generations only 1 cell clone will be. Bozic et al. (10) have also calculated that a selective advantage of 0.4% would be sufficient for clonal replacement and, presumably, the reduction or loss of diversity.
Non-Darwinian vs. Darwinian Evolution Within and Between Tumors.
Darwinian evolution within tumors has often been suggested without a formal test against a null model. The selectionism–neutralism debate in the last 50 y showed how difficult it is to catch selection in action by studying genetic variants within populations (26, 66). “Good” mutations are rare and, after emergence, are expected to spread through the population rapidly. Although Darwinian selection has been shown to be a main driving force of genic evolution between species (28, 67), variations within populations appear minimally affected by Darwinian selection (66, 68). Evolution at the cellular level may require even more rigorous analyses than those used in the studies of natural populations. For example, the hallmark of selection is Ka/Ks < 1, which has been observed among all organisms studied (1, 3). [Ka/Ks is the ratio of the nonsynonymous rate of changes over the synonymous rate (1)]. Whereas Ka/Ks ∼ 0.3 in the human/primate lineage, it is ∼1 in almost all cancers in the TCGA data (36). A Ka/Ks ratio of 1 indicates that selection on nonsynonymous changes is no greater than on synonymous changes. Indeed, the absence of recombination in large genomes, as in tumors, makes selection inefficient when it is in operation (69, 70).
Finally, in the companion study (31), we study 12 additional multinodule HCCs. Instead of comparing the observed clone sizes with the predictions, we compare each descendant clone with the parental one to test the neutral prediction. Under neutrality, the younger clones are expected to be smaller than the older parental clones. This approach permits a more comprehensive and sensitive test of the non-Darwinian model of evolution. Tao et al. (31) focus on the evolution between tumors whereas this study focuses on the evolution within tumors.
Supplementary Material
Acknowledgments
We are grateful to Nick Navin, Steve Frank, Nelly Polyak, Carlo Maley, Robert Gatenby, Rick Durrett, Thomas Nagylaki, Tian Xu, Jianzhi (George) Zhang, Andy Clark, Xionglei He, and Yang Shen for inputs in various phases of this work. This study was supported by the National Basic Research Program (973 Program) of China (2014CB542006 to C.-I.W.), Research Programs of the Chinese Academy of Sciences (XDB13040300 and KJZD-EW-L06-1 to X.L. and C.-I.W.), the National Science Foundation of China (91131903 to X.L., 31301036 and 91231204 to Z.Y.), the National High-Tech R&D Program (863 Program) of China (2012AA022502 to X.L.), and the 985 Project (33000-18811202 to C.-I.W.) and Science Foundation of State Key Laboratory of Biocontrol (SKLBC15A37 to C.-I.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence data reported in this paper have been deposited in the genome sequence archive of Beijing Institute of Genomics, Chinese Academy of Sciences, gsa.big.ac.cn (accession no. PRJCA000091).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519556112/-/DCSupplemental.
References
- 1.Wen-Hsiung L. 1997. Molecular evolution (Sinauer Associates Inc., Sunderland, MA)
- 2.Ewens WJ. Mathematical Population Genetics 1: Theoretical Introduction. Springer; New York: 2010. [Google Scholar]
- 3.Hartl DL, Clark AG. Principle of Population Genetics. 4th Ed Sinauer; Sunderland, MA: 2006. [Google Scholar]
- 4.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 5.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38(4):468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 6.Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6(12):924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk A, Polyak K. Tumor heterogeneity: Causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 9.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 12.Tao Y, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci USA. 2011;108(29):12042–12047. doi: 10.1073/pnas.1108715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155(3):1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher RA. The Genetical Theory of Natural Selection. Clarendon; Oxford: 1930. [Google Scholar]
- 21.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23(1):23–35. [PubMed] [Google Scholar]
- 22.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105(11):4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durrett R. Population genetics of neutral mutations in exponentially growing cancer cell populations. Ann Appl Probab. 2013;23(1):230–250. doi: 10.1214/11-aap824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; Cambridge, UK: 1984. [Google Scholar]
- 27.Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- 28.Fay JC, Wyckoff GJ, Wu C-I. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415(6875):1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 29.Woo YH, Li W-H. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun. 2012;3:1004. doi: 10.1038/ncomms1982. [DOI] [PubMed] [Google Scholar]
- 30.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao Y, et al. 2015. Further genetic diversification in multiple tumors and an evolutionary perspective on therapeutics. BioRxiv:025429.
- 32.Schulze K, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255(5505):197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 34.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 2014;10(3):e1004239. doi: 10.1371/journal.pgen.1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read AF, Day T, Huijben S. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10871–10877. doi: 10.1073/pnas.1100299108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Catenacci DVT. Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity. Mol Oncol. 2015;9(5):967–996. doi: 10.1016/j.molonc.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leder K, et al. Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell. 2014;156(3):603–616. doi: 10.1016/j.cell.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loven D, Hasnis E, Bertolini F, Shaked Y. Low-dose metronomic chemotherapy: From past experience to new paradigms in the treatment of cancer. Drug Discov Today. 2013;18(3-4):193–201. doi: 10.1016/j.drudis.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 41.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69(11):4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva AS, et al. Evolutionary approaches to prolong progression-free survival in breast cancer. Cancer Res. 2012;72(24):6362–6370. doi: 10.1158/0008-5472.CAN-12-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson DE, et al. SomaticSniper: Identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth A, et al. JointSNVMix: A probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics. 2012;28(7):907–913. doi: 10.1093/bioinformatics/bts053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu X, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46(3):287–293. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 52.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162(4):2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durrett R. Branching Process Models of Cancer. Springer; Cham, Germany: 2015. pp. 1–63. [Google Scholar]
- 55.Poleszczuk J, Enderling H. A high-performance cellular automaton model of tumor growth with dynamically growing domains. Appl Math. 2014;5(1):144–152. doi: 10.4236/am.2014.51017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525(7568):261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11(12):R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harbers M, Kahl G, Kahl G. Tag-Based Next Generation Sequencing. Wiley; Weinheim, Germany: 2012. [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Csilléry K, Blum MGB, Gaggiotti OE, François O. Approximate Bayesian computation (ABC) in practice. Trends Ecol Evol. 2010;25(7):410–418. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 61.Csilléry K, François O, Blum MGB. abc: An R package for approximate Bayesian computation (ABC) Methods Ecol Evol. 2012;3:475–479. [Google Scholar]
- 62.Iwasa Y, Michor F. Evolutionary dynamics of intratumor heterogeneity. PLoS One. 2011;6(3):e17866. doi: 10.1371/journal.pone.0017866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nei M. Mutation-Driven Evolution. Oxford Univ Press; Oxford: 2013. [Google Scholar]
- 67.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351(6328):652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 68.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 2007;89(5-6):311–336. doi: 10.1017/S001667230800949X. [DOI] [PubMed] [Google Scholar]
- 70.Gerrish PJ, Colato A, Perelson AS, Sniegowski PD. Complete genetic linkage can subvert natural selection. Proc Natl Acad Sci USA. 2007;104(15):6266–6271. doi: 10.1073/pnas.0607280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.