Significance
NMDA receptors (R) are ion channels that participate in many important physiological and pathophysiological processes in the brain. Recent studies suggest that NMDARs can also act in a manner that does not require ion flow. One way this could occur is for agonist binding to the NMDAR to produce conformational movement of its cytoplasmic domain in the absence of ion flow. Here we use FRET imaging of genetically encoded fluorescent proteins linked to the cytoplasmic domain of NMDAR subunits to monitor directly agonist-driven movement within the cytoplasmic domain of NMDARs. This study demonstrates that transmembrane information transfer through the NMDAR is possible in the absence of ion flow through the receptor.
Keywords: conformational change, FRET-FLIM, antibody infusion, metabotropic, receptor cross-linking
Abstract
The NMDA receptor (R) plays important roles in brain physiology and pathology as an ion channel. Here we examine the ion flow-independent coupling of agonist to the NMDAR cytoplasmic domain (cd). We measure FRET between fluorescently tagged cytoplasmic domains of GluN1 subunits of NMDARs expressed in neurons. Different neuronal compartments display varying levels of FRET, consistent with different NMDARcd conformations. Agonist binding drives a rapid and transient ion flow-independent reduction in FRET between GluN1 subunits within individual NMDARs. Intracellular infusion of an antibody targeting the GluN1 cytoplasmic domain blocks agonist-driven FRET changes in the absence of ion flow, supporting agonist-driven movement of the NMDARcd. These studies indicate that extracellular ligand binding to the NMDAR can transmit conformational information into the cell in the absence of ion flow.
The NMDA receptor (R) is a ligand-gated ion channel precisely positioned at synapses to act as a coincidence detector (1, 2), which may permit this molecule to mediate the association of memories in the brain (3, 4). Despite the importance of the NMDAR in the development, function, and dysfunction of the brain (5–8), the molecular link between its agonist-driven activation to its signaling function is not well understood. Recent studies suggested that activation of NMDARs can transmit signals in the absence of ion flux through the channel (9–12). An important test of this view is to measure directly agonist-driven changes in the cytoplasmic domain of NMDARs in the absence of ion flow through the NMDAR.
Transmembrane signaling by transmembrane receptors in the absence of ion flux is common in cells; less common are classic ion channels with a secondary metabotropic function. G protein-coupled receptors (GPCRs) act by transducing extracellular agonist binding to a conformational change that activates signaling molecules controlling intracellular physiology (13). Such conformational changes induced by agonist binding to GPCRs have been measured directly using spectroscopic methods, including FRET (14, 15). For the NMDAR, a tetrameric receptor composed of two GluN1 and two GluN2 subunits in the hippocampus (7, 16), intrareceptor FRET signals from fluorescent molecules connected to the extracellular domain of subunits have been measured and used to monitor the assembly of the receptors (17). However, detection of transmembrane transduction that couples agonist binding to conformational changes at the cytoplasmic domain of the NMDAR has not been attempted.
Results
FRET Between Fluorescent Proteins Placed at the NMDAR Cytoplasmic Domain.
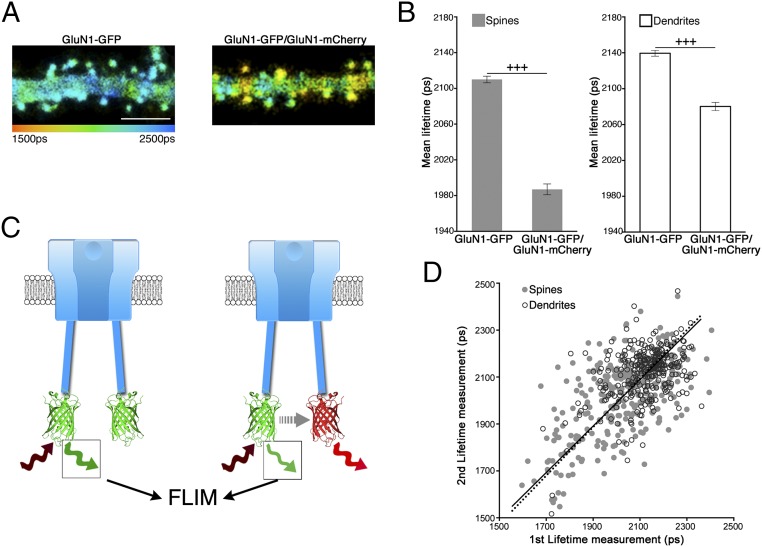
To monitor conformational changes in the cytoplasmic domain of the NMDA receptor (NMDARcd), we cotransfected carboxyl terminally tagged GluN1-GFP and GluN1-mCherry into primary cultured hippocampal neurons. Untagged GluN2B was also cotransfected to ensure delivery of recombinant functional tetrameric NMDAR to synapses (18). We chose to use GluN1 C-terminus tagging, rather than tagging of GluN2B, because the latter affects NMDAR trafficking (18). We used fluorescence lifetime imaging microscopy (FLIM) to measure FRET between GluN1-GFP and GluN1-mCherry within the NMDAR (the GluN1-mCherry cDNA transfected was threefold that of GluN1-GFP, to minimize GluN1-GFP/GluN1-GFP pairings). The fluorescence decay time, or lifetime, of the FRET donor GFP is highly sensitive to the proximity of mCherry, the FRET acceptor (19). Hence, we observed that GluN1-GFP lifetime was significantly shorter in dendritic spines (sites of synaptic communication) and nearby dendritic segments when GluN1-mCherry/GluN2B were coexpressed (1,986 ± 6 ps; n = 470), compared with only GluN1-GFP/GluN2B expression (2,110 ± 4 ps; n = 711, P < 0.0001), indicating successful FRET between GFP and mCherry (Fig. 1 A and B). The level of FRET we observed is comparable to values measured in other studies (SI Appendix, Table S1) and is consistent with a distance of 8.3 nm between carboxyl terminally tagged GluN1-GFP and GluN1-mCherry, assuming random orientation between fluorophores (Materials and Methods). This distance is well within the crystal structure dimensions obtained for individual NMDARs (20).
Fig. 1.
FRET between GluN1 subunits at individual NMDARs. (A) FLIM of dendrite and spines expressing indicated constructs (along with GluN2B in all figures). Pseudocolor (scale below) indicates GFP lifetime at each pixel. (Scale bar, 5 µm.) (B) Average GluN1-GFP lifetime for spines and corresponding dendritic segments (located under the spines) expressing indicated constructs; n > 20 neurons; > 400 spines (for each condition); +++P < 0.001 (Mann–Whitney). Error bars indicate SEM in all figures. (C) Model of intrareceptor FRET between GluN1-GFP and GluN1-mCherry in NMDAR; GluN2 cytoplasmic domain not shown for clarity (GluN2 N-terminal domain is dark blue). Two-photon excitation (brown), emission (green, red), and FRET (gray) energies indicated. (D) Plot of first vs. second mean lifetime measurement of GluN1-GFP/GluN1-mCherry/GluN2 expressing spines (gray circles) and dendrites (white circles with black outline). Black line is best fit minimizing both x and y distances (spines: full line; dendrites: dotted line); n = 309 spines, or dendritic segments, 17 neurons.
The amount of FRET between GluN1-GFP and GluN1-mCherry was higher in spines than in nearby dendritic compartments (Fig. 1 A and B), suggesting that the conformation of the NMDARcd in spines, which contain numerous synaptic proteins (21), differs from that of extrasynaptic NMDARs, which are thought to transduce different signals (22). Although there was considerable variation in FRET among spines (GluN1-GFP lifetime SD = 162 ps; n = 309) (Fig. 1B), the GluN1-GFP lifetime for individual spines and dendritic segments was significantly correlated when measured a second time (for spines: R = 0.62; n = 309; P < 0.0001; Pearson’s r test) (Fig. 1C). Furthermore, despite a reduced mean value, the variance in GluN1-GFP lifetime was significantly greater when GluN1-mCherry was coexpressed than when not coexpressed (F statistic = 1.80; P < 0.0001; F test). These data support the view that the variation in GluN1-GFP lifetime in neurons expressing GluN1-GFP/GluN1-mCherry/GluN2B is a result of biological differences among spines, as well as between spines and dendrites, producing different NMDARcd conformations, rather than noise in lifetime measurements.
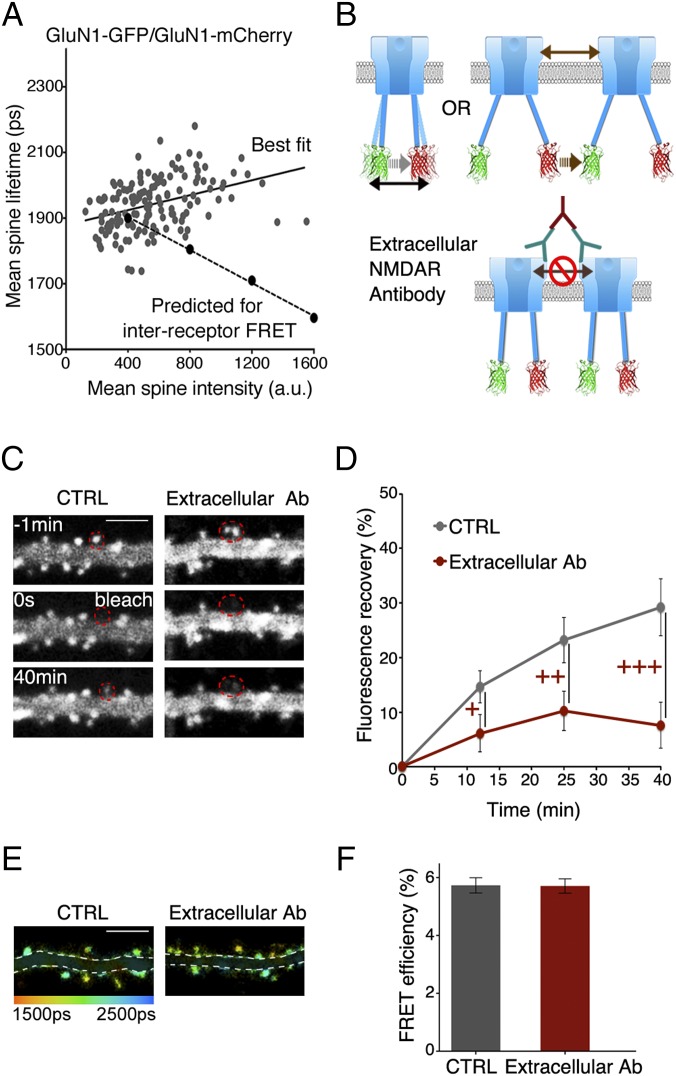
A number of observations indicate that the FRET measured is caused by interactions between GluN1-GFP and GluN1-mCherry on individual NMDARs; that is, intrareceptor rather than interreceptor between fluorophores on different NMDARs. We first noted that the estimate of the distance between fluorophores (8.3 nm) is considerably smaller than the average distance estimated between NMDARs on a synapse (∼100 nm) (23), and that not all molecules colocalized at a synapse (e.g., GluN1-GFP and Homer-mCherry) display FRET (24). Nevertheless, receptor clustering could still produce interreceptor FRET. To test experimentally if any FRET was caused by interreceptor interactions, we first examined if the GFP lifetime in spines expressing GluN1-GFP, GluN1-mCherry, and GluN2B was reduced in spines containing more recombinant receptors, as would be expected for increased receptor concentration (Fig. 2A). However, there was no reduction in GFP lifetime as a function of fluorescence intensity; the data are significantly different from what would be expected if there were interreceptor FRET (P < 0.0001) (Materials and Methods).
Fig. 2.
GluN1-GFP/GluN1-mCherry FRET occurs within individual NMDARs and is unaffected by induced receptor crosslinking. (A) Plot of lifetime vs. fluorescence intensity with best fit line (solid) and with interreceptor FRET expected line (dashed). Each data point is mean of all spines from one neuron expressing GluN1-GFP/GluN1-mCherry; n = 150 neurons (Materials and Methods). (B) Ligand-induced lifetime increase should be unaffected by extracellular cross-linking for intrareceptor FRET, but blocked for interreceptor FRET. (C) Representative fluorescence intensity images of neurons expressing GluN1-GFP/GluN1-mCherry in indicated conditions at indicated times before and after photobleaching spines encircled in red. (Scale bar, 5 µm.) (D) FRAP curves for indicated conditions; n >18 neurons; > 22 spines +++P < 0.001; ++P < 0.01; +P < 0.05; error bars, SEM. (E) Representative FLIM images of neurons in indicated conditions in 7CK. (Scale bar, 5 µm.) (F) Plot of FRET efficiency in spines with indicated treatments; n > 30 neurons; > 550 spines per condition.
As a second test to distinguish between interreceptor and intrareceptor FRET, we extracellularly applied antibodies (primary antibody to GluN1 extracellular domain along with a secondary antibody to the primary antibody) to cross-link NMDARs (25) (Fig. 2B). We used a similar procedure as what was used for AMPA receptors (25) and confirmed immobilization of surface receptors by fluorescence recovery after photobleaching experiments (FRAP) (26) (Fig. 2 C and D). Extracellular antibody incubation reduced the mobile fraction of recombinant receptors by more than 70% (Materials and Methods). If a significant amount of FRET were a result of interaction between different receptors, one would expect that cross-linking different receptors would increase FRET efficiency between GluN1-GFP and GluN1-mCherry. However, no change in FRET efficiency was observed (Fig. 2 E and F). Therefore, these data (also see below) support the FRET signal occurring because of interactions between GluN1-GFP and GluN1-mCherry at individual NMDARs (Fig. 2).
Agonist Binding Drives Reduced FRET Within NMDAR Cytoplasmic Termini.
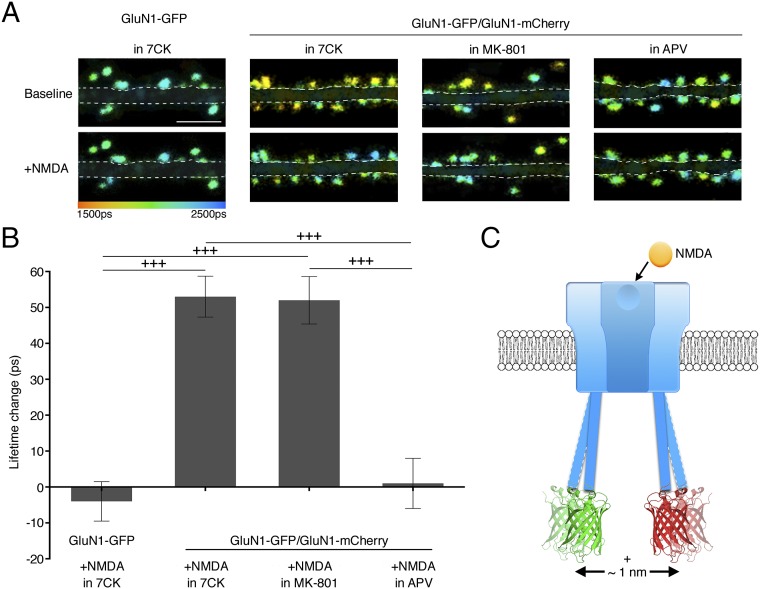
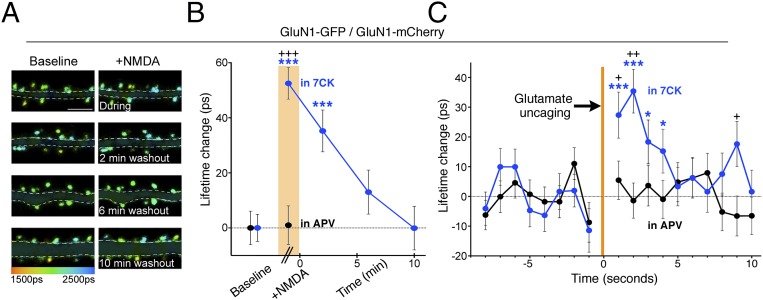
At nonchannel transmembrane receptors that produce intracellular signaling, agonist-induced conformational changes in the cytoplasmic domain can be detected by intramolecular FRET (27). We thus investigated if agonist binding to NMDARs produced conformational changes in the NMDARcd in the absence of ion flux through the receptors. GluN1-GFP, GluN1-mCherry, and GluN2B were transfected into dissociated cultured hippocampal neurons and allowed to express for several days. FLIM measurements were obtained before and during bath application of NMDA; for each spine we measured the difference in lifetime (spine lifetime in NMDA – spine lifetime before NMDA) and then averaged these values. The bath also contained (before and during agonist application) the noncompetitive NMDAR antagonist 7CK or the NMDAR ion-channel blocker MK-801. In both cases, upon addition of NMDA the GluN1-GFP lifetime increased (in 7CK: 52 ± 6 ps; n = 588; P < 0.0001; in MK-801: 47 ± 7 ps; n = 481; P < 0.0001) (Fig. 3 A and B), corresponding to lower FRET efficiency (EFRET) (from 5.8 ± 0.2% to 3.3 ± 0.3% in 7CK and 5.5 ± 0.3% to 3.1 ± 0.3% in MK-801) comparable in magnitude to FRET-FLIM studies on other molecular processes (SI Appendix, Table S1). This reduced FRET efficiency indicates an increase in the separation of GFP and mCherry by ∼1 nm (Materials and Methods and Fig. 3C) and/or a change in their relative orientation, demonstrating the sensitivity of FRET-FLIM in detecting small changes in protein conformation. Similar agonist-induced results were obtained if NMDAR conductance was blocked with a lower concentration of 7CK or if all excitatory transmission was blocked with 7CK, NBQX, and TTX, or if glutamate was used as agonist (SI Appendix, Fig. S1); permitting ion passage did not block the effect (SI Appendix, Fig. S2). Smaller, yet significant, changes were seen in dendritic segments (SI Appendix, Fig. S3). In contrast, an NMDA-induced increase in GluN1-GFP lifetime was not observed when APV was present during the NMDA application (Fig. 3 A and B), nor if NMDA was applied in the presence of 7CK to neurons expressing only GluN1-GFP and GluN2B (Fig. 3 A and B) nor if mCherry (alone) was coexpressed along with GluN1-GFP and GluN2B. These results indicate that agonist binding to the NMDAR produces a conformational change in the NMDARcd in the absence of ion flux through the receptor.
Fig. 3.
Agonist binding induces conformational change in NMDAR cytoplasmic domain without ion flux. (A) Representative FLIM images of neurons expressing indicated constructs (with GluN2B in all figures) before and in 25 µM NMDA. (Scale bar, 5 µm.) Dendritic segments are masked for clarity (see SI Appendix, Fig. S3 for unmasked segments). (B) Average NMDA-induced spine GluN1-GFP lifetime change for indicated constructs and conditions, n > 20 neurons, > 495 spines for each condition; +++P < 0.001; error bars SEM; Mann–Whitney U test. (C) Model consistent with FRET changes in NMDAR.
We next tested if the observed agonist-driven change in FRET between GluN1-GFP and GluN1-mCherry is a result of changes within individual NMDARcds rather than between different NMDARcds. Primary antibodies directed to the extracellular domain of the NMDA receptor, along with secondary antibodies, were applied to neurons expressing GluN1-GFP, GluN1-mCherry, and GluN2B. Such treatment effectively cross-linked different NMDARs with each other, as indicated by FRAP experiments (Fig. 2 B and C). NMDA was applied to these neurons and a similar increase in GluN1-GFP lifetime was observed compared with interleaved control neurons that were not incubated with antibodies (with extracellular antibody in 7CK: 41 ± 7 ps; n = 634; control neurons in 7CK: 45 ± 7 ps; n = 577; P = 0.67). Because extracellular antibody immobilized NMDARs along the surface membrane, and ligand induced a comparable FRET reduction, we can conclude that the observed change in FRET cannot be because of a ligand-driven modification of clustering of distinct NMDARs.
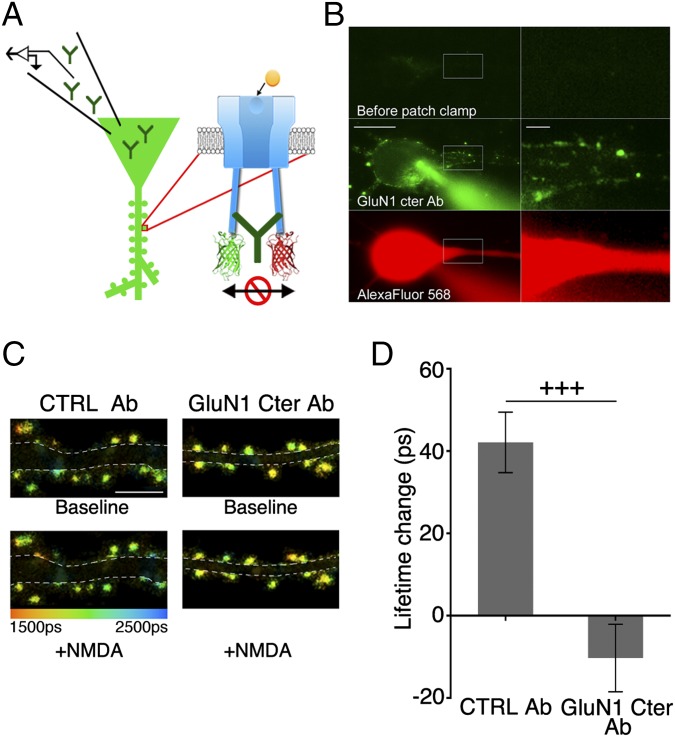
To test further that the ligand-driven FRET reduction was because of movement within individual NMDARcds, we designed an experiment to block NMDARcd movement [notably, the downstream effects of NMDARcd movement, described in the companion paper (28), were also blocked by this method]. Neurons were infused with a patch pipette containing an antibody targeting the GluN1cd (or an anti-rabbit antibody as a control) (Fig. 4A), which would be expected to bind—and immobilize—two nearby GluN1cds. Indeed, after allowing GluN1cd antibody to diffuse into the neuron (Fig. 4B), NMDA application in the presence of 7CK failed to produce a change in GluN1-GFP lifetime in neurons expressing GluN1-GFP/GluN1-mCherry/GluN2B. In contrast, after infusion of neurons with a control antibody, NMDA application in the presence of 7CK did produce a significant increased lifetime in neurons (Fig. 4 C and D). The basal amount of FRET between GluN1 subunits was unaffected by GluN1cd antibody infusion (GluN1cd antibody EFRET = 6.2 ± 0.3%; n = 478; control antibody EFRET = 5.9 ± 0.3%; n = 378; P = 0.4, unpaired t test), suggesting that this procedure is not affecting NMDARcd basal conformation. Thus, intracellular delivery of a GluN1cd antibody blocked agonist-driven FRET reduction, supporting the view that agonist binding leads to movement of the NMDARcd. Importantly, intracellular GluN1-cd antibody infusion had no effect on the mobile fraction of NMDARs measured with FRAP (with GluN1cd antibody: 28 ± 7%; n = 30; control: 29 ± 5%; n = 27; P = 0.88, unpaired t test), indicating this antibody treatment (which contained only primary antibody) produced intrareceptor immobilization rather than interreceptor immobilization (which was achieved above with extracellularly applied primary and secondary antibodies).
Fig. 4.
NMDA-induced FRET changes are blocked by intracellular infusion of GluN1 C-ter antibody. (A) GluN1 C terminus (GluN1 Cter) antibody introduced in neurons expressing GluN1-GFP/GluN1-mCherry to restrain NMDARcd movement. (B) Representative images of a neuron infused with Alexa Fluor 488 conjugated GluN1 Cter Ab, along with red dye. [Scale bars, 10 μm (Left); 2 μm (Right).] (C) Representative FLIM images of neurons expressing GluN1-GFP/GluN1-mCherry in 7CK and indicated conditions, infused with indicated antibody 30–60 min before imaging. (Scale bar, 5 μm.) (D) Average spine GluN1-GFP lifetime change for conditions in C. N, CTRL Ab (16 neurons, 378 spines), GluN1 Cter Ab (21 neurons, 478 spines). +++P < 0.001; error bars SEM; unpaired t test.
Transient Agonist Binding Drives Transient FRET Changes Within NMDAR Cytoplasmic Termini.
We next sought to determine the temporal dynamics of the NMDARcd conformational change observed during agonist binding. In neurons expressing GluN1-GFP/GluN1-mCherry/GluN2B, NMDA was briefly (∼6 min) bath-applied in the presence of 7CK and lifetime changes were measured in spines during NMDA application and at fixed intervals during NMDA washout. GluN1-GFP lifetime increased in the presence of NMDA and returned to baseline levels (Fig. 5 A and B) with a time course consistent with the gradual decrease in NMDA concentration in the bath; measurements using a fluorescent dye to mimic NMDA washout dynamics indicate an agonist half-decay time of 2.7 ± 0.4 min in the bath (n = 4). This result suggests that (i) NMDARcd conformational change persists only while extracellular agonist is bound, and (ii) after removal of the agonist the NMDARcd returns to the prestimulus conformation, although we cannot exclude the possibility that the NMDARcd returns to a different conformation that preserves the initial GluN1-GFP/GluN1-mCherry FRET.
Fig. 5.
Time course of NMDA-driven conformational changes of NMDAR cytoplasmic domain. (A) Representative FLIM images of neurons expressing indicated constructs before (Left) during and after (Right) NMDA application in 7CK (Materials and Methods). (Scale bar, 5 μm.) (B) Plot of GluN1-GFP spine lifetime change, relative to baseline, in 7CK (blue) or APV (black) before, during (orange bar) and after NMDA application; n > 300–600 spines, > 13 neurons per condition; ***P < 0.001 compared with baseline value (Wilcoxon); +++P < 0.001 compared with value in APV (Mann–Whitney). (C) Plot of change, relative to baseline, in GluN1-GFP lifetime induced by glutamate uncaging (orange bar). Blue, in 7CK; black, in APV; n > 235 spines, > 35 neurons per condition; +P < 0.05, ++P < 0.01, comparing values in 7CK and APV (Mann–Whitney); *P < 0.05, ***P < 0.001 compared with baseline value (Wilcoxon).
To examine how quickly agonist-dependent conformational changes in the NMDARcd occur, we delivered rapid localized pulses of increased glutamate near dendritic spines using one-photon uncaging of caged glutamate (RuBi-glutamate) (29). Individual spines were imaged at high magnification and photons were captured over 1 s time windows to generate fluorescence decay curves (SI Appendix, Fig. S4). First, to confirm effective activation of NMDARs by uncaging of caged glutamate, calcium entry through NMDARs (in the absence of antagonists) was measured. Neurons were loaded with Oregon-Green-BAPTA (OGB), which shows increased fluorescence intensity, as well as fluorescence lifetime, upon binding calcium ions (30). The fluorescence intensity and lifetime of OGB inside a spine increased significantly immediately after glutamate uncaging near a spine, followed by a return to basal levels within a few seconds (SI Appendix, Fig. S5). This increase was blocked by adding 7CK to the perfusion, indicating that calcium entry through NMDARs is effectively blocked by 7CK (SI Appendix, Fig. S5). Next, GluN1-GFP, GluN1-mCherry, and GluN2B were expressed and FRET was monitored before and after glutamate uncaging in the presence of 7CK. A rapid increase in GluN1-GFP fluorescence lifetime was detected after glutamate uncaging in the presence of 7CK, and not in the presence of APV, that returned to baseline levels within a few seconds (Fig. 5C). Moreover, the lifetime change decays from peak to one-half value in ∼1 s; which is within ∼one-half order-of-magnitude of the NMDA current (which is an underestimate of the ligand binding lifetime because there is some desensitization that reduces current, but not ligand binding). These results indicate that conformational changes in the NMDARcd occur within 1 s of agonist binding and decay within a few seconds, suggesting that NMDARcd conformational change persists only while extracellular agonist is bound.
Discussion
Ion channels transmit information by transmembrane passage of ions. In addition, a few reports suggest transmembrane information transfer by ion channels in the absence of ion flow (11, 12, 18, 31–34). Here we show directly that agonist binding to the NMDAR, a ligand-gated ion channel, leads to a rapid conformational change in its cytoplasmic domain. Because NMDA (and glutamate) binds the GluN2 subunit, and here we monitor fluorophores on the GluN1cd, there must be conformational changes propagating between subunits, which have been proposed (35, 36).
It is notable that FRET between GluN1-GFP and GluN1-mCherry displays considerable variation among different spines. This finding suggests that the NMDARcd is in different conformations in different spines, possibly because of interactions with a different pool of proteins. Indeed, distinct classes of synapses on individual neurons based on different MAGUK. protein content have been reported (37). Different MAGUK. proteins, which bind the NMDARcd (38), may produce different NMDARcd conformations that could confer different properties (e.g., transmission, plasticity, trafficking, and so forth) to synapses. Further studies on this issue may elucidate the mechanism underlying variation among spines.
Our data indicate that transmission of information to the NMDARcd triggered by extracellular agonist binding is rapid, as it begins within 1 s and decays within seconds. Notably, the changes in the NMDARcd are triggered by stimuli that induce long-term depression (LTD) in cultured neurons, and display the same pharmacology as brain slice LTD of synaptic transmission (11) and long-term dendritic spine structural plasticity (12): they are blocked by APV but not by 7CK or MK-801.
In general, our findings indicate that agonist binding to the NMDAR produces a conformational change in the NMDARcd in the absence of ion flow through the receptor, consistent with the view that NMDARs can transmit agonist-driven information to the inside of a cell in the absence of ion flow through the receptor.
Materials and Methods
Procedures involving constructs used, chemicals and reagents, primary culture preparation, transfection, image analysis, prediction of interreceptor versus intrareceptor FRET (Fig. 2A), and statistics are detailed in SI Appendix.
Fluorescence Lifetime Imaging.
Fluorescence lifetime imaging was performed on a SliceScope two-photon microscope (Scientifica) using a 60× water-immersion objective (LUMPLFLN 60XW, NA = 1.0; Olympus). A Chameleon Ultra II IR laser (Coherent) (80-MHz repetition rate, 100- to 150-fs pulses) tuned at 930 nm was used for the excitation of GluN1-GFP. ScanImage r3.8 was used to control the scanning mirrors (39). Fluorescence emission was detected with a hybrid PMT detector (HPM-100–40, Becker and Hickl) between 490 and 540 nm by means of a GFP emission filter (ET 515/50, Chroma). The acquisition of fluorescence lifetimes was synchronized by a TCSPC module (SPC-150, Becker and Hickl). The following parameters were kept constant for all acquired images: pixel size (80 nm; all 512 × 512 pixels), pixel dwell time (3.2 μs), laser excitation intensity (3 mW after the microscope objective), FLIM acquisition time (∼120 s per image), and number of time bins (256) in the fluorescence decay curves.
Fluorescence lifetime images were analyzed with SPCImage (40). To ensure sufficient photons in the regions of interest, a binning factor between 6 and 10 pixels was used; this corresponds to ∼1.2 μm, which is around the size of a spine (one average lifetime value is then extracted from each spine). To minimize lifetime calculation errors, we used a minimum threshold of 10 photons at the peak time bin (corresponding to ∼1,000 photons per pixel summed across the 256 time bins) and used the same calculated instrumental response function for each set of experiments. FLIM images were analyzed with a single exponential model because the number of photons in the pixels located in synaptic regions was ∼1,000–4,000 per pixel, which is insufficient for reliable multiexponential analysis (40) but is sufficient for a single exponential analysis (19, 40). Curve offset and shift were not fixed; this allowed for better goodness of fit values, particularly in images that had a broad range of pixel intensities. For further analysis, each FLIM image was exported as a matrix of lifetimes, photon counts, and goodness-of-fit values (χ2); see SI Appendix, Image Analysis for details.
FRET Calculations.
To calculate FRET efficiency the following formula was used: EFRET= 1 − TDA/TD (TDA = lifetime of the donor in the presence of the acceptor; TD = average lifetime of the donor alone) (41). To make sure that the shorter lifetimes we measured in the presence of overexpressed acceptors was not because of random or collisional FRET, we used the lifetime of GluN1-GFP coexpressed with mCherry [which gave at least double the fluorescence of GluN1-mCherry, PP1-mCherry, and CaMKII-mCherry (28)] as the donor lifetime in all FRET efficiency calculations (SI Appendix, Table S1). The GluN1-GFP alone experiment shown in Fig. 1 A and B was repeated both with and without mCherry overexpression with the same results. The average distance between the GluN1 tagged NMDARs was calculated using this formula: r = R0 [(1/EFRET) – 1]1/6, where r = distance separating the fluorophores, R0 = Förster radius for the GFP and mCherry FRET pair (41); 5.4 nm was used (42).
Extracellular Antibody Treatment.
For experiments shown in Fig. 2 C–F, when specified, neurons were incubated with an antibody binding to GluN1 extracellular domain (MAB363, Millipore); we note that this antibody allows for live labeling of surface NMDARs (43, 44) for 60 min in normal culture media (10 μg/mL). Neurons were then washed once in culture media then incubated for 30–60 min with a secondary antibody (10 μg/mL; GAM-AF647, Life Technologies); both incubations were carried out at 37 °C in the incubator. Finally, neurons were washed once more in culture media then placed in the perfusion solution for imaging.
FRAP.
FRAP experiments were performed on the same microscope as for FLIM using similar settings. A 60× water objective was used to acquire 23 × 23-μm images of 256 × 256 pixels. For each neuron, a baseline image was first acquired. One or two spines were then photobleached using the imaging laser (930 nm) at a high-zoom magnification (2 × 2-μm images) for 2–3 min. This method reduced the fluorescence intensity of the photobleached spines by 80 ± 10% (error indicates SD) compared with their baseline values, a reduction that was similar across control, extracellular antibody-treated, and fixed neurons. This value was then normalized to 100% (or 0% fluorescence intensity) to measure fluorescence recovery. Next, fluorescence images were acquired immediately after photobleaching, and at 12, 25, and 40 min later. FRAP analysis was done in ImageJ. For each dendritic segment, three nearby spines (not receiving photobleaching) were used to normalize the fluorescence intensity of the photobleached spines and a background region of interest the same size as the photobleached spine was substracted from all integrated fluorescence-intensity measurements. To evaluate nonbiological fluorescence recovery, we fixed a group of neurons (10 min in methanol at −20 °C) before performing FRAP experiments (25). In these fixed cells, we observed a small and gradual return of fluorescence (5%, 7%, and 10% at 12, 25, and 40 min, respectively). This finding should be because of the partially reversible nature of GFP photobleaching (45); these values were thus subtracted from the control, extracellular antibody-treated, and GluN1 C-terminal antibody-infused neurons fluorescence recovery values, respectively, for each time point (Fig. 2D). The GluN1-GFP mobile fraction, assessed 40 min after photobleaching [similar to previously reported value for NMDARs (26)], was significantly reduced by extracellular antibody cross-linking by 73%.
Infusion of GluN1 Cterminal Antibody into Neurons.
To immobilize the NMDAR C-terminal domain, we used an antibody binding to the GluN1 C-terminal domain (MAB1570) that was introduced into neurons via a patch pipette. The cesium-based internal solution (containing: 115 mM cesium methanesulfonate, 20 mM CsCl, 10 mM Hepes, 2.5 mM MgCl2, 4 mM Na2ATP, 0.4 mM Na3GTP, 10 mM sodium phosphocreatine, 0.6 mM EGTA, and 0.1 mM spermine, pH 7.25) was supplemented with the GluN1 C-terminal antibody or the control antibody (GAR-AF647) (Fig. 4) to a 20-μg/mL concentration and the osmolality was adjusted (to ∼290 mOsm/kg) to compensate for the higher osmolality of the antibody solution. For the experiments shown in Fig. 4 C and D, neurons were patched under the FLIM microscope for ∼10 min and were imaged after 30–60 min to allow for both increased antibody diffusion along the dendrites and the ability to image more than one neuron per experiment (maximum of three neurons) (SI Appendix, Fig. S4A). To visualize antibody diffusion into neurons (Fig. 4B), we used a GluN1cd antibody conjugated with Alexa Fluor 488 (Millipore, # 05–432A4); a water-soluble red dye (Alexa Fluor 568 Hydrazide, Life Technologies; dissolved in water and used at a final concentration of 5 μM) was also included in the internal solution.
One-Photon Uncaging.
One-photon uncaging of RuBi-Glutamate was done using a BML-450-30FLD 450-nm blue diode laser (Lasermate). Laser excitation intensity was <0.2 mW under the objective. Suprasaturating levels of photons from the laser could be detected by the PMT detector even through the emission filter, so a shutter (Edmund Optics, no. 87208) was used to prevent detector overload during the uncaging stimulation. Spine lifetime curves were obtained using high magnification low-resolution spine imaging (SI Appendix, Fig. S4B); all photons within a 1-s window were used to generate the lifetime curve. An ∼15-s baseline imaging period was captured for each spine. Uncaging pulses were controlled by TriggerSync (Prairie Technologies). Two stimulation protocols were used: (i) three 10-ms pulses, each separated by 200 ms (3 × 10 ms); (ii) 100 1-ms pulses (tetanic stimulation), each separated by 9 ms (100 × 1 ms). Spines were imaged for ∼20 s following glutamate uncaging.
Fluorescence lifetimes were analyzed using MATLAB. For each experiment, the time point of maximum fluorescence was first determined by taking an average of the lifetime curves. Each curve was then fitted using a single exponential beginning with the time point of maximum fluorescence. Lifetimes were then tabulated in Excel and analyzed. Uncaging experiments were rejected if: (i) the total intensity in each curve was too low (<1,000 photons per curve; corresponding to a <0.90 adjusted R2 goodness-of-fit statistic reported by MATLAB); (ii) spine lifetimes were too low (<1.5 ns, indicating structures identified as spines were likely autofluorescent structures that were occasionally observed in lifetime images as well); (iii) spines suffered from significant bleaching during the course of the experiment (>20% loss of fluorescence intensity during the last ∼10 s of imaging). Results from both stimulation protocols were similar and therefore averaged in Fig. 5C.
Supplementary Material
Acknowledgments
We thank Paul de Koninck and Luo Jian-hong for constructs; and Drs. J. Isaacson, A. Newton, M. Kunkel, N. Spitzer, D. Berg, and members of the R.M. laboratory for constructive comments. J.A. is supported by a scholarship from the Agency for Science, Technology and Research (A-STAR), Singapore.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520023112/-/DCSupplemental.
References
- 1.Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309(5965):261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- 2.Bourne HR, Nicoll R. Molecular machines integrate coincident synaptic signals. Cell. 1993;72(Suppl):65–75. doi: 10.1016/s0092-8674(05)80029-7. [DOI] [PubMed] [Google Scholar]
- 3.Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: An evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Nabavi S, et al. Engineering a memory with LTD and LTP. Nature. 2014;511(7509):348–352. doi: 10.1038/nature13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368(6467):144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258(5082):597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- 7.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14(6):383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 8.Sheng M, Sabatini BL, Südhof TC. Synapses and Alzheimer’s disease. Cold Spring Harb Perspect Biol. 2012;4(5):a005777. doi: 10.1101/cshperspect.a005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayford M, Wang J, Kandel ER, O’Dell TJ. CaMKII regulates the frequency-response function of hippocampal synapses for the production of both LTD and LTP. Cell. 1995;81(6):891–904. doi: 10.1016/0092-8674(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 10.Nabavi S, Fox R, Alfonso S, Aow J, Malinow R. GluA1 trafficking and metabotropic NMDA: Addressing results from other laboratories inconsistent with ours. Philos Trans R Soc Lond B Biol Sci. 2014;369(1633):20130145. doi: 10.1098/rstb.2013.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabavi S, et al. Metabotropic NMDA receptor function is required for NMDA receptor-dependent long-term depression. Proc Natl Acad Sci USA. 2013;110(10):4027–4032. doi: 10.1073/pnas.1219454110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein IS, Gray JA, Zito K. Non-ionotropic NMDA receptor signaling drives activity-induced dendritic spine shrinkage. J Neurosci. 2015;35(35):12303–12308. doi: 10.1523/JNEUROSCI.4289-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9(1):60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 14.Ghanouni P, et al. Functionally different agonists induce distinct conformations in the G protein coupling domain of the beta 2 adrenergic receptor. J Biol Chem. 2001;276(27):24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 15.Granier S, Kim S, Fung JJ, Bokoch MP, Parnot C. FRET-based measurement of GPCR conformational changes. Methods Mol Biol. 2009;552:253–268. doi: 10.1007/978-1-60327-317-6_18. [DOI] [PubMed] [Google Scholar]
- 16.Traynelis SF, et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62(3):405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao JY, et al. Transmembrane region of N-methyl-D-aspartate receptor (NMDAR) subunit is required for receptor subunit assembly. J Biol Chem. 2011;286(31):27698–27705. doi: 10.1074/jbc.M111.235333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barria A, Malinow R. Subunit-specific NMDA receptor trafficking to synapses. Neuron. 2002;35(2):345–353. doi: 10.1016/s0896-6273(02)00776-6. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr Opin Neurobiol. 2006;16(5):551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Lee CH, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511(7508):191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rácz B, Weinberg RJ. Microdomains in forebrain spines: An ultrastructural perspective. Mol Neurobiol. 2013;47(1):77–89. doi: 10.1007/s12035-012-8345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons MP, Raymond LA. Extrasynaptic NMDA receptor involvement in central nervous system disorders. Neuron. 2014;82(2):279–293. doi: 10.1016/j.neuron.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 23.Santucci DM, Raghavachari S. The effects of NR2 subunit-dependent NMDA receptor kinetics on synaptic transmission and CaMKII activation. PLOS Comput Biol. 2008;4(10):e1000208. doi: 10.1371/journal.pcbi.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doré K, Labrecque S, Tardif C, De Koninck P. FRET-FLIM investigation of PSD95-NMDA receptor interaction in dendritic spines; control by calpain, CaMKII and Src family kinase. PLoS One. 2014;9(11):e112170. doi: 10.1371/journal.pone.0112170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashby MC, Maier SR, Nishimune A, Henley JM. Lateral diffusion drives constitutive exchange of AMPA receptors at dendritic spines and is regulated by spine morphology. J Neurosci. 2006;26(26):7046–7055. doi: 10.1523/JNEUROSCI.1235-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: Long-term regulation by synaptic NMDA receptor activation. Mol Cell Neurosci. 2006;31(4):702–712. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Granier S, et al. Structure and conformational changes in the C-terminal domain of the beta2-adrenoceptor: insights from fluorescence resonance energy transfer studies. J Biol Chem. 2007;282(18):13895–13905. doi: 10.1074/jbc.M611904200. [DOI] [PubMed] [Google Scholar]
- 28.Aow J, Dore K, Malinow R. Conformational signaling required for synaptic plasticity by the NMDA receptor complex. Proc Natl Acad Sci USA. 2015;112:14711–14716. doi: 10.1073/pnas.1520029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fino E, et al. RuBi-glutamate: Two-photon and visible-light photoactivation of neurons and dendritic spines. Front Neural Circuits. 2009;3:2. doi: 10.3389/neuro.04.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagolla K, Löhmannsröben HG, Hille C. Time-resolved fluorescence microscopy for quantitative Ca2+ imaging in living cells. Anal Bioanal Chem. 2013;405(26):8525–8537. doi: 10.1007/s00216-013-7290-6. [DOI] [PubMed] [Google Scholar]
- 31.Schneider MF, Chandler WK. Voltage dependent charge movement of skeletal muscle: A possible step in excitation-contraction coupling. Nature. 1973;242(5395):244–246. doi: 10.1038/242244a0. [DOI] [PubMed] [Google Scholar]
- 32.Vissel B, Krupp JJ, Heinemann SF, Westbrook GL. A use-dependent tyrosine dephosphorylation of NMDA receptors is independent of ion flux. Nat Neurosci. 2001;4(6):587–596. doi: 10.1038/88404. [DOI] [PubMed] [Google Scholar]
- 33.Nong Y, et al. Glycine binding primes NMDA receptor internalization. Nature. 2003;422(6929):302–307. doi: 10.1038/nature01497. [DOI] [PubMed] [Google Scholar]
- 34.Rozas JL, Paternain AV, Lerma J. Noncanonical signaling by ionotropic kainate receptors. Neuron. 2003;39(3):543–553. doi: 10.1016/s0896-6273(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 35.Regalado MP, Villarroel A, Lerma J. Intersubunit cooperativity in the NMDA receptor. Neuron. 2001;32(6):1085–1096. doi: 10.1016/s0896-6273(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 36.Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol. 2013;20(4):477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]
- 37.Elias GM, et al. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52(2):307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16(7):2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker W, et al. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc Res Tech. 2004;63(1):58–66. doi: 10.1002/jemt.10421. [DOI] [PubMed] [Google Scholar]
- 41.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; New York: 2006. [Google Scholar]
- 42.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9(10):1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer DK, Lindemeyer AK, Wilmes T, Sobottka H, Nörenberg W. GluA and GluN receptors regulate the surface density of GluN receptor subunits in cultured neocortical interneurons. J Neurochem. 2012;121(4):597–606. doi: 10.1111/j.1471-4159.2012.07719.x. [DOI] [PubMed] [Google Scholar]
- 44.Lin LH, Moore SA, Jones SY, McGlashon J, Talman WT. Astrocytes in the rat nucleus tractus solitarii are critical for cardiovascular reflex control. J Neurosci. 2013;33(47):18608–18617. doi: 10.1523/JNEUROSCI.3257-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaminathan R, Hoang CP, Verkman AS. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: Cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys J. 1997;72(4):1900–1907. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.