Significance
Noninvasive prenatal testing (NIPT) using sequencing of fetal cell-free DNA from maternal plasma has allowed accurate prenatal diagnosis of aneuploidy without requirement of an invasive procedure; this approach has gained increasing clinical acceptance. In this manuscript, we evaluate whether NIPT using semiconductor sequencing platform (SSP) could reliably detect subchromosomal deletions/duplications in women carrying a high-risk fetus. We show that detection of fetal subchromosomal abnormalities is a viable extension of NIPT using SSP. Given the recent strong interest in NIPT using cell-free DNA, these results should be of broad interest and immediate clinical importance, even beyond the NIPT field, such as for detection of cell-free tumor DNA.
Keywords: noninvasive prenatal testing, NIPT, maternal plasma DNA, cell-free DNA, semiconductor sequencing
Abstract
Noninvasive prenatal testing (NIPT) using sequencing of fetal cell-free DNA from maternal plasma has enabled accurate prenatal diagnosis of aneuploidy and become increasingly accepted in clinical practice. We investigated whether NIPT using semiconductor sequencing platform (SSP) could reliably detect subchromosomal deletions/duplications in women carrying high-risk fetuses. We first showed that increasing concentration of abnormal DNA and sequencing depth improved detection. Subsequently, we analyzed plasma from 1,456 pregnant women to develop a method for estimating fetal DNA concentration based on the size distribution of DNA fragments. Finally, we collected plasma from 1,476 pregnant women with fetal structural abnormalities detected on ultrasound who also underwent an invasive diagnostic procedure. We used SSP of maternal plasma DNA to detect subchromosomal abnormalities and validated our results with array comparative genomic hybridization (aCGH). With 3.5 million reads, SSP detected 56 of 78 (71.8%) subchromosomal abnormalities detected by aCGH. With increased sequencing depth up to 10 million reads and restriction of the size of abnormalities to more than 1 Mb, sensitivity improved to 69 of 73 (94.5%). Of 55 false-positive samples, 35 were caused by deletions/duplications present in maternal DNA, indicating the necessity of a validation test to exclude maternal karyotype abnormalities. This study shows that detection of fetal subchromosomal abnormalities is a viable extension of NIPT based on SSP. Although we focused on the application of cell-free DNA sequencing for NIPT, we believe that this method has broader applications for genetic diagnosis, such as analysis of circulating tumor DNA for detection of cancer.
Genomic disorders are defined by loss, gain, or translocation of chromosomal material. Deletion/duplication syndromes are known to be associated with a wide range of structural and functional abnormalities (1), such as Cri du Chat Syndrome (5p deletion) (2) and DiGeorge Syndrome (22q11.2 deletion) (3). Such deletion/duplication syndromes can be reliably diagnosed prenatally from the DNA of fetal cells; fetal DNA may be assessed for chromosomal abnormalities by karyotyping, FISH, comparative genomic hybridization (CGH), and array-based technologies (4). G-banded karyotyping is the predominant technique for diagnosis of chromosomal abnormalities, but it is limited to resolution of 5–10 Mb (5, 6). Genomic disorders of a smaller size are more reliably detected by chromosomal microarray analysis (CMA), of which array CGH (aCGH) is an example.
According to The American Society of Human Genetics, CMA has replaced the standard metaphase karyotype in postnatal assessment of individuals with developmental delay, intellectual disability, congenital anomalies, and autism (7). In December of 2013, The American Congress of Obstetricians and Gynecologists and the Society of Maternal Fetal-Medicine recommended prenatal CMA as a first-line test in the case of abnormal ultrasound findings (8). This method identifies structural anomalies of chromosomes with great accuracy in prenatal and postnatal stages alike and increases detection of clinically significant copy number changes, even when the conventional metaphase karyotype appears normal. However, CMA or any of the other above-mentioned conventional techniques requires that an invasive procedure, such as amniocentesis, chorionic villus sampling (CVS), or umbilical cord blood collection, be performed to obtain a source of fetal genomic DNA. These procedures carry small but well-validated rates of pregnancy loss that may approach 1% (9) and can be performed only by specially trained obstetricians. These invasive diagnostic tests are most often performed after a positive screening test or abnormal ultrasound. Widely used trisomy 21 screening programs at best detect 95% of cases with a false-positive rate of 5%, implying that the risk of pregnancy loss is greater than 1/10,000 among normal pregnancies that undergo screening with subsequent invasive testing. There are no serum-based tests in clinical use that specifically screen for subchromosomal abnormalities.
Recently published international guidelines (10) recommend the clinical use of noninvasive prenatal testing (NIPT) for aneuploidy screening only among pregnant women whose fetuses are deemed at high risk; NIPT has become increasing adopted for this purpose. Studies, such as the study by Bianchi et al. (11), have shown the potential of using cell-free DNA (cfDNA) for fetal aneuploidy testing in average-risk women, in particular because of reductions in the false-positive rate compared with standard prenatal testing. However, NIPT has not yet been adopted as standard practice for average-risk women, and concerns persist regarding the relatively high cost per detected case in this population (10). NIPT methods currently in clinical use depend primarily on Illumina sequencing technologies. We recently published a study (12) that showed the viability of using a semiconductor sequencing platform (SSP) for NIPT. SSP offers the potential advantages of lower cost and faster sequencing, and the small footprints of these machines may make them easier to deploy in clinical laboratories.
NIPT through either platform offers several advantages over current screening practices that rely on serum and sonographic markers. NIPT has the potential to avoid most invasive antenatal diagnostic procedures performed secondary to positive trisomy screen, abnormal ultrasound, or advanced maternal age because of greatly reduced false-positive rates. NIPT can generally be performed in the late first trimester or around 12–13 wk (13), and the sequencing turnaround may be faster than methods relying on cell culture. Thus, a pregnant woman who chooses to terminate a pregnancy based on the result will have the option to do so earlier in her pregnancy compared with the most accurate standard screening (10). Elective termination at an earlier gestational age is safer for the woman (14) as well as enables greater privacy and less emotional distress.
Previous studies have shown the potential of extending NIPT to detect fetal deletion/duplication syndromes from maternal plasma. Peters et al. (15) reported that maternal plasma sequencing with 243 million reads could identify a 4-Mb deletion at 35 wk of gestation, and more recently, Srinivasan et al. (16) identified subchromosomal duplications and deletions, translocations, mosaicism, and trisomy 20 by maternal plasma sequencing in seven cases, including a 300-kb microdeletion. Unfortunately, both of these studies required deep sequencing, increasing both cost and time required, and they were limited to only a handful of patients (15, 16). No study has yet been published that evaluates the potential of NIPT to detect subchromosomal abnormalities in a large population of pregnant women.
The fetal DNA fraction is a critical parameter affecting the accuracy of NIPT using cfDNA in maternal plasma. Sequencing SNPs was considered as the primary method of determining fetal DNA fraction (17). Recently, Lo et al. (18) developed a novel method that relies on the fact that circulating fetal DNA molecules are generally shorter than the corresponding maternal DNA molecules to estimate the fraction of fetal DNA in maternal plasma. This approach can be adapted for an SSP, which is capable of sequencing entire molecules of fetal DNA to obtain the size of the DNA fragments.
Based on the potential for NIPT to provide a fetal karyotype, we investigated whether we could further develop our technique of NIPT with SSP to detect subchromosomal deletions/duplications in a population of women with high-risk pregnancies. The sliding window strategy has been reported to accurately detect copy number variations in the human genome with relativity low-coverage whole-genome sequencing (19). Thus, this strategy could enable the detection of deletions/duplications in fetal cfDNA from maternal plasma using less sequencing depth and at lower cost. Our study highlights the prospect of universal and practical noninvasive screening for deletions/duplications in the fetal genome that can be incorporated into the current program of noninvasive prenatal detection of fetal aneuploidy.
Results
Optimization of Fetal Deletion/Duplication Detection by SSP.
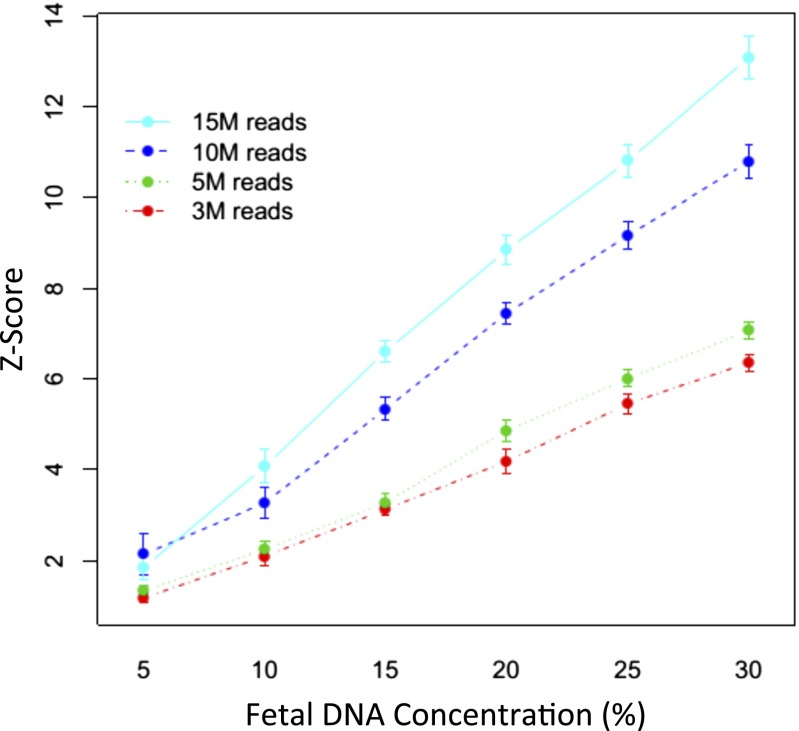
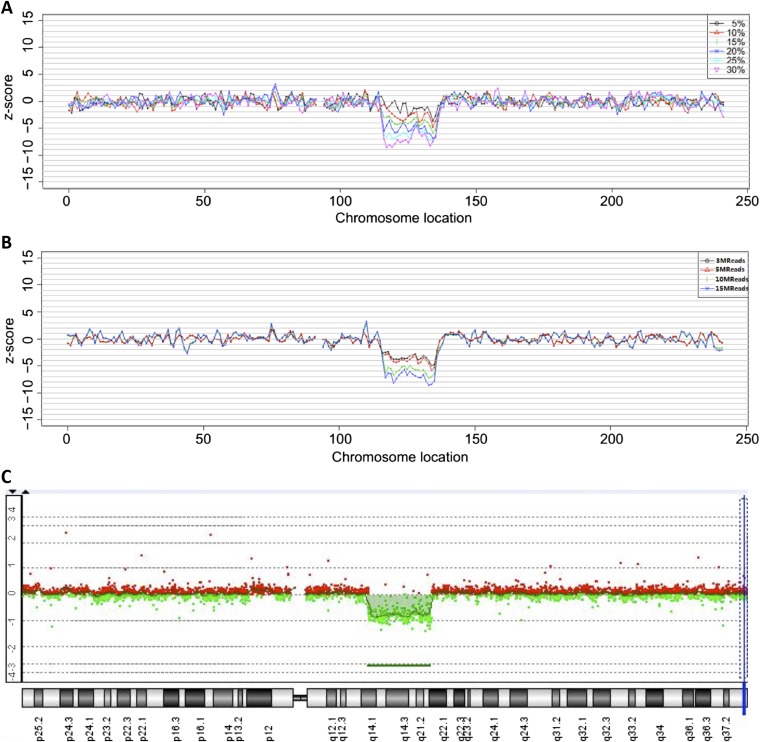
To test the accuracy of fetal deletion/duplication detection by SSP, DNA derived from the blood of 19 newborns with known deletions/duplications diagnosed by aCGH (Table S1) was mixed with DNA from normal samples at a range of concentrations [5%, 10%, 15%, 20%, and 30% (mass/mass)]. Each mixed sample was sequenced at various depths (3.5, 5, 10, or 15 million reads). Samples 14,452, 14,095, and 14,181 harbored two deletions or duplications each, whereas the remaining samples had one deletion/duplication. In total, 22 deletions/duplications were analyzed. Stouffer’s Z scores were generated as described in Materials and Methods for each sample at every concentration and sequencing depth; Z scores increased with both concentration and sequencing depth, with the average Z scores for all samples shown in Fig. 1. The Z scores of sample 14,945, with DNA that contains a 21-Mb deletion, are shown in Fig. S1 A and B as an example. Fig. S1B shows that, at a constant sequencing depth of 5 million reads, Z-score magnitude increases with greater abnormal DNA concentration. Fig. S1B shows increasing Z-score magnitude with increasing sequencing depth at a constant 15% abnormal DNA concentration. The aCGH plot for sample 14,945 is shown in Fig. S1C as a comparison.
Table S1.
aCGH results of newborns with DNA deletions and duplications
| Sample identification | Dup/del | Chr | Start | End | Size (Mb) | Syndrome annotation |
| 14,437 | Del | Chr16 | 14956252 | 16157108 | 1.2 | 16p13.11 Microdeletion (neurocognitive disorder susceptibility locus) |
| 14,394 | Del | Chr1 | 145031367 | 146253330 | 1.2 | Thrombocytopenia-absent radius syndrome |
| 14,416 | Dup | Chr22 | 48219551 | 49451468 | 1.2 | Phelan–Mcdermid Syndrome/22q13 deletion syndrome |
| 14,452 | Del | Chr5 | 204737 | 1726099 | 1.5 | Cri du Chat Syndrome (5p deletion) |
| 14,944 | Del | Chr19 | 11693072 | 13505798 | 1.8 | Uncertain |
| 14,095 | Del | Chr21 | 44565059 | 46880878 | 2.3 | Uncertain |
| 14,368 | Del | Chrx | 31012571 | 33514518 | 2.5 | Xp21 deletion syndrome |
| 14,209 | Del | Chr17 | 15549649 | 18845678 | 3.3 | Potocki–Lupski Syndrome (17p11.2 duplication syndrome) |
| 14,181 | Del | Chr2 | 32444 | 3562673 | 3.5 | Uncertain |
| 14,777 | Del | Chr22 | 17900000 | 22200000 | 4.3 | DiGeorge Syndrome/22q11.2 deletion syndrome |
| 14,095 | Del | Chr16 | 46271 | 4904686 | 4.9 | Rubinstein–Taybi syndrome |
| 14,540 | Del | Chr15 | 21250794 | 26199055 | 4.9 | Prader–Willi/Angelman syndrome |
| 14,883 | Del | Chr3 | 137461910 | 143694820 | 6.2 | Uncertain |
| 14,157 | Del | Chr13 | 96437228 | 103424298 | 7 | Uncertain |
| 14,119 | Del | Chr18 | 50413206 | 58403399 | 8 | 18q Deletion syndrome |
| 14,159 | Del | Chr10 | 183492 | 11035280 | 10.9 | Hypoparathyroidism, sensorineural deafness, and renal disease (HDRS) |
| 14,115 | Del | Chr4 | 133413 | 13498201 | 13.4 | Wolf–Hirschhorn syndrome |
| 14,873 | Dup | Chr2 | 161423992 | 176132164 | 14.7 | 2q31.1 Duplication syndrome |
| 14,452 | Dup | Chr15 | 80043155 | 100282878 | 20.2 | 15q25 Deletion syndrome |
| 14,945 | Del | Chr1 | 115862191 | 136892070 | 21 | Uncertain |
| 14,798 | Del | Chr1 | 156153073 | 182952784 | 26.8 | Uncertain |
| 14,181 | Dup | Chr9 | 261257 | 31357372 | 31.1 | 9p Deletion syndrome |
Chr, chromosome; Del, deletion; Dup, duplication.
Fig. 1.
Average absolute Z scores of subchromosomal abnormalities for all mixed abnormal and normal DNA samples at various concentrations and sequencing depths. Error bars represent ±1.
Fig. S1.
Characteristic Z scores of a section of DNA encompassing a deletion in sample 14,945 at (A) varying concentrations of abnormal DNA at 5 million reads and (B) varying sequencing depths at 15% concentration of abnormal DNA. (C) The aCGH plot of sample 14,945 is shown for comparison.
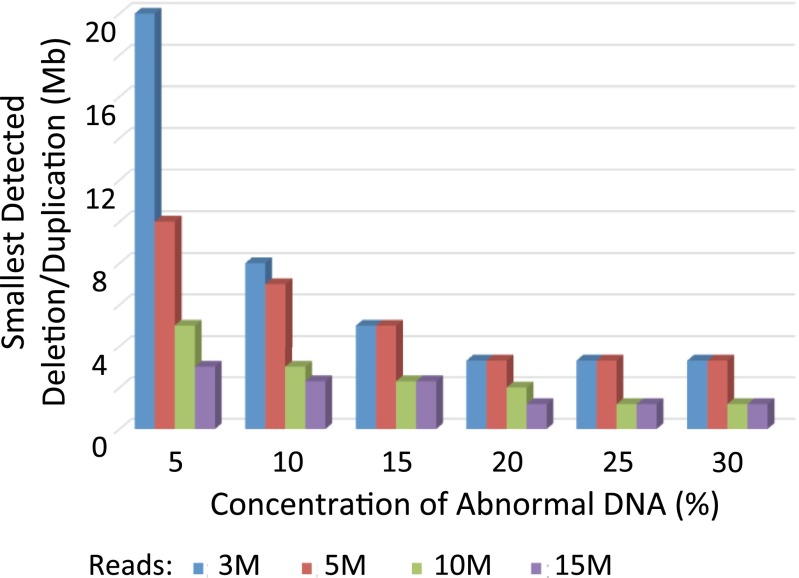
Increasing the fraction of abnormal DNA as well as increasing the sequencing depth resulted in improved detection and delineation of subchromosomal abnormalities (Fig. 2). At the lowest concentration of 5% and 3.5 million reads, the smallest abnormality detected was 20 Mb. Increasing sequencing depth at this concentration dramatically improved detection of deletions/duplications to as small as 3 Mb with 15 million reads. The rates of false-positive and false-negative events decreased with increasing concentration as well as increasing sequencing depth. These results show the feasibility of using Stouffer’s Z scores generated with a sliding window strategy on SSP to detect subchromosomal abnormalities.
Fig. 2.
Smallest size of deletions/duplications detected in mixed abnormal and normal DNA samples at various sequencing depths and concentrations of abnormal DNA.
Fetal Fraction Estimation in Maternal Plasma DNA by Size Analysis.
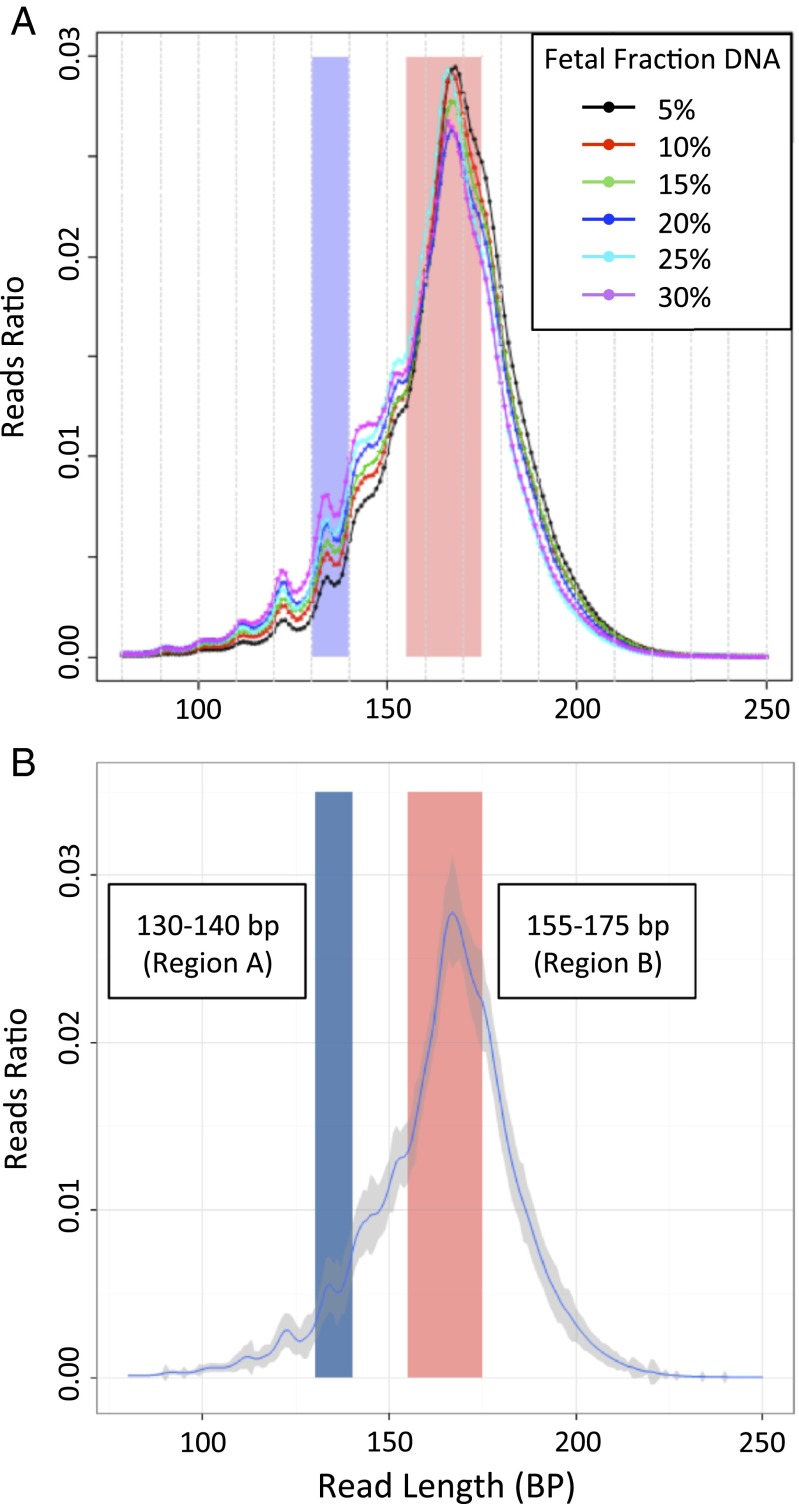
Given that the concentration of fetal DNA seems to affect the accuracy of NIPT, we investigated a technique using sequencing to estimate the fraction of fetal cfDNA in maternal plasma. Because fetal cfDNA is generally shorter than maternal cfDNA, maternal plasma samples with a higher fetal DNA concentration would have a higher proportion of short DNA fragments. We postulated that, if the proportions of short and long DNA fragments in maternal plasma were correlated with the fetal DNA fraction, we would be able to determine the fetal DNA fraction using maternal plasma DNA size analysis alone. We analyzed plasma samples from 1,456 pregnant women, 712 of whom carried a male fetus, performing SSP at 400 flows to sequence the entire length of fetal DNA fragments. The overall DNA fragment size distribution for each maternal plasma sample was generated. Characteristic size distributions from six individual maternal plasma samples with various fetal DNA fractions are shown in Fig. 3A. The aggregated distributions of all samples are shown in Fig. 3B as the average reads ratio at each fragment size, with the variance around each point shown by the gray region.
Fig. 3.
Size distribution of maternal plasma cfDNA fragments sequenced by SSP in a population of pregnant women. The blue region encompasses 130–140 bp (region A), and the red region encompasses 155–175 bp (region B). (A) Representative examples from maternal plasma with different fetal DNA fractions. (B) Aggregate of all samples. The blue line represents the mean read ratio of all samples, and the gray region represents ±1 SD.
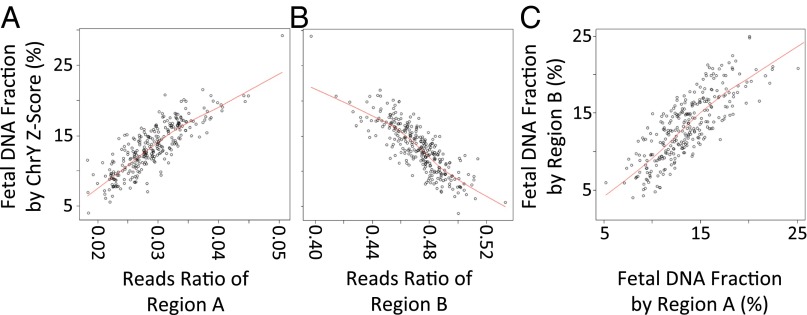
Samples from 712 women carrying male fetuses were randomly divided into a training group and a validation group, each containing 356 samples. The fetal DNA concentration was determined from the proportion of Y-chromosome sequences in maternal plasma. We then examined the relationship between the reads ratio of different fragment sizes and fetal DNA fraction in the training group. We observed a correlation between the fetal DNA fraction estimated by the Z score of Y chromosome and the reads ratio of two DNA fragment size regions: 130–140 bp (region A in Fig. 4A) and 155–175 bp (region B in Fig. 4B). Fig. 3 shows regions A and B in relation to the size distributions of the maternal cfDNA fragments denoted by vertical blue or red bands, respectively. As described in Materials and Methods, we used this correlation to derive an equation for estimating the fetal DNA concentration. Next, we estimated the fetal DNA fractions for 356 samples in the validation group using the regression equation obtained from the training group. For each size region, the size-deduced fetal DNA fractions were highly concordant with those determined using the proportion of Y chromosome (region A: r = 0.839, P < 2.2e-16 and region B: r = −0.83, P < 2.2e-16 by linear regression). The estimated fetal fraction was highly concordant between each region as well (Fig. 4C) (r = 0.818, P < 2.2e-16 by linear regression). As described in Materials and Methods, the estimated fetal DNA fraction was considered unreliable for a small number of samples, in which the estimated fetal fraction was inconsistent between the different size regions.
Fig. 4.
Estimation of the fetal DNA concentration in maternal plasma using the size distribution of cfDNA fragments. (A and B) Correlation between the reads ratio of DNA fragments in (A) the 130- to 140-bp region or (B) the 155- to 175-bp region and the fetal DNA concentration predicted by the Z score of Y chromosome. (C) Correlation between fetal DNA concentration estimated by regions A and B.
Performance of SSP for Detecting Fetal Deletions/Duplications.
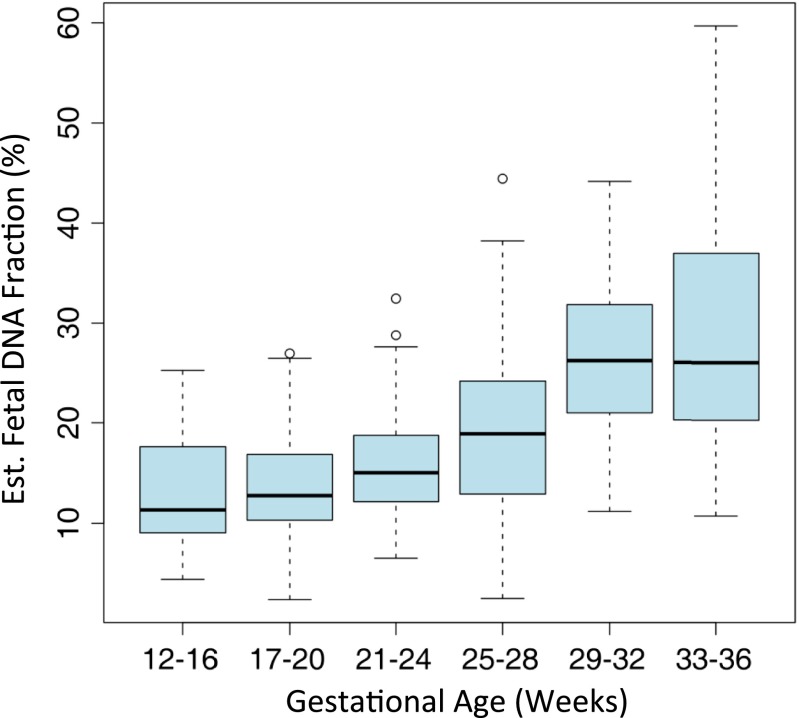
We prospectively recruited a cohort of 1,476 pregnant women presenting with a fetal indication of structural abnormalities detected on ultrasonography for invasive prenatal testing. The average maternal age of the cohort was 29.7 ± 5.4 y old (range = 17–46 y old), and the average gestational age was 24 ± 6 wk (range = 12–37 wk). Maternal plasma was collected concurrently with fetal DNA obtained using an invasive procedure, such as amniocentesis or chorionic villous sampling. Invasively collected fetal DNA was tested by aCGH, whereas DNA derived from maternal plasma was sequenced by SSP. We applied our method described above to estimate the fetal DNA fraction in maternal plasma for the entire cohort (Fig. S2); 88.5% samples were estimated to have a fetal DNA fraction of >10%. Although the average fetal DNA fraction was greater than 10% for all gestational ages, fetal DNA fraction increased significantly with gestational age (r = 0.5863, P value = 3.0190e-27); 66.7% of samples with a gestational age of 12–16 wk had a fetal DNA fraction greater than 10% compared with 95% of samples with gestational age greater than 20 wk.
Fig. S2.
Distribution of fetal DNA concentrations estimated by NIPT of maternal plasma in a cohort of 1,476 pregnant women at various gestational ages.
The same sequencing depth of 3.5 million reads was used like in our previous study on trisomy 21, 18, and 13 and sex chromosome aneuploidies (12). To confirm that our technique provided results consistent with our previous study, we first assessed the ability of SSP to detect aneuploidy in our cohort. In 1,476 samples, 73 cases of chromosomal aneuploidy were identified, with the extra chromosome clearly detected in the 1-Mb bin curve of the SSP analysis. Compared with aCGH performed on invasively derived fetal DNA, the false-positive rate for aneuploidy was 0.63% (nine false-positive samples), and the false-negative rate was 0%, consistent with our previous study and other previously published results (11–13, 20). The most common etiologies of false-positive results in NIPT for aneuploidy are placental mosaicism and a demised twin fetus (21, 22). Small duplications present in maternal DNA may also lead to relative overrepresentation of a chromosome in cfDNA, leading to a false-positive result (23), although we did not detect any instances of this in our study.
We then applied our method to detect subchromosomal deletions and duplications (Table S2). Seventy-eight subchromosomal deletions and duplications ranging from 0.52 to 84 Mb in size were identified by aCGH in 57 samples, many corresponding to known deletion or duplication syndromes; 56 of 78 (71.8%) abnormalities were detected by our method at 3.5 million reads. The size of the deletion/duplication seemed to be the major determinant of the performance of our method; 42 of 45 (93.3%) deletions/duplications larger than 5 Mb and 35 of 35 (100%) larger than 10 Mb in size were detected at 3.5 million reads. In contrast, only 14 of 34 (41.2%) of samples less than 5 Mb in size and none less than 1 Mb in size were detected at 3.5 million reads. However, for abnormalities greater than 1 Mb in size, sensitivity improved to 69 of 73 (94.5%) with increasing sequencing depth up to 10 million reads.
Table S2.
Comparison of NIPT and aCGH in clinical samples
| Sample identification | aCGH result | Syndrome associated with region* | NIPT result | |||||||
| Dup/del | Location predicted by aCGH | Size (Mb) | FC (%) | Location predicted by NIPT | 3 Million reads | 5 Million reads | 10 Million reads | 15 Million reads | ||
| A0587 | Del | Chr16:29673954–30198600 | 0.52 | 16p11.2 Microduplication syndrome | 17.01 | — | No | No | No | No |
| A0133 | Dup | Chr19:327273–863312 | 0.54 | 2.01 | — | No | No | No | No | |
| A0553 | Del | Chr19:3806228–4343253 | 0.54 | 11.61 | — | No | No | No | No | |
| A0519 | Del | Chr3:27275–853692 | 0.83 | 3p Syndrome | 9.57 | — | No | No | No | No |
| A0957 | Del | Chr7:157683596–158602499 | 0.92 | 7q36.1–36.3 Deletion | 10.34 | — | No | No | No | No |
| A0281 | Del | Chr17:674618–1859251 | 1.18 | 17p13.1 Deletion syndrome | 9.76 | — | No | No | No | No |
| A1005 | Dup | Chr22:22069437–23318455 | 1.25 | 22q11.2 Deletion syndrome | 19.82 | Chr22:22000000–23000000 | No | No | Yes | Yes |
| A1102 | Dup | Chr2:88015932–89387655 | 1.37 | 20.53 | Chr2:88000000–90000000 | Yes | Yes | Yes | Yes | |
| A1444 | Del | Chr17:72364514–73777326 | 1.41 | 17q25 Duplication | 16.93 | Chr17:71000000–73000000 | No | No | Yes | Yes |
| A0187 | Del | Chrx:6500000–8000000 | 1.5 | Steroid sulphatase deficiency (STS) | NA | Chrx:5000000–7000000 | Yes | Yes | Yes | Yes |
| A0197 | Dup | Chrx:6500000–8000000 | 1.5 | Steroid sulphatase deficiency (STS) | 9.61 | Chrx:6000000–7000000 | Yes | Yes | Yes | Yes |
| A0403 | Del | Chrx:6490000–8040000 | 1.55 | Steroid sulphatase deficiency (STS) | 10.58 | Chrx:5000000–7000000 | Yes | Yes | Yes | Yes |
| A1042 | Dup | Chr18:587754–2180664 | 1.59 | 18p11.32-p11.31 Duplication | 18.42 | Chr18:0–2000000 | No | No | Yes | Yes |
| A1138 | Dup | Chr18:36319629–37915204 | 1.60 | 18q12.2–21.1 Deletion | 17.27 | Chr18:35000000–37000000 | No | No | Yes | Yes |
| A0495 | Dup | Chr16:14408492–16089759 | 1.68 | 16p13.11 Microdeletion | 20.54 | Chr16:14000000–16000000 | Yes | Yes | Yes | Yes |
| A1285 | Del | Chr17:31461588–33242217 | 1.78 | 21.65 | Chr17:31000000–33000000 | No | No | Yes | Yes | |
| A0844 | Dup | Chr8:1.44e+08–1.46e+08 | 1.85 | 8q22.1-qter Duplication syndrome | 10.42 | — | No | No | No | No |
| A1034 | Del | Chr4:27770182–29952659 | 2.18 | 14q11-q22 Deletion syndrome | 15.77 | Chr4:27000000–29000000 | No | No | Yes | Yes |
| A0248 | Del | Chr4:91399112–93621545 | 2.22 | 25.90 | Chr4:90000000–92000000 | Yes | Yes | Yes | Yes | |
| A0129 | Dup | Chr4:1.57e+08–1.60e+08 | 2.42 | 4q32.1-q32.2 Triplication syndrome | 14.98 | Chr4:157000000–159000000 | Yes | Yes | Yes | Yes |
| A0769 | Del | Chr22:17299942–19770514 | 2.47 | DiGeorge Syndrome/22q11.2 deletion syndrome | NA | — | No | No | No | No |
| A0286 | Dup | Chr22:18919942–21440514 | 2.52 | DiGeorge Syndrome/22q11.2 deletion syndrome | 13.05 | Chr22:20000000–22000000 | Yes | Yes | Yes | Yes |
| A0686 | Del | Chr6:170426–2753293 | 2.58 | 6pter-p24 Deletion syndrome | NA | Chr6:0–3000000 | No | No | Yes | Yes |
| A0901 | Del | Chr13:65054495–67810584 | 2.76 | 28.55 | Chr13:65000000–67000000 | No | No | Yes | Yes | |
| A0786 | Dup | Chr21:44077514–46847409 | 2.77 | 22.78 | Chr21:44000000–47000000 | No | No | Yes | Yes | |
| A0301 | Del | Chr2:1.72e+08–1.75e+08 | 3 | 31.85 | Chr2:171000000–174000000 | Yes | Yes | Yes | Yes | |
| A0310 | Del | Chr22:17096855–20311763 | 3.21 | DiGeorge Syndrome/22q11.2 deletion syndrome | 14.74 | Chr22:26000000–30000000 | Yes | Yes | Yes | Yes |
| A0580 | Dup | Chr11:1.31e+08–1.35e+08 | 3.39 | Jacobsen Syndrome | 8.68 | Chr11:131000000–135000000 | No | No | Yes | Yes |
| A1052 | Dup | Chry:6688691–10511314 | 3.82 | 17.85 | Chry:6000000–10000000 | Yes | Yes | Yes | Yes | |
| A0001 | Del | Chr1:814245–4882747 | 4.07 | 1p36 Microdeletion syndrome | 19.05 | Chr1:1000000–3000000 | Yes | Yes | Yes | Yes |
| A0109 | Dup | Chr17:16429920–20667174 | 4.24 | Smith–Magenis Syndrome | 18.88 | Chr17:16000000–19000000 | Yes | Yes | Yes | Yes |
| A0142 | Del | Chr22:17900000–22200000 | 4.3 | DiGeorge Syndrome/22q11.2 deletion syndrome | 11.83 | Chr22:170000000–22000000 | No | Yes | Yes | Yes |
| A1012 | Del | Chr1:749625–5619192 | 4.87 | 1p36.33–36.32 Deletion | 14.72 | Chr1:0–5000000 | Yes | Yes | Yes | Yes |
| A0844 | Del | Chr21:41935392–46880878 | 4.95 | 10.42 | Chr21:41000000–47000000 | No | No | Yes | Yes | |
| A0901 | Dup | Chry:17801068–22916805 | 5.12 | AZFb | NA | Chry:20000000–23000000 | Yes | Yes | Yes | Yes |
| A0433 | Dup | Chr15:23076361–28436403 | 5.36 | 15q11.2 Deletion syndrome | 20.75 | Chr15:23000000–27000000 | Yes | Yes | Yes | Yes |
| A0899 | Del | Chr6:1.65e+08–1.71e+08 | 5.37 | Microdeletion 6q27 anosmia | 8.93 | Chr6:164000000–171000000 | No | Yes | Yes | Yes |
| A0947 | Dup | Chr15:20627802–26109998 | 5.48 | Prader–Willi Syndrome (type 2) | 14.76 | Chr15:20000000–27000000 | Yes | Yes | Yes | Yes |
| A0133 | Del | Chr14:1.01e+08–1.07e+08 | 6.06 | 2.01 | — | No | No | No | No | |
| A0001 | Dup | Chr19:51529057–59092570 | 7.56 | 19.05 | Chr19:50000000–57000000 | Yes | Yes | Yes | Yes | |
| A0202 | Dup | Chr22:17096855–25153910 | 8.06 | DiGeorge Syndrome/22q11.2 deletion syndrome | 24.87 | Chr22:17000000–23000000 | Yes | Yes | Yes | Yes |
| A0894 | Dup | Chr3:1.90e+08–1.99e+08 | 9.13 | 3q29 Microdeletion syndrome | 28.29 | Chr3:190000000–199000000 | Yes | Yes | Yes | Yes |
| A0786 | Del | Chr4:61552–9237101 | 9.18 | Wol–Hirschhorn Syndrome | 22.78 | Chr4:0–9000000 | Yes | Yes | Yes | Yes |
| A0107 | Dup | Chry:12571053–22916805 | 10.35 | AZFa | 23.31 | Chry:5000000–8000000 | Yes | Yes | Yes | Yes |
| A0107 | Del | Chry:1091–10379571 | 10.38 | Sex-determining region Y/SRY | 23.31 | Chry:5000000–8000000 | Yes | Yes | Yes | Yes |
| A0011 | Del | Chr4:61660–10975146 | 10.91 | Wolf–Hirschhorn Syndrome | 20.89 | Chr4:0–12000000 | Yes | Yes | Yes | Yes |
| A0256 | Dup | Chr12:45001–11278012 | 11.23 | 12p13.33 Microdeletion syndrome | 19.27 | Chr12:0–13000000 | Yes | Yes | Yes | Yes |
| A0578 | Del | Chr18:10001–11497100 | 11.49 | 18p Deletion syndrome | 14.16 | Chr18:3000000–11000000 | Yes | Yes | Yes | Yes |
| A0676 | Del | Chr4:1.78e+08–1.91e+08 | 12.6 | ASD; small omphalocele, anteriorly placed anus, cleft palate | 8.54 | Chr4:180000000–189000000 | Yes | Yes | Yes | Yes |
| A0248 | Del | Chr4:75392078–88436655 | 13.04 | 4q21 Deletion syndrome | 25.90 | Chr4:75000000–92000000 | Yes | Yes | Yes | Yes |
| A0577 | Dup | Chr18:10001–13279511 | 13.27 | 18p Deletion syndrome | 11.13 | Chr18:0–8000000 | Yes | Yes | Yes | Yes |
| A0856 | Del | Chr13:1.01e+08–1.14e+08 | 13.29 | 17.84 | Chr13:101000000–113000000 | Yes | Yes | Yes | Yes | |
| A0322 | Dup | Chr17:87009–13501809 | 13.41 | 17p13.1 Deletion syndrome | NA | Chr17:0–13000000 | Yes | Yes | Yes | Yes |
| A0359 | Del | Chr18:142096–13885315 | 13.74 | 18p Deletion syndrome | 15.81 | Chr18:0–13000000 | Yes | Yes | Yes | Yes |
| A0652 | Dup | Chr13:99103482–1.13e+08 | 14.32 | 22.09 | Chr13:50000000–95000000 | Yes | Yes | Yes | Yes | |
| A1092 | Dup | Chr18:4316–14918854 | 14.91 | 18p11.32-p11.31 Duplication | 12.48 | Chr18:0–15000000 | Yes | Yes | Yes | Yes |
| A0580 | Del | Chr15:83800036–99660791 | 15.86 | 15q25 Deletion syndrome | 8.68 | Chr15:69000000–100000000 | Yes | Yes | Yes | Yes |
| A0310 | Dup | Chr11:1.17e+08–1.35e+08 | 18.14 | Jacobsen syndrome | 14.74 | Chr11:116000000–131000000 | Yes | Yes | Yes | Yes |
| A0953 | Del | Chr4:61552–18242617 | 18.18 | 4p16.3–16.1 Deletion | 9.45 | Chr4:0–18000000 | Yes | Yes | Yes | Yes |
| A0676 | Dup | Chr3:93949–19556862 | 19.46 | 3p Syndrome | 8.54 | Chr3:0–20000000 | Yes | Yes | Yes | Yes |
| A0856 | Dup | Chr5:204737–20929211 | 20.72 | Cri du Chat Syndrome (5p deletion) | 17.84 | Chr5:7000000–21000000 | Yes | Yes | Yes | Yes |
| A0686 | Dup | Chr10:1.14e+08–1.35e+08 | 21.26 | 10q26 Deletion syndrome | NA | Chr10:113000000–133000000 | Yes | Yes | Yes | Yes |
| A0224 | Dup | Chr7:1.37e+08–1.59e+08 | 22.01 | 17.61 | Chr7:135000000–157000000 | Yes | Yes | Yes | Yes | |
| A1073 | Dup | Chr2:161989273–185124629 | 23.14 | 2q31.1 Duplication syndrome | 9.78 | Chr2:163000000–180000000 | Yes | Yes | Yes | Yes |
| A0202 | Dup | Chr7:1.36e+08–1.59e+08 | 23.16 | 24.87 | Chr7:136000000–157000000 | Yes | Yes | Yes | Yes | |
| A0359 | Del | Chr18:53764221–77982126 | 24.22 | 18q Deletion syndrome | 15.81 | Chr18:53000000–76000000 | Yes | Yes | Yes | Yes |
| A0302 | Dup | Chr13:90000001–1.15e+08 | 25.17 | 19.03 | Chr13:91000000–113000000 | Yes | Yes | Yes | Yes | |
| A0644 | Del | Chr13:37474970–65394654 | 27.92 | 13q14 Deletion syndrome | 14.70 | Chr13:40000000–66000000 | Yes | Yes | Yes | Yes |
| A0224 | Dup | Chr13:80744674–1.13e+08 | 32.68 | 17.61 | Chr13:82000000–113000000 | Yes | Yes | Yes | Yes | |
| A0894 | Del | Chr13:74724554–1.14e+08 | 39.35 | 28.29 | Chr13:81000000–113000000 | Yes | Yes | Yes | Yes | |
| A0256 | Dup | Chr14:20472548–64879219 | 44.41 | 14q11-q22 Deletion syndrome | 19.27 | Chr14:20000000–62000000 | Yes | Yes | Yes | Yes |
| A0519 | Dup | Chr18:33381218–783231 17 | 44.94 | 18q Deletion syndrome | 9.57 | Chr18:33000000–76000000 | Yes | Yes | Yes | Yes |
| A0093 | Dup | Chr5:0–46100000 | 46.1 | Cri du Chat Syndrome (5p deletion) | NA | Chr5:1000000–45000000 | Yes | Yes | Yes | Yes |
| A0403 | Dup | Chr21:1–48129895 | 48.13 | Early-onset Alzheimer disease with cerebral amyloid angiopathy | 10.58 | Chr21:15000000–46000000 | Yes | Yes | Yes | Yes |
| A0322 | Del | Chrx:61529–48771280 | 48.71 | Xp11.22-p11.23 microduplication | NA | Chrx:3000000–48000000 | Yes | Yes | Yes | Yes |
| A0899 | Dup | Chr2:1.90e+08–2.43e+08 | 53.11 | 2q33.1 Deletion syndrome | 8.93 | Chr2:188000000–241000000 | Yes | Yes | Yes | Yes |
| A0971 | Del | Chrx:90374470–155190083 | 64.82 | Xq26-28 deletion | 11.03 | Chrx:93000000–150000000 | Yes | Yes | Yes | Yes |
| A0632 | Dup | Chr9:261257–84260002 | 84 | 9p Deletion syndrome | 14.90 | Chr9:0–37000000 | Yes | Yes | Yes | Yes |
ASD, atrial septal defect; AZFa, azoospermia factor region a; AZFb, azoospermia factor region b; Chr, chromosome; Del, deletion; Dup, duplication; FC, fetal DNA concentration; NA, not applicable.
Syndrome is associated with either deletion or duplication in the region but may not match the observed DNA abnormality.
Several false-negative samples (A0133, A0947, 0899, A0310, A0686, and A0769) had a low estimated fetal DNA fraction, or the fraction could not be reliably calculated, likely making detection of abnormalities more difficult. Furthermore, certain regions of the genome, primarily those with highly repetitive DNA sequences, such as near centrosomes, showed significantly greater covariance and thus, made detection of true abnormalities in these regions more challenging. In our analysis, we masked some repetitive sequences before read mapping to increase accuracy of read counting in each bin, which may have resulted in false negatives in these repetitive sequence-rich regions. For example, the deletion in sample A0769 (chr22: 17299942–19770514) is close to centrosome, and the deletion in A0133 (chr14: 1.01E+08–1.07E+08) is located at the end of chromosome 14, resulting in false negatives, despite increased sequencing depth.
Our assay produced 58 false positives from 55 samples at a sequencing depth of 3.5 million reads for a false-positive rate of 3.8%; 39 of 58 (67.2%) false positives predicted a deletion/duplication less than 5 Mb in size. We hypothesized that many of these false positives might be caused by deletions/duplications in maternal DNA and therefore, would be detected in the maternal plasma. To investigate this hypothesis, we performed aCGH on all 55 samples of maternal DNA to assess for the presence of deletions and duplications in the maternal DNA; 35 of 55 (63.6%) samples were validated as maternal deletions/duplications, suggesting that the majority of false positives using this method can be accounted for by abnormalities present in the mother’s genome. This finding has significant implications for the clinical utility of NIPT to detect subchromosomal abnormalities, because a positive test using this assay may require a follow-up test to rule out deletions and duplications in the maternal DNA before the fetus can be diagnosed. For the remaining 20 false-positive samples, repeat sequencing and analysis revealed no repeat false-positive results. Thus, these false positives seem to be the random result of our sequencing technique and statistical model.
Detection of subchromosomal abnormalities would most likely be applied as an extension of current methods of NIPT to detect aneuploidy. When results for aneuploidy and subchromosomal abnormalities are considered together, our assay detected 129 of 151 karyotype abnormalities of all sizes in 1,476 samples at 3.5 million reads for an overall sensitivity of 85.4% compared with aCGH. There were 64 false-positive samples total for an overall specificity of 95.7%. If size is restricted to abnormalities greater than 5 Mb, 115 of 117 abnormalities in 1,476 samples were detected for an overall sensitivity and specificity of 98.3% and 98.1%, respectively. These results show that detection of subchromosomal abnormalities is a viable extension of NIPT using SSP.
Discussion
This study shows that sequencing maternal plasma DNA can reliably produce a fetal karyotype of chromosomal deletions and duplications with relatively high resolution while avoiding the necessity of an invasive procedure. Although our NIPT method is less accurate than CMA, especially for deletions/duplications smaller than 5 Mb, it offers the advantage of unbiased testing and is substantially more accurate than current blood-based screening tests for aneuploidy. In our study population, we detected 75% more abnormal karyotypes than detection of aneuploidy alone at a sequencing depth of 3.5 million reads, with greater sequencing depth significantly increasing sensitivity. Thus, because no specific prenatal screening exists for subchromosomal abnormalities, the ability of a single test to detect these abnormalities in addition to aneuploidy represents a powerful extension of NIPT.
Our false-positive rate of 3.8% for subchromosomal abnormalities was relatively high and would potentially limit the clinical utility of our method as a stand-alone screening test. However, 64% of false positives were found to be because of abnormalities present in maternal DNA, suggesting that the false-positive rate could be greatly reduced with a reflex follow-up test of maternal DNA collected simultaneously with maternal plasma. A reflex test of maternal DNA would also identify inherited variants, which may represent a significant fraction of true positives as well. Most of these variants are polymorphisms and unlikely to be detrimental for the mother. Further lowering of the false-positive rate may be possible with greater sequencing depth; additional study is required to investigate these possibilities adequately.
Our study population was drawn from pregnancies with anatomic abnormalities found on ultrasound to generate a high incidence of karyotype abnormalities, resulting in tested pregnancies ranging in gestational age from 12 to 37 wk, with an average age of 24 wk, much later than NIPT would ideally be applied. In a future study, detection of subchromosomal abnormalities could be applied to a population with lower but still elevated risk (i.e., advanced maternal age) at a gestational age of 12–16 wk. This timing would offer the advantage of safer, less expensive, and more private elective termination and would be consistent with current prenatal screening practices. In addition, many governments limit the legal gestational age at which an elective termination may be performed. Therefore, accurate diagnosis of chromosomal abnormalities at earlier gestational ages is an important goal of prenatal testing. Because the fetal concentration of DNA is lower at a gestational age of 12–16 wk, with only 66.7% having a fetal DNA fraction greater than 10% in our study, deeper sequencing than 3.5 million reads is likely required for accurate detection of subchromosomal abnormalities. Increasing sequencing depth is likely the most direct way of improving accuracy of diagnosis in general and may be achieved in the near future with additional reductions in sequencing costs over time. However, certain parts of the genome may not be amenable to adequate coverage because of the inherent nature of chromosome structure or features of DNA (for example, heterochromatin regions or repetitive DNA sequences).
One crucial challenge is that our technique may identify deletions and duplications of unknown clinical significance, with incomplete penetrance, or with mild or unpredictable clinical consequences. Many of these abnormalities may be normal inherited variants. Thus, if our extension of NIPT is put into clinical practice, great care must be taken in presenting results and providing appropriate counseling to patients. We expect that the utility of NIPT for subchromosomal abnormalities will increase with greater understanding of genomic disorders. Databases compiling information on these deletions/duplications are being developed, such as DECIPHER (24). With increased sample sizes and additional clinical correlations, such databases will undergo validation and become more useful in the future. At minimum, detecting such abnormalities will alert the child’s providers to the need for close surveillance for sequelae, such as neurodevelopmental delay or the consequences of a known genomic syndrome.
NIPT using SSP is able to reliably estimate the fetal DNA fraction in maternal plasma, which we showed with a large cohort of pregnant women with normal fetuses. We applied this technique to identify samples with a lower fetal DNA fraction that, thus, might provide a less reliable result, although the presence of a subchromosomal abnormality was correctly identified in many samples with fetal DNA fractions less than 10%. As noted above, this effect may be more significant and therefore, estimation of the fetal fraction may be more useful in trials studying women receiving NIPT at earlier gestational ages. In a clinical setting, this analysis may identify samples that require either deeper sequencing, which may ameliorate the effect of a low fetal DNA fraction, or repeat testing at a later gestational age when the fetal fraction of DNA has increased.
The current cost of sequencing a human genome with 0.5× coverage by single reads, approximately the coverage in our study at 3.5 million reads, is comparable with the cost of CMA while avoiding an invasive procedure. This depth allowed excellent detection of deletions/duplications greater than 5 Mb in size in our study. Deeper sequencing allows for finer resolution at an additional cost. Already, many obstetric practices in the United States are offering NIPT as a replacement for invasive testing for detection of common chromosomal aneuploidies and select common deletions/duplications. Our method offers unbiased detection and greater resolution relative to current NIPT methods, with the potential for lower costs because of lower sequencing requirements. SSP has the potential to be a cheaper and faster technology to use for this purpose compared with Illumina technology; however, both technologies continue to improve, with increasing speed and accuracy and falling costs. Crucially, this type of analysis has the potential to be implemented for all high-risk pregnancies and eventually, as standard of care prenatal screening for all women, replacing current screening methods.
The methods developed here can be applied to any mixed biological sample to determine the subchromosomal abnormalities in the minor component, even when this fraction represents only a small percentage of the total DNA. In prenatal diagnostics, samples obtained from chorionic villi could be analyzed for mosaic karyotypes or maternal contamination. Furthermore, many different cancers have been associated with copy number changes that could potentially be detected from serum cfDNA or a solid tumor sample that contains both normal and cancer cells (25, 26). Because the cost of next generation sequencing continues to drop, we expect that this technique will become applicable for a broad range of diseases.
Materials and Methods
Subject Recruitment.
This study was approved by the Institutional Review Boards of Guangdong Women and Children Hospital. Informed consent was obtained from all participants. Women were offered participation in our study if they presented with a singleton gestation, in which either CVS or amniocentesis was indicated for structural anomalies detected on ultrasonography. Those choosing to participate provided written informed consent after discussion of the potential advantages and risks of chromosomal microarray testing, including the possibility of findings of uncertain clinical significance and the identification of genetic variants in the fetus or a parent that cause adult-onset disorders. CVS was performed in the usual fashion. For women undergoing amniocentesis, an additional 10 mL amniotic fluid was retrieved.
Microarray CGH Laboratory Procedures.
After DNA extraction, an aCGH analysis was performed using Agilent’s 8 × 60K commercial arrays (Agilent Technologies) according to the procedures described in our previous paper (12). The data were analyzed with Agilent Genomic Workbench Lite Edition 6.5.0.18 software (Agilent Technologies). Aberrations were identified using the Agilent Genomic Workbench Lite Edition 6.5.0.18 software through the Aberration Detection Method-2 Algorithm, with a sensitivity threshold of 6.0 and a data filter that rejected aberrations that did not include at least five probes with a log2 set of 0.25. All quality control metrics passed.
cfDNA Preparation and Sequencing.
Preparation and sequencing of cfDNA obtained from maternal plasma were performed as previously described (12). To obtain complete sequences of cfDNA fragments, sequencing was performed using an Ion Proton Sequencer (Life Technologies) at 400 flows, and BaseCalling options were modified as follows: BaseCaller–disable-all-filters off–keypass-filter off–phasing-residual-filter=2.0–trim-qual-cutoff 100–trim-qual-window-size 30–trim-adapter-cutoff 16–num-unfiltered 1000.
Data Preprocessing.
Reads mapping and filtering procedures were performed according to our previous paper (12). The retained reads were aligned to the human genomic reference sequences (hg19) using the BWA (27). Reads that were unmapped or had multiple primary alignment records were filtered by “FLAG” field in the alignment file using an in-house Perl script. Duplicate reads were identified by Picard (broadinstitute.github.io/picard/) with the parameters java -jar MarkDuplicates.jar M=picard_duplication_metrics REMOVE_DUPLICATES=true ASSUME_SORTED=true 1 and removed by an in-house Perl script. The remaining reads were considered unique reads for additional analysis.
Calculation of Fetal DNA Fraction.
To identify the full sequences of cfDNA, we used in-house Perl scripts to exclude incomplete reads and determine the size distributions of plasma cfDNA molecules. Fetal DNA is generally shorter than maternal DNA (28), resulting in the feasibility of predicting the fetal DNA concentration in maternal plasma (18). Plasma samples with a higher fetal DNA fraction would have a higher proportion of short plasma DNA fragments (∼130–140 bp; region A) and a lower proportion of long plasma DNA fragments (∼155–175 bp; region B). Locally weighted scatterplot smoothing (LOESS) regression was applied to fit the fetal fraction against reads ratio in features A and B. We obtain the LOESS fit-predicted fetal fraction for feature A and for feature B. Because both and predict the fetal DNA fraction, and should also closely correlate. Therefore, we used reference samples to compare and and thus, identify instances of poor correlation. If is larger than 0.40 (larger than 99% normal samples), and are inconsistent, and the fetal fraction is considered unpredictable. Otherwise, we calculated the final predicted fetal fraction using .
Identification of Subchromosomal Abnormalities.
After reads mapping, guanosine-cytosine (GC) correction was performed according to our previous paper (12). In brief, the number of unique reads from each 20-kb bin on each chromosome was counted. To eliminate the effect of GC bias in different samples, an integrated GC correction method was applied, in which LOESS regression was used to compute the corrected number of unique reads in each 20-kb bin depending on the GC content of the genomic sequence in the corresponding bin. The corrected reads number of each chromosome was determined by summing the weighted values of all 20-kb bins of each specific chromosome in a sample.
Other than GC bias, some genome blocks behaved similarly. Straver et al. (29) applied a “within-sample” comparison method to similar chromosome regions to detect subchromosomal aberrations. Thus, reads ratio values were obtained from 500 normal samples as within-sample reference bins. Corrected reads ratio for each 1-Mb region in each sample and then, the SD, mean, and coefficient of variation (CV) values for each 1-Mb region were calculated. Consequently, for block i and sample j, we obtain the Z score (). For block i of sample j, we obtain a Z score in each independent test. Combining the Z scores of adjacent blocks would increase the precision to detect subchromosome aberrations using Stouffer’s Z-score method (29): . When Stouffer’s Z score is larger than 3, we classify it as microduplication, whereas when it is less than −3, we classify it as microdeletion. We tested different k values and found that k = 7 performed best. Lastly, if the Stouffers’ Z scores of adjacent blocks are similar, we would merge these blocks together. If the subchromosomal aberration region is larger than 1 Mb, we can use the Stouffer’s Z score to enlarge the aberration signal and detect abnormalities. To avoid false positives, the Z scores of each bin were filtered by a threshold based on fetal DNA fraction and sequencing depth. We used the formula =. In the formula, CV (CV in each bin) was related to sequencing depth. We found that the precision of subchromosomal deletion/duplication detection is closely related to the fetal DNA fraction in maternal plasma. When the fetal DNA fraction is less than 10%, may be less than 3, resulting in a false negative.
Acknowledgments
This work was supported, in part, by the National Science Foundation for Young Scholars of China Grant 81000255, National Natural Science Foundation of China.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518151112/-/DCSupplemental.
References
- 1.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8(8):639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 2.Lejeune J, et al. 3 Cases of partial deletion of the short arm of a 5 chromosome. C R Hebd Seances Acad Sci. 1963;257:3098–3102. [PubMed] [Google Scholar]
- 3.Lammer EJ, Opitz JM. The DiGeorge anomaly as a developmental field defect. Am J Med Genet Suppl. 1986;2:113–127. doi: 10.1002/ajmg.1320250615. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi DW. From prenatal genomic diagnosis to fetal personalized medicine: Progress and challenges. Nat Med. 2012;18(7):1041–1051. doi: 10.1038/nm.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CN, et al. Clinical utility of array comparative genomic hybridisation for prenatal diagnosis: A cohort study of 3171 pregnancies. BJOG. 2012;119(5):614–625. doi: 10.1111/j.1471-0528.2012.03279.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Bernier F, Lauzon J, Lowry RB, Chernos J. Application of microarray-based comparative genomic hybridization in prenatal and postnatal settings: Three case reports. Genet Res Int. 2011;2011:976398. doi: 10.4061/2011/976398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller DT, et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.American College of Obstetricians and Gynecologists Committee on Genetics Committee Opinion No. 581: The use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol. 2013;122(6):1374–1377. doi: 10.1097/01.AOG.0000438962.16108.d1. [DOI] [PubMed] [Google Scholar]
- 9.Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: A systematic review. Obstet Gynecol. 2007;110(3):687–694. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 10.Benn P, et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn. 2013;33(7):622–629. doi: 10.1002/pd.4139. [DOI] [PubMed] [Google Scholar]
- 11.Bianchi DW, et al. CARE Study Group DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 12.Liao C, et al. Noninvasive prenatal diagnosis of common aneuploidies by semiconductor sequencing. Proc Natl Acad Sci USA. 2014;111(20):7415–7420. doi: 10.1073/pnas.1321997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu RW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartlett LA, et al. Risk factors for legal induced abortion-related mortality in the United States. Obstet Gynecol. 2004;103(4):729–737. doi: 10.1097/01.AOG.0000116260.81570.60. [DOI] [PubMed] [Google Scholar]
- 15.Peters D, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011;365(19):1847–1848. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92(2):167–176. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 18.Yu SC, et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci USA. 2014;111(23):8583–8588. doi: 10.1073/pnas.1406103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13(11):913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 21.Liao C, Zhengfeng X, Zhang K. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;371(6):577–578. doi: 10.1056/NEJMc1405486. [DOI] [PubMed] [Google Scholar]
- 22.Lutgendorf MA, Stoll KA, Knutzen DM, Foglia LM. Noninvasive prenatal testing: Limitations and unanswered questions. Genet Med. 2014;16(4):281–285. doi: 10.1038/gim.2013.126. [DOI] [PubMed] [Google Scholar]
- 23.Snyder MW, et al. Copy-number variation and false positive prenatal aneuploidy screening results. N Engl J Med. 2015;372(17):1639–1645. doi: 10.1056/NEJMoa1408408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Firth HV, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karachaliou N, et al. Spanish Lung Cancer Group Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC Trial. JAMA Oncol. 2015;1(2):149–157. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 26.Bianchi DW, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314(2):162–169. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan KC, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50(1):88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 29.Straver R, et al. WISECONDOR: Detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014;42(5):e31. doi: 10.1093/nar/gkt992. [DOI] [PMC free article] [PubMed] [Google Scholar]