Abstract
Neurons live for decades in a postmitotic state, their genomes susceptible to DNA damage. Here we survey the landscape of somatic single-nucleotide variants (SNVs) in the human brain. We identified thousands of somatic SNVs by single-cell sequencing of 36 neurons from the cerebral cortex of three normal individuals. Unlike germline and cancer SNVs, which are often caused by errors in DNA replication, neuronal mutations appear to reflect damage during active transcription. Somatic mutations create nested lineage trees, allowing them to be dated relative to developmental landmarks and revealing a polyclonal architecture of the human cerebral cortex. Thus, somatic mutations in the brain represent a durable and ongoing record of neuronal life history, from development through postmitotic function.
Ongoing random mutation of DNA ensures that no two cells in an individual are genetically identical (1). Some of these “somatic” mutations cause cancer or, when they occur in progenitors in the developing human brain, neurological diseases such as epilepsy and developmental brain malformations (2). Whereas mature neurons can survive for the life of the individual, DNA is buffeted by mutagens such as oxygen free radicals, electromagnetic radiation, and endogenous transposable elements. These forces have the potential to induce somatic mutations throughout the life of a neuron, and they may contribute to normal aging and neurodegenerative disease (3). High-throughput sequencing of DNA isolated from thousands of cells from “bulk” tissue, although useful, is unable to detect mutations present in one or small numbers of cells, and determining whether mutations exist together within one cell requires single-cell analyses (4, 5). Although previous analyses of single neurons in the human brain have demonstrated occasional somatic copy-number variants (6, 7) and L1 retrotransposon insertions (8, 9), SNVs are a major source of germline and cancer variation (10, 11), and likely represent a more substantial source of the overall somatic mutation burden in neurons.
To investigate rates and patterns of somatic SNVs, we analyzed high-coverage (~40×) whole-genome sequencing (WGS) data from 36 single neurons from postmortem brain tissue of three neurotypical individuals: a 15-year-old female (UMB4638), a 17-year-old male (UMB1465) (9), and a 42-year-old female (UMB4643) (sources designated brains A, B, and C, respectively). Using fluorescenceactivated nuclear sorting (FANS), we purified single NeuN+ cortical neuronal nuclei from the prefrontal cortex of postmortem human brain tissue; we then amplified the DNA by multiple-displacement amplification (MDA) (8) (Fig. 1A). We subjected this amplified DNA to high-throughput sequencing, achieving ≥10× average coverage per neuron at 82 to 87% of loci (table S1). After analysis of overall quality (9), false-positive and false-negative variant calls, allelic balance, and locus/allele dropout (figs. S1 and S2; supplementary text), we used these single-cell MDA WGS data to identify somatic mutations, focusing on SNVs.
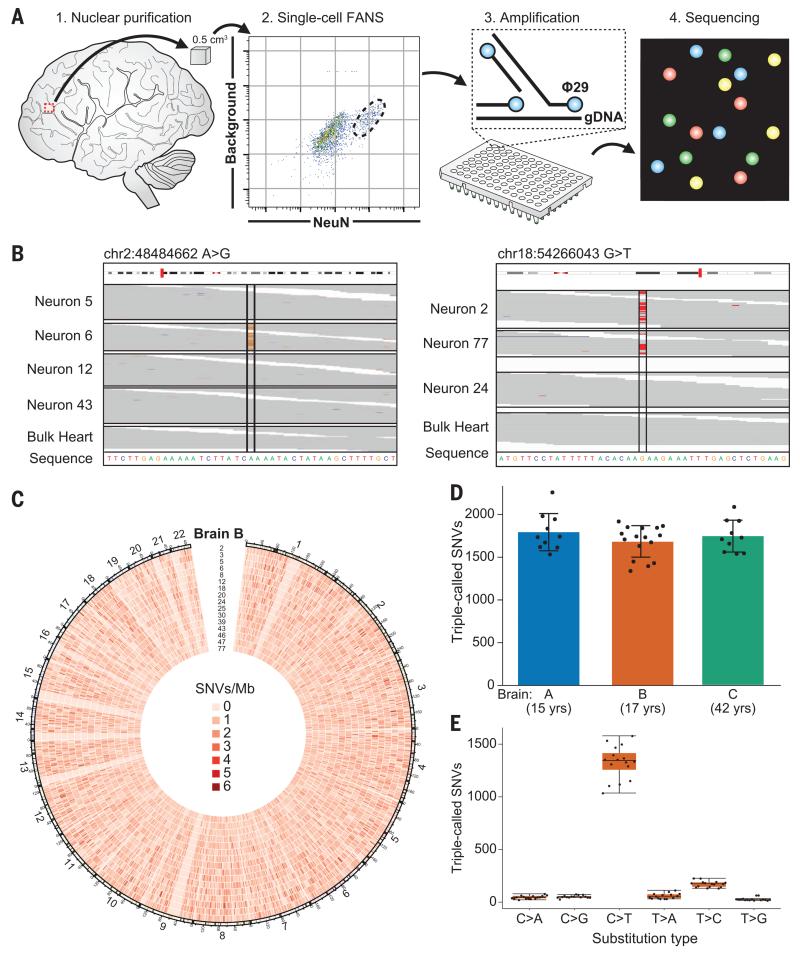
Fig. 1. Somatic SNVs are detected by single-neuron whole-genome sequencing.
(A) Schematic of approach. Nuclei are isolated from a 0.5-cm3 piece of frozen postmortem tissue by fluorescence-activated nuclear sorting (FANS), and DNA is amplified by Φ29 polymerase-mediated multiple-displacement amplification (MDA) and subjected to whole-genome sequencing (WGS). (B) Sample sequencing alignment tracks from brain B. One SNV is called uniquely in one sample (left), and one is shared between neurons 2 and 77 but absent from other single cells and heart (right). (C) Circos plot of SNV rate per megabase in brain B across human autosomes demonstrates that somatic mutations are distributed broadly. (D) SNVs per neuron are tightly correlated within each individual. (E) Most single-neuron SNVs in brain B are C>T transitions. Error bars in (D) and (E) denote SD; data points from individual neurons are spread horizontally for visibility.
We identified single-cell SNV candidates by means of three established mutation-calling algorithms (see materials and methods), thereby defining a conservative list of somatic SNVs as those identified by all three callers in at least one single neuron but absent from DNA isolated from bulk tissue (e.g., heart) from the same individual (Fig. 1B). These “triple-called” SNVs were confirmed by Sanger sequencing at a very high rate (92%) in the single-cell DNA sample from which they were identified (table S2 and fig. S3). Single neurons from the three brains averaged 1685 to 1793 triple-called SNVs (Table 1, Fig. 1D, table S3, and fig. S4). Across all neurons, we observed a mean of 8.3% allelic dropout (loss of one allele of a heterozygous locus) and 3.3% locus dropout (fig. S1). With an estimated 23% false-discovery rate, our results suggest that each neuron from these individuals may have contained 1458 to 1580 somatic SNVs. Additional model-based analyses resulted in a similar range of SNVs identified (fig. S5; supplementary text). These rates of mutation per neuron are consistent with single-cell sequencing of other normal cell types (12) but are lower than somatic SNV rates in normal skin cells, which are exposed to damaging ultraviolet light (13), and in several tumor types (11, 12, 14).
Table 1. SNVs found in 36 single human cortical neurons isolated from three normal individuals.
| SNV | Brain A (n = 10) |
Brain B (n = 16) |
Brain C (n = 10) |
|||
|---|---|---|---|---|---|---|
| Count | Percentage | Count | Percentage | Count | Percentage | |
| Coding | 23.7 | 1.3% | 23.4 | 1.4% | 24.4 | 1.4% |
|
| ||||||
| Silent | 8.3 | 0.5% | 7.1 | 0.4% | 6.2 | 0.4% |
|
| ||||||
| Missense | 14.1 | 0.8% | 14.4 | 0.9% | 16.8 | 1.0% |
|
| ||||||
| Truncating | 1.3 | 0.1% | 1.8 | 0.1% | 1.3 | 0.1% |
|
| ||||||
| Noncoding | 254.3 | 14.2% | 243.5 | 14.5% | 243.4 | 13.9% |
|
| ||||||
| Untranslated region | 22.0 | 1.2% | 21.0 | 1.2% | 26.5 | 1.5% |
|
| ||||||
| Noncoding RNA | 232.3 | 13.0% | 222.5 | 13.2% | 216.9 | 12.4% |
|
| ||||||
| Intronic | 710.0 | 39.5% | 660.1 | 39.2% | 693.6 | 39.7% |
|
| ||||||
| Splice | 0.3 | 0.0% | 0.4 | 0.0% | 0.6 | 0.0% |
|
| ||||||
| Other intronic | 709.7 | 39.5% | 659.7 | 39.2% | 693.0 | 39.7% |
|
| ||||||
| Intergenic | 805.0 | 44.9% | 757.6 | 45.0% | 785.5 | 45.0% |
|
| ||||||
| Mean | 1793.0 | 1685.0 | 1747.0 | |||
The molecular profile of neural SNVs is quite distinct from cancer and germline mutations, and reveals mutagenic forces that affect a neuron during its life. Cancer and germline mutation rates correlate with DNA replication (15, 16), with late-replicating DNA more susceptible to mutation, and they negatively correlate with transcription (15, 17). In contrast, single-neuron SNVs did not correlate with replication timing (fig. S6 and table S4) and were enriched in coding exons (Fig. 2A) with a strand bias (purine-purine transitions enriched on the nontemplate strand), as expected for transcription-associated mutations (18, 19) (Fig. 2B and fig. S7). We also observed a signature of methylated cytosine (meC) to thymine (T) transitions (fig. S8), which can occur as a result of replication-independent deamination of meC, in single-neuron SNVs. Taken together, these data demonstrate that replication-independent mutational mechanisms generate more SNVs than does replication in human neurons, which are postmitotic and live long, transcriptionally active lives.
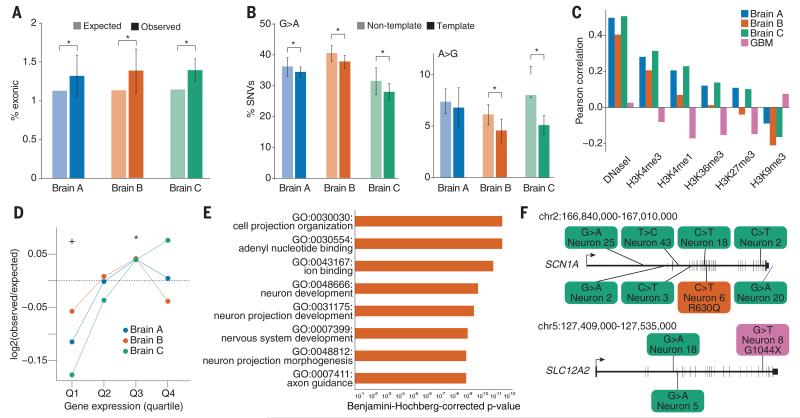
Fig. 2. Somatic SNVs occur at loci that are expressed in the brain and associated with nervous system function and disease.
(A) Coding exons are enriched for somatic SNVs. *P < 0.05, combined binomial test, Bonferroni-corrected. (B) SNVs in transcribed regions display a strand bias, suggesting that transcriptional damage influences the somatic SNV rate. *P < 0.05, analysis of variance with Sidak’s correction for multiple testing. Error bars in (A) and (B) denote SD. (C) Single-neuron SNVs correlate with epigenetic marks of transcription in the fetal brain and are depleted in heterochromatin, in opposition to the pattern observed in glioblastoma multiforme (GBM) SNVs. (D) Single-neuron SNVs occur in genes expressed in the cerebral cortex. Genes in the lowest expression quartile were significantly depleted, and those in the third quartile were significantly enriched, for single-neuron SNVs. *P < 0.05, +P < 0.05, Fisher combined one-tailed Poisson P value for enrichment and depletion of SNVs, respectively. (E) Gene ontology categories associated with nervous system development and function are enriched for mutated genes across single neurons from brain B. (F) Examples of genes implicated in human disease that were mutated in single brain B neurons (for full list, see table S8). Green SNVs occurred in introns or downstream of the coding region, the orange SNV occurred in an exon and induced a missense mutation, and the magenta SNV is a nonsense mutation.
Integration of our SNV data with gene expression in the prefrontal cortex from public data-bases (20) revealed that highly expressed genes (in particular, those in the third quartiles) were enriched for single-neuron SNVs with statistical significance (Fig. 2D, fig. S9, and table S5), and, unlike mutations in cancers (such as glioblastoma multiforme), single-neuron SNVs correlated with chromatin markers of transcription (21) from fetal brain (Fig. 2C and table S6). Neural-related gene sets were enriched for somatic SNVs (Fig. 2E, fig. S10, and table S7), and single neurons harbored heterozygous coding SNVs in genes that, when a single copy is mutated in the germline, confer a high risk of neurological disease (table S8). For example, SCN1A (seizure disorder) (22) and SLC12A2 (schizophrenia) (23) both contained coding mutations in neurons from brain B (Fig. 2F and table S8). Thus, genes active in human neurons and critical for their function are vulnerable to somatic mutation, and even the normal brain contains individual neurons with disruptive mutations.
Mutations shared by multiple neurons, which must necessarily have arisen during development, revealed surprising lineage relationships in the human brain. We genotyped shared somatic variants in brain B, including 15 SNVs (table S9), a TG-dinucleotide expansion on chromosome 4 (see materials and methods), and two L1 retrotransposon insertions (8, 9), in 210 amplified single-neuron genomes isolated from the same region of cortex as the original 16 cells from brain B; 136 of 226 (60%) single-neuron genomes contained at least one clonal SNV (fig. S11), and the results suggested serial mutations over the course of development. For example, three SNVs showed distinct mosaic profiles: SNV C1 was present in 23% (51/226) of the neurons tested (suggesting that it occurred early in development), C8 was present only in three of these 51 neurons, and C10 only in two of this set of three, neurons 2 and 77 (Fig. 3A and fig. S11). SNVs identified mutually exclusive sets of neurons such that, for example, cells marked by variants discovered in neurons 39 and 47 never contained SNVs present in neurons 6 and 18, 2 and 77, or 3 and 12 (except one anomalous cell; fig. S11). These data placed 9/16 sequenced cells, and 60% (136/226) of analyzed cortical neurons, into four separate clades (pink, green, blue, and purple in Fig. 3A), which suggests that at least five distinct clades—the four marked, plus at least one additional unmarked clade—gave rise to the 226 neurons in this sample.
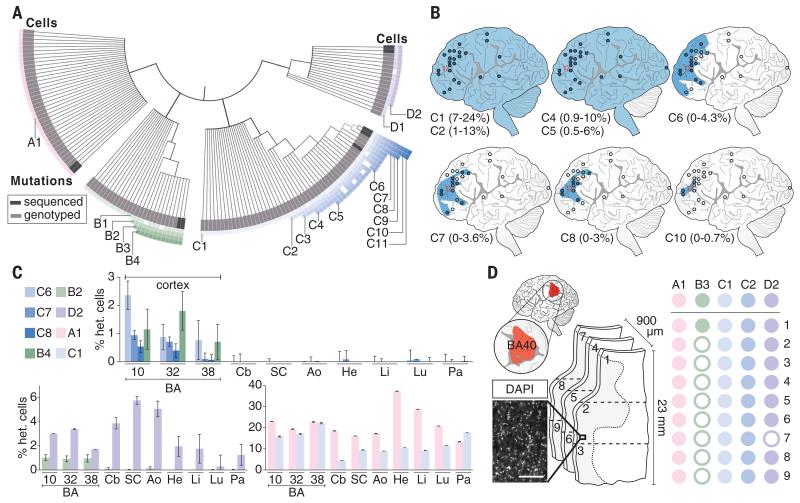
Fig. 3. Somatic mutations are shared between multiple neurons and demonstrate lineage relationships.
(A) Lineage map of 136 human cortical neurons from brain B derived from 18 clonal somatic mutations, including SNVs, long interspersed nuclear element (LINE) insertions, and a TG-dinucleotide expansion. Neurons are placed into four distinct nested clades (pink, green, blue, purple) defined by one or more independent mutations. Cells are ordered within clades according to the presence of multiple somatic mutations. A few cells in each clade fail to manifest individual SNVs shared by other cells of the same clade (indicated by open squares), likely representing incomplete amplification (fig. S2). Dark gray boxes represent cells analyzed by WGS; light gray represents cells analyzed by Sanger-based genotyping. Genomic locations of somatic mutations are given in fig. S11. (B) Ultradeep sequencing of mutated loci across the cortex of brain B. Clonal SNVs from a single clade are progressively regionally restricted to frontal cortex and become progressively rarer in bulk tissue, reflecting their later origin during development and neurogenesis. Blue circle, mutation present; empty circle, mutation absent; blue shading, likely spatial distribution of mutation. Percentage range of heterozygous cells is indicated for each SNV. (C) Ultradeep sequencing of mutated loci across the brain and body. Some variants are brain-specific (top) and others are shared across germ layers (bottom). Samples sequenced are prefrontal cortex [Brodmann area (BA) 10/BA46], cingulate cortex (BA32/BA8), temporal cortex (BA38), cerebellum (Cb), spinal cord (SC), aorta (Ao), heart (He), liver (Li), lung (Lu), and pancreas (Pa). (D) Genotyping shared variants in small sections of human cortex. Left: 4′,6-diamidino-2-phenylindole (DAPI) stain of segment of representative section; scale bar, 200 μm. Center: Three consecutive 300-μm coronal sections from BA40 (red, upper left) were dissected into three axial regions each (1 to 9). Right: Genotyping results for dissected sections. Solid circles denote presence of mutation in indicated sample; open circles denote absence. Mutations with high allele fractions are present in all or virtually all regions, whereas only the least prevalent somatic variant (present in <0.5% of cells) is present in one region but not most regions.
Genotyping DNA samples from across the brain, using ultradeep sequencing of a targeted panel of clonal somatic SNVs, showed that although some SNVs were present at low mosaic fractions (up to 4% of cells) only in restricted regions of frontal cortex, all four major clades dispersed across the cortex at surprisingly low levels of mosaicism (green clade, 0.2 to 2%; blue clade, 0.5 to 6%; purple clade, 1 to 7%; pink clade, 18 to 32%) (Fig. 3B, fig. S12, and table S10). For example, SNVs C8 and C10 were restricted to <3% of cells in the middle frontal gyrus surrounding the site of sequencing, whereas C1 marked deeper branches of this same clade and was found throughout the cortex in 7 to 24% of cells, as well as in the cerebellum and spinal cord (Fig. 3B). Analysis of non-brain tissue of individual B showed that variants present in >5 to 10% of his brain cells were generally detected widely outside the brain, in tissues derived from endoderm, ectoderm, and mesoderm (Fig. 3C, fig. S13, and table S10), which suggests that these SNVs arose before the three germ layers segregated at gastrulation, yet still marked a minority of neurons.
Our data demonstrate that individual clones intermingle widely in the human cortex and that neurons from a given cortical region constitute no fewer than five distinct clades of cells that trace their lineage back to separate mutation-marked pluripotent founder cells in the pregastrulation embryo. A given cortical neuron marked by SNV C1, for example, shares a more recent common cellular ancestor with a cardiomyocyte marked by this same somatic mutation than it does with approximately 75% of neighboring cortical neurons, with which no clonal connection is evident as far back as gastrulation. To confirm this polyclonal derivation, we dissected three consecutive 300-μm coronal sections from Brodmann area (BA) 40 and axially divided these sections into three regions, each 8 mm wide (Fig. 3D). Each region contained contributions from at least three, and usually four, of the major clades identified by single-neuron sequencing. Our data suggest that although late divisions of cortical progenitors likely generate neurons within a relatively restricted cortical zone (24, 25), a given anatomical column of cortex (26) derives from overlapping, intermingled clones from at least four, and likely more, developmental lineages (fig. S14).
Our results show that each human cortical neuron has a profoundly distinctive genome, harboring as many as 1458 to 1580 somatic SNVs, in addition to large CNVs and occasional retroelement insertions, as previously reported (6-9). These estimates are likely to improve with better sequencing and amplification methods as well as more samples. Similar SNV rates were seen at ages 15, 17, and 42, but whether older age is associated with increased SNV rates remains to be explored. These SNVs display signatures of mutagenic processes, such as transcription-associated DNA damage and a preponderance of meC>T deamination. SNVs in coding regions of genes involved in nervous system development and mature neuronal function suggest a “use it and lose it” scenario, in which the very genes used for the function of a neuron are those most likely to be damaged during its life.
Our work demonstrates that somatic mutations can be used to reconstruct the developmental lineage of neurons, suggesting a potential “population genetics” of brain cells and representing a durable record of the series of cell divisions that gives rise to the human brain. This clonal mosaicism in the brain, an organ with exquisite arealization of function, may buffer the brain against deleterious clonal mutations that inevitably arise during development (2). Somatic mutations are also likely to modify the penetrance of germline neurological mutations to generate variable phenotypic effects of germline mutations in different family members, or even between identical twins.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Rozzo, R. S. Hill, H. Lehmann, and W. Paolella for assistance; J. Macklis and N. Sestan for helpful comments on the manuscript; the Dana-Farber Cancer Institute Hematologic Neoplasia Flow Cytometry Core; and the Research Computing group at Harvard Medical School for computing resources, including the Orchestra computing cluster (partially provided through National Center for Research Resources grant 1S10RR028832-01). Human tissue was obtained from the NIH NeuroBioBank at the University of Maryland. We thank R. Johnson of the NeuroBioBank for assistance with tissues, and we thank the donors and their families for their invaluable donations for the advancement of scientific understanding. Figure 3A was illustrated by K. Probst (Xavier Studio). Supported by National Institute on Aging grant T32 AG000222 (M.A.L.), the Leonard and Isabelle Goldenson Research Fellowship (M.B.W.), National Institute of General Medical Sciences (NIGMS) grant T32 GM007753 and the Louis Lange III Scholarship in Translational Research (G.D.E.), NIGMS grants T32 GM007753 and T32 GM007226 (A.M.D.), the Eleanor and Miles Shore Fellowship (E.L.), National Institute of Mental Health grant P50 MH106933 (P.J.P.), and National Institute of Neurological Disorders and Stroke grants R01 NS032457, R01 NS079277, and U01 MH106883 and the Manton Center for Orphan Disease Research (C.A.W.). C.A.W. is a Distinguished Investigator of the Paul G. Allen Family Foundation and an Investigator of the Howard Hughes Medical Institute. Supplement contains additional data. Sequencing data have been deposited in the NCBI SRA under accession numbers SRP041470 and SRP061939.
Footnotes
www.sciencemag.org/content/350/6256/94/suppl/DC1
Figs. S1 to S14
Tables S1 to S10
Materials and Methods
References (27–56)
REFERENCES AND NOTES
- 1.Frank SA. Proc. Natl. Acad. Sci. U.S.A. 2010;107(suppl. 1):1725–1730. doi: 10.1073/pnas.0909343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poduri A, Evrony GD, Cai X, Walsh CA. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De S. Trends Genet. 2011;27:217–223. doi: 10.1016/j.tig.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Dumanski JP, Piotrowski A. Methods Mol. Biol. 2011;838:249–272. doi: 10.1007/978-1-61779-507-7_12. [DOI] [PubMed] [Google Scholar]
- 5.Behjati S, et al. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConnell MJ, et al. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai X, et al. Cell Reports. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evrony GD, et al. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evrony GD, et al. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abecasis GR, et al. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan CW, et al. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, et al. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martincorena I, et al. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo G, et al. Nat. Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence MS, et al. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatoyannopoulos JA, et al. Nat. Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgkinson A, Eyre-Walker A. Nat. Rev. Genet. 2011;12:756–766. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- 18.Green P, Ewing B, Miller W, Thomas PJ, Green ED. Nat. Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 19.Polak P, Arndt PF. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JA, et al. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kundaje A, et al. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parihar R, Ganesh S. J. Hum. Genet. 2013;58:573–580. doi: 10.1038/jhg.2013.77. [DOI] [PubMed] [Google Scholar]
- 23.Morita Y, et al. J. Neurosci. 2014;34:4929–4940. doi: 10.1523/JNEUROSCI.1423-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao P, et al. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid CB, Liang I, Walsh C. Neuron. 1995;15:299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 26.Kornack DR, Rakic P. Neuron. 1995;15:311–321. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.