Abstract
Currently, various long noncoding RNAs (lncRNAs) have been identified as key regulators of multiple cancers. However, cancer stem cell (CSC)-related lncRNAs have rarely been reported. In this study, we found an lncRNA that is a promoter upstream transcript of hypoxia-inducible factor-2α (HIF-2α), and we named it “lncRNA-HIF2PUT”. The function of HIF-2α is closely connected with “stem cell-like” properties, and the function of PROMPTs is often associated with the adjacent protein-coding transcripts. Herein, we showed that the expression of lncRNA-HIF2PUT was significantly correlated with HIF-2α in colorectal cancer (CRC) tissues. Knockdown of lncRNA-HIF2PUT blocked the HIF-2α expression and inhibited the CSC properties in CRC cell lines DLD-1 and HT29. LncRNA-HIF2PUTsmall interfering RNA transfection resulted in decreased stemness genes expression, impaired colony formation, and spheroid formation ability, retarded migration, and invasion of the cells. These data suggest that lncRNA-HIF2PUT may be a regulator of HIF-2α and a mediator of CSCs in CRC.
Keywords: HIF-2α, long noncoding RNA, colorectal cancer, stem cell properties
Introduction
Colorectal cancer (CRC) is one of the most common malignancies and a leading cause of cancer-related deaths worldwide. The treatment of CRC has achieved certain advances in recent years. However, for the patients with metastasis and tumor recurrence, prognosis remains poor. Current opinion is that malignant tumors may derive from a small subset of cancer stem cells (CSCs) responsible for tumor aggressiveness and recurrence, and new therapies targeting these CRC cancer stem cells (CRC-CSCs) may remarkably improve clinical treatment of CRC.1–4
In recent years, more advanced genome analyses have revealed that more than 90% of the human genome is pervasively transcribed into noncoding RNAs (ncRNAs). In addition, long noncoding RNAs (lncRNAs) are the RNA molecules that are larger than 200 nt and that lack an open reading frame.5 Currently, numerous cancer-related lncRNAs have been discovered.6 Some lncRNAs have been found to be involved in stem cell properties like pluripotency and differentiation.7 Nevertheless, CSC-related lncRNAs have rarely been investigated.
A large number of lncRNA transcripts originate from transcription at promoters or other nearby location of protein-coding genes. These “lncRNA/messenger RNA (mRNA) gene pairs” interact with each other in expression and in their function, which is related to stem cell properties.8 Through bioinformatics analysis, we found a lncRNA TCONS_00004241 (data from USCS website), which is located at 2p21 on the upstream side of Hypoxia-inducible factor-2α (HIF-2α) promoter and is transcribed from the antisense direction of it. This lncRNA belongs to a class of ncRNAs called promoter upstream transcripts (PROMPTs).9,10 Therefore, we renamed it as the “lncRNA-HIF-2α promoter upstream transcript (HIF2PUT)”. The expression and function of PROMPTs are often associated with adjacent protein-coding transcripts.11,12
The function of HIF-2α, also named EPAS1, has been demonstrated to be in association with “stem cell-like” properties in stem cells and CSCs.13–15 Recently, we have already demonstrated that lncRNA-HIF2PUT is functionally associated with the CSC properties in osteosarcoma.16 Therefore, we speculated that lncRNA-HIF2PUT may also regulate the stem cell-like characteristics of CRC-CSCs by regulating HIF-2α. Based on these findings, in the current study, we investigated the expression pattern of the lncRNA/mRNA gene pairs of lncRNA-HIF2PUT and HIF-2α in 23 CRC specimens. The results showed that the expression of lncRNA-HIF2PUT was positively correlated with HIF-2α. In addition, in CRC cell lines DLD-1 and HT29, we found knockdown of lncRNA-HIF2PUT resulted in decreased HIF-2α expression as well as impaired CSC properties, including proliferation, self-renewal, migration, and invasion abilities.
Materials and methods
Patient samples
Patients with CRC who underwent initial surgery at the Chinese People’s Liberation Army (PLA) General Hospital, Beijing, from 2011 to 2013 were retrospectively selected for this study. No patient had received therapy before resection. The utilization of tumor material for research was approved by the ethical committee of the PLA General Hospital. Written informed consent was obtained from each participant prior to tumor samples collection.
Cancer cell lines
Human CRC cell lines SW620, SW480, HT29, HCT116, CACO2, DLD-1, LS174T, and RKO, and an epithelial cell line FHC, which was derived from normal human colon, were all purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). The SW620 and SW480 cells were cultured in Leibovitz’s L-15 Medium (Thermo Fisher Scientific, Waltham, MA, USA), FHC cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/F-12 Medium, and the other CRC cell lines were cultured in DMEM medium (Thermo Fisher Scientific), all containing 10% fetal bovine serum (Thermo Fisher Scientific) at 37°C with 5% CO2. Before CSC–related experiments, we enriched the CSCs in DLD-1 and HT29. The DLD-1 and HT29 cells were transferred into the stem cell culturing medium, the serum-free DMEM/F12 medium with 20 ng/mL human epidermal growth factor (Peprotech Inc, Rocky Hill, NJ, USA), 20 ng/mL human basic fibroblast growth factor (Peprotech Inc), and 1% N2 supplement (Thermo Fisher Scientific).
Real-time quantitative polymerase chain reaction
Total RNA was isolated from CRC tumor tissues, matched adjacent normal tissues, and CRC cells using TRIzol® Total RNA reagent (Thermo Fisher Scientific). Complementary DNA (cDNA) synthesis was performed with 2 μg total RNA using the RevertAid™ H Minus First Strand cDNA synthesis kit (Thermo Fisher Scientific). The primers were obtained from Sangon biotech (Shanghai, People’s Republic of China), and the sequences are shown in Table 1.
Table 1.
Primers for real-time PCR analysis
| Gene name | Forward | Reverse |
|---|---|---|
| β-Actin | 5′-CCACTGGCATCGTGATGGA-3′ | 5′-CGCTCGGTGAGGATCTTCAT-3′ |
| HIF2PUT | 5′-CGGAGGTGTTCTATGAGCTGG-3′ | 5′-AGCTTGTGTGTTCGCAGGAA-3′ |
| HIF-2α | 5′-TGGGATCTAACAGGAACAGC-3′ | 5′-CTAAATAGCCAGACAAGGGT-3′ |
| OCT4 | 5′-TATGCAAAGCAGAAACCCTCGTGC-3′ | 5′-TTCGGGCACTGCAGGAACAAATTC-3′ |
| SOX2 | 5′-GCCGAGTGGAAACTTTTGTCG-3′ | 5′-GGCAGCGTGTACTTATCCTTCT-3′ |
| NANOG | 5′-TCCAGCAGATGCAAGAACTCTCCA-3′ | 5′-CACACCATTGCTATTCTTCGGCCA-3′ |
| KLF4 | 5′-CCCACATGAAGCGACTTCCC-3′ | 5′-CAGGTCCAGGAGATCGTTGAA-3′ |
| CD44 | 5′-CTGCCGCTTTGCAGGTGTA-3′ | 5′-CATTGTGGGCAAGGTGCTATT-3′ |
| CD133 | 5′-AGTCGGAAACTGGCAGATAGC-3′ | 5′-GGTAGTGTTGTACTGGGCCAAT-3′ |
Abbreviations: PCR, polymerase chain reaction; HIF-2α, hypoxia-inducible factor-2 alpha.
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using the SYBR® PrimeScript™ RT-PCR kit (Thermo Fisher Scientific) in a 7500 Fluorescent qPCR System (Thermo Fisher Scientific). The reaction mixtures were incubated at 95°C for 30 seconds, followed by 40 amplification cycles of 95°C for 5 seconds and 60°C for 34 seconds. The comparative ΔCt method was used to quantify relative expression of mRNA and lncRNA. Expression level of housekeeping gene β-actin was used to normalize gene-of-interest expression. The expression level of a gene in a patient was calculated as the ratio of target in tumor tissue/target in nontumorous tissue (R [C/N]).
Western blot analysis
Cells were washed twice with ice-cold phosphate-buffered saline (PBS), and cell lysates were harvested by the addition of lysis buffer (40 nM Tris [pH 7.4], 150 mM NaCl, 10 mM ethylene diamine tetra-acetic acid, 10% glycerol, 1% Triton X-100, 10 mM glycerophosphate, 1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride) supplemented with protease inhibitor (Hoffman-La Roche Ltd, Basel, Switzerland). Exactly 30 µg of protein lysates were separated on a NuPAGE® 4%–12% Bis-Tris Gel (Thermo Fisher Scientific), and the separated proteins were transferred onto a polyvinylidene difluoride membrane (Thermo Fisher Scientific). After blocking for 60 minutes with 5% nonfat dry milk, membranes were incubated with the primary antibody overnight at 4°C, followed by incubation with the corresponding secondary antibody for 60 minutes at room temperature. The membranes were developed using enhanced chemiluminescence solutions (Thermo Fisher Scientific).
Transfection of small interfering RNA
For small interfering RNA (siRNA) analysis, siRNA for the lncRNA-HIF2PUT sequence and nontargeting siRNA were obtained from GenePharma (Shanghai, People’s Republic of China). The lncRNA-HIF2PUT sense strand was 5′-CAGCCAUCAUGAUGGUACU-3′, and the antisense strand was 5′-AGUACCAUCAUGAUGGCUG-3′. Approximately 5% DLD-1 and HT29 cells were plated to each well of 12-well plates for at least 24 hours before transfection to achieve 30%–50% confluency. SiRNA transfection was done with X-tremeGENE™ transfection reagent (Hoffman-La Roche Ltd) according to the manufacturer’s instructions. Cells were collected after transfection for RNA isolation, cell clonogenic survival assay, and spheroid formation assay.
Colony formation assay
Cells were trypsinized, counted, and then seeded at low density on six-well culture plates and were allowed to grow undisturbed at 37°C in 5% CO2 for 10 days; cells were then stained with crystal violet.
Spheroid formation assay
The capability of self-renewal was assessed using ultra-low attachment surface 96-well culture dishes (Corning Incorporated, Corning, NY, USA). Cells in the nonsense siRNA group or in the lncRNA-HIF2PUT siRNA group were resuspended in 200 μL serum-free medium DMEM/F12 with 20 ng/mL human epidermal growth factor, 20 ng/mL human basic fibroblast growth factor, and 1% N2 supplement at a density of 200 cells in each well. Phase-contrast images were obtained 7 days later.
Scratch wound healing assay
Uniform wounds were scraped in DLD-1 and HT29 epithelial monolayers grown on plastic six-well plates using a pipette tip before transfection. The initial gap length (0 hours) and the residual gap length of 24 hours after wounding were calculated from photomicrographs.
Matrigel invasion assays
A cell invasion assay was carried out using modified Boyden chambers consisting of transwell-precoated matrigel membrane filter inserts with 8 mm pores in 24-well tissue culture plates (BD Biosciences, San Jose, CA, USA). DMEM containing 10% fetal bovine serum in the lower chamber served as the chemoattractant.
Statistical analysis
Differences between groups were analyzed using the Student’s t-test. Correlation between gene expressions was studied by using Pearson’s correlation. Statistical analyses were performed using SPSS version 18.0 (SPSS Inc, Chicago, IL, USA). For all statistical analyses, P<0.05 was considered statistically significant.
Results
Expression of lncRNA-HIF2PUT was correlated with HIF-2α in CRC tissue
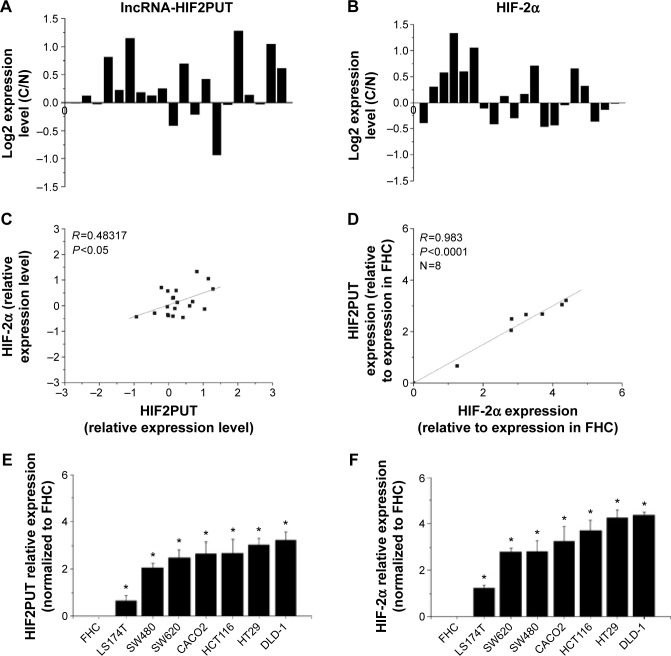
LncRNA-HIF2PUT and HIF-2α expression levels were assessed in a group of 20 patients with CRC. For each patient, lncRNA was isolated from cancerous tissues and adjacent nontumorous CRC tissues. The results showed that the expression of lncRNA-HIF2PUT was significantly correlated with that of HIF-2α in CRC tissue samples (R=0.5793, P<0.0001; Figure 1A–C). LncRNA-HIF2PUT and HIF-2α expression were also investigated in a normal colon epithelial cell line FHC and seven different CRC cell lines. The results revealed that the expression of lncRNA-HIF2PUT was coincident with that of HIF-2α in all eight cell lines (R=0.983, P<0.0001; Figure 1D, E). Moreover, expression of both lncRNA-HIF2PUT and HIF-2α were significantly higher in CRC cell lines than in FHC (Figure 1E). Also, the expression of both lncRNA-HIF2PUT and HIF-2α was higher in DLD-1 and HT29 cells than the other cell lines (Figure 1E). Therefore, we chose DLD-1 and HT29 as our candidate cell lines for a lncRNA-HIF2PUT knockdown experiment.
Figure 1.
IncRNA-HIF2PUT and HIF-2α expression levels were analyzed by real-time PCR in CRC tissue samples and in eight cell lines.
Notes: (A) The expression levels of lncRNA-HIF2PUT and (B) HIF-2α were calculated as the ratio of targets in cancerous tissue/targets in adjacent normal tissue (R [C/N]). (C) The expression of lncRNA-HIF2PUT was significantly correlated with that of HIF-2α (P<0.0001). (D) The expression of lncRNA-HIF2PUT was coincident with that of HIF-2α in the eight cell lines (P<0.0001). (E and F) The expression of both lncRNA-HIF2PUT and HIF-2α were significantly higher in each of the seven CRC cell lines than in the normal colon epithelial cell line FHC. The expression of lncRNA-HIF2PUT and HIF-2α was highest in DLD-1 and HT29 cells. *P<0.05.
Abbreviations: lncRNA-HIF2PUT, long noncoding RNA that is a promoter upstream transcript (PROMPT) of hypoxia-inducible factor-2α; HIF-2α, hypoxia-inducible factor-2 alpha; CRC, colorectal cancer; PCR, polymerase chain reaction.
Knockdown of lncRNA-HIF2PUT downregulated HIF-2α expression
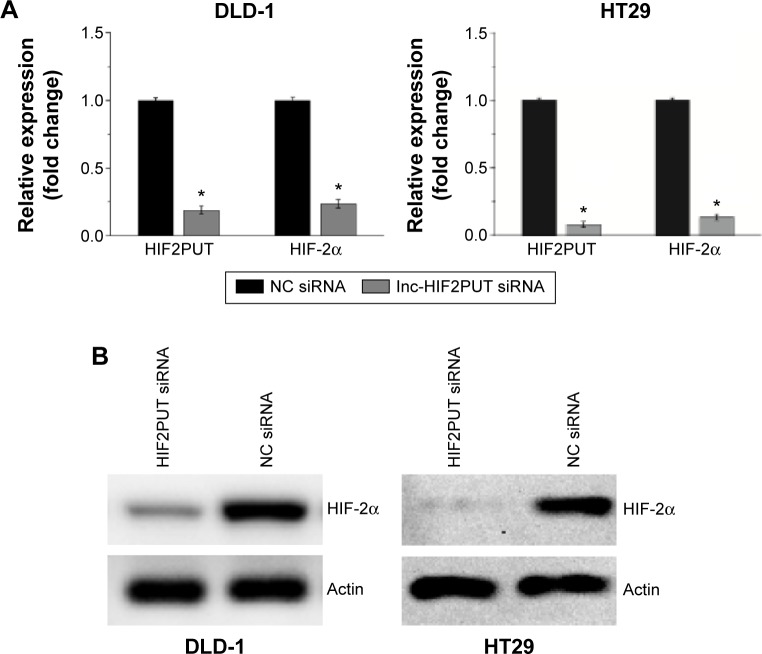
LncRNA-HIF2PUT siRNA was transfected into DLD-1 and HT29. Quantification analysis showed that lncRNA-HIF2PUT expression level in the lncRNA-HIF2PUT siRNA group was significantly knocked down in DLD-1 and HT29 cells (Figure 2A). Using RT-qPCR and Western blots, we showed that HIF-2α expression was also markedly decreased in both mRNA level and protein level (Figure 2A, B).
Figure 2.
Knockdown of lncRNA-HIF2PUT inhibited the expression of HIF-2α.
Notes: (A) Quantification analysis by real-time PCR showed that the expression levels of both lncRNA-HIF2PUT and HIF-2α in lncRNA-HIF2PUT siRNA groups were significantly knocked down in DLD-1 and HT29 cell lines. (B) Western blot showed that the HIF-2α protein expression was also markedly decreased. *P<0.05.
Abbreviations: lncRNA-HIF2PUT, long noncoding RNA that is a promoter upstream transcript (PROMPT) of hypoxia-inducible factor-2α; HIF-2α, hypoxia-inducible factor-2 alpha; PCR, polymerase chain reaction; siRNA, small interfering RNA; NC, noncancerous.
CSC properties were impaired by lncRNA-HIF2PUT knockdown
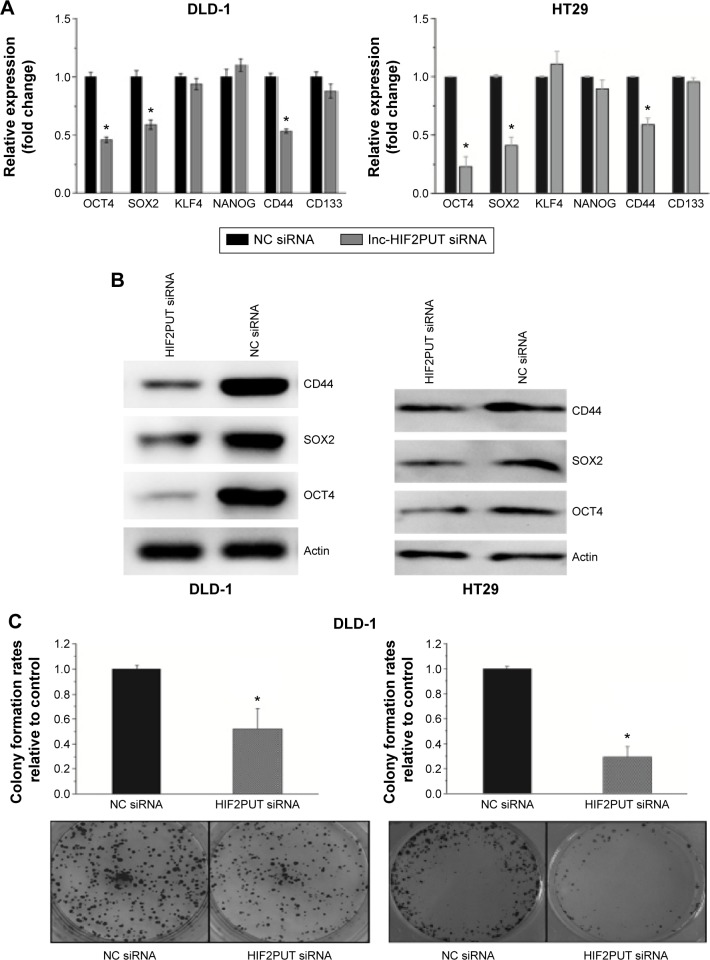
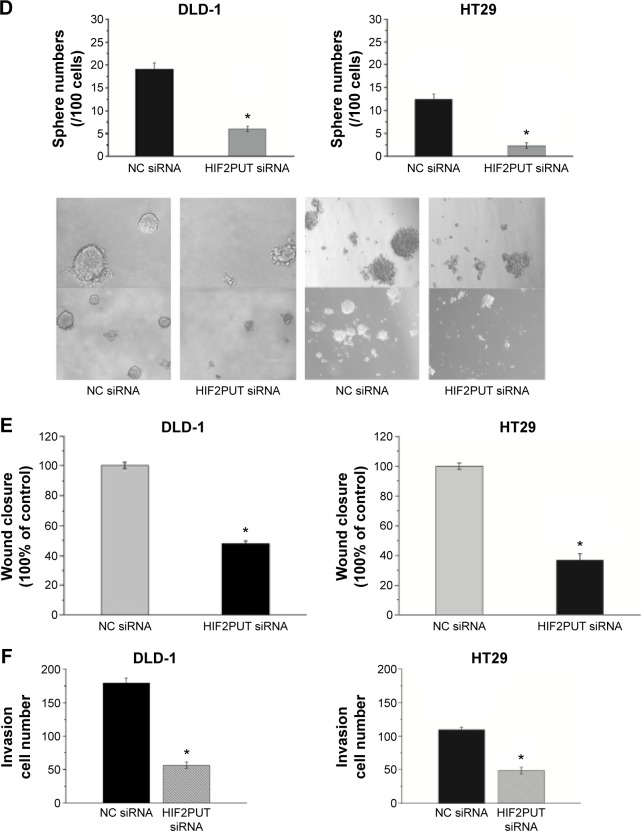
Next, we determined the effects of lncRNA-HIF2PUT knockdown on stem cell-like properties in DLD-1 and HT29 cells in vitro. After lncRNA-HIF2PUT siRNA treatment, the stemness genes OCT4, SOX2, and CD44 decreased significantly (Figure 3A, B), which was coincident with decreased colony formation rates in medium containing fetal bovine serum, and spheroid formation ability in serum-free medium (Figure 3C, D). To further identify the functional role of lncRNA-HIF2PUT RNA in cell migration and invasion, the scratch wound healing assay and matrigel invasion assay were performed in DLD-1 and HT29 cells in vitro. The wound healing assay showed remarkable cell migration inhibition in DLD-1 and HT29 siRNA groups compared with the cells in the noncancerous group (Figure 3E). The matrigel invasion assay also showed significant cell invasion inhibition in the lncRNA-HIF2PUT siRNA group compared with the noncancerous groups in the DLD-1 and HT29 cell lines (Figure 3F).
Figure 3.
Knockdown of lncRNA-HIF2PUT impairs CSC properties.
Notes: (A) HIF2PUT lncRNA level and six stemness genes’ mRNA levels were measured by real-time PCR in DLD-1 and HT29 cells transfected with lncRNA-HIF2PUT siRNA and negative control siRNA. The expression of OCT4, SOX2, and CD44 was significantly downregulated. (B) Western blot showed that the expression of OCT4, SOX2, and CD44 protein was also significantly reduced in lncRNA-HIF2PUT siRNA groups compared with negative control groups. (C) Colony formation assay showed that CSC proliferation was retarded by lncRNA-HIF2PUT siRNA (P<0.05). (D) Spheroid formation assay showed that the self-renewal capacity in lncRNA-HIF2PUT siRNA groups was remarkably impaired. (E) The wound healing assay showed remarkable cell migration inhibition by lncRNA-HIF2PUT siRNA. (F) The matrigel invasion assay also showed significant cell invasion inhibition by lncRNA-HIF2PUT siRNA. *P<0.05.
Abbreviations: lncRNA-HIF2PUT, long noncoding RNA that is a promoter upstream transcript (PROMPT) of hypoxia-inducible factor-2α; HIF-2α, hypoxia-inducible factor-2 alpha; PCR, polymerase chain reaction; siRNA, small interfering RNA; NC, noncancerous; mRNA, messenger RNA; CSC, cancer stem cell.
Discussion
Hypoxia in the stem cell niche is very important in maintaining the undifferentiated phenotype of normal stem cells and CSCs.17,18 HIF-2α is a pivotal hypoxia-activated factor in this process, which can interact with multiple CSC-related pathways, including c-Myc and OCT4, and plays a role in regulating stem cell proliferation, differentiation, and pluripotency.14,15,19,20
Li et al demonstrated that in glioma stem cells, HIF-2α protein may be preferentially expressed under both normoxic and hypoxic conditions.13 Interestingly, we also detected substantial HIF-2α protein expression in CSC-enriched CRC cells, even under normoxic conditions. A recent study revealed that transcription of most lncRNA genes is coordinated with transcription of adjacent protein-coding genes.8 HIF-2α, together with its PROMPT, which we named HIF2PUT, is exactly such a lncRNA/mRNA pair.
In the current study, we first investigated the expression of this lncRNA/mRNA pair in 20 cases of CRC specimens; we found that the expressions of HIF-2α and lncRNA- HIF2PUT were positively correlated with each other in tumor tissues and adjacent normal tissues of CRC. Also, in eight different cell lines, the expressions of HIF-2α and lncRNA-HIF2PUT were coincident with each other, and both were elevated in CRC cell lines compared with the normal colon epithelial cell line FHC.
Accordingly, when the expression of lncRNA-HIF2PUT was knocked down by siRNA, the expression of HIF-2α was significantly suppressed. These results indicated that lncRNA-HIF2PUT may regulate the expression of HIF-2α in CRC.
Moreover, before this study, the function of lncRNA had rarely been linked with CSC properties, including the expression of stemness genes, self-renewal, proliferation, migration, and invasion abilities. Herein, we showed that lncRNA-HIF2PUT was functionally associated with CSC properties. Knockdown of lncRNA-HIF2PUT by siRNA resulted in significantly decreased expression of OCT4, SOX2, and CD44, and impaired colony formation ability, spheroid formation ability, and migration and invasion abilities in vitro.
Taken together, these results indicate that lncRNA- HIF2PUT is likely to regulate the characteristics of CRC-CSCs by regulating HIF-2α expression. Further work targeting this new lncRNA in CRC will be very useful for the cancer clinic in future.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8(5):511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi S, Kamiyama T, Tomaru U, et al. Frequency and pattern of expression of the stem cell marker CD133 have strong prognostic effect on the surgical outcome of colorectal cancer patients. Oncol Rep. 2010;24(5):1201–1212. doi: 10.3892/or_00000973. [DOI] [PubMed] [Google Scholar]
- 4.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18(5):460–406. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sigova AA, Mullen AC, Molinie B, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110(8):2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preker P, Nielsen J, Kammler S, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 10.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 11.Preker P, Almvig K, Christensen MS, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39(16):7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8(15):2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11(4):335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covello KL, Kehler J, Yu H, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20(5):557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Yao J, Meng H, et al. A novel long non-coding RNA, hypoxia-inducible factor-2α promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. 2015;11(4):2534–2540. doi: 10.3892/mmr.2014.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang CP, Zhu LL, Zhao T, et al. Characteristics of neural stem cells expanded in lowered oxygen and the potential role of hypoxia-inducible factor-1Alpha. Neurosignals. 15(5):2006–2007. 259–265. doi: 10.1159/000103385. [DOI] [PubMed] [Google Scholar]
- 18.McCord AM, Jamal M, Shankavaram UT, Lang FF, Camphausen K, Tofilon PJ. Physiologic oxygen concentration enhances the stem-like properties of CD133+ human glioblastoma cells in vitro. Mol Cancer Res. 2009;7(4):489–497. doi: 10.1158/1541-7786.MCR-08-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazumdar J, Dondeti V, Simon MC. Hypoxia-inducible factors in stem cells and cancer. J Cell Mol Med. 2009;13(11–12):4319–4328. doi: 10.1111/j.1582-4934.2009.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathieu J, Zhang Z, Zhou W, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71(13):4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]