Abstract
Disruption of SMIM1, encoding Small Integral Membrane Protein 1, is responsible for the Vel-negative blood type, a rare but clinically-important blood type. However, the exact nature of the Vel antigen and how it is presented by SMIM1 are poorly understood. Using mass spectrometry we found several sites of phosphorylation in the N-terminal region of SMIM1 and we found the initiating methionine of SMIM1 to be acetylated. Flow cytometry analyses of human erythroleukemia cells expressing N- or C-terminally Flag-tagged SMIM1, several point mutants of SMIM1, and a chimeric molecule between Kell and SMIM1 demonstrated that SMIM1 carries the Vel antigen as a type II membrane protein with a predicted C-terminal extracellular domain of only 3 to 12 amino acids.
Keywords: type II membrane protein, SMIM, blood group, mass spectrometry, phosphorylation
1. Introduction
Small integral membrane proteins (SMIMs) constitute a collection of currently 24 unrelated proteins with a single predicted transmembrane domain and a low theoretical molecular mass (6.9 to 18.4 kDa). The genes encoding SMIMs were recently included in the annotation of the human genome by the Human Gene Organization (HUGO) given experimental evidence emerged for the existence of their encoded proteins, primarily by mass spectrometry. Outside of the field of peptide hormone research, very small proteins encoded by the genome have been rather neglected, with most SMIMs remaining as uncharacterized proteins. The subcellular location of SMIMs and their orientation in the membrane currently rely on prediction tools that are unfortunately not as efficient as those that predict the presence of a transmembrane domain.
The SMIM1 gene, previously called LOC388588, encodes a 78 amino acid protein that was recently found by our and two other research teams to carry the long-sought-after Vel blood group antigen [1–4]. Using a biochemical approach we purified the carrier of the Vel antigen from red cell membranes, and using mass spectrometry identified it to be the hypothetical protein LOC588388 a few weeks before it was renamed SMIM1 by the HUGO Gene Nomenclature Committee. Analyzing a cohort of 70 Vel-negative individuals we found that the Vel-negative blood type was associated almost exclusively with a homozygous deletion of 17 nucleotides causing a frameshift in the SMIM1 coding sequence (c.64_80del17fs) [1]. Coincidentally the same conclusion was reached at about the same time by Storry et al. [2] and Cvejic et al. [3] using genetic approaches. Together, these studies established SMIM1 as responsible for a new blood group system that was named VEL by the International Society of Blood Transfusion (ISBT). The Vel-negative blood type (officially named VEL:-1) is now reliably typed in reference immunohematology laboratories by checking for the homozygosity of the 17 nucleotide deletion in SMIM1. Still, only a few features regarding the biochemistry of SMIM1 and how it might present the Vel antigen have been revealed.
First, SMIM1 can form oxidation-dependent multimers. During the characterization and purification of the Vel antigen carrier from red cell membranes, we found that it did not run at the theoretical molecular mass of SMIM1 (8.7 kDa) during SDS-PAGE. Rather, the Vel antigen carrier behaved as a ~18 kDa protein under reducing conditions and primarily as a ~32 kDa protein under non-reducing conditions [1]. Storry et al. [2] also reported oxidation-dependent multimerization of SMIM1. Furthermore, we found the multimerization of SMIM1 was reversible as the reduced ~18 kDa form could once again run at ~32 kDa when the reducing agent was removed [1]. These data suggest that SMIM1 can form multimers via disulfide bonds. SMIM1 contains two cysteines in the N-terminal half (Cys35 and Cys43) and one cysteine near its C-terminus (Cys77). As noted by Storry et al. [2], oxidation-independent dimerization might also be mediated by a GxxxG motif present toward the C-terminus of SMIM1 (Gly67-x-x-x-Gly71) [5,6].
Second, SMIM1 appears not to contain a canonical signal sequence. The identification of the Vel antigen carrier by mass spectrometry revealed a peptide harboring the initiating methionine of SMIM1 [1], suggesting that SMIM1 was likely a type II membrane protein with no cleavable signal peptide. Single-spanning membrane proteins of type II display their N-terminus on the cytoplasmic side of the membrane in contrast to the more common type I membrane proteins. However, computational analyses give variable predictions regarding the membrane orientation of SMIM1 as well as the exact location of its transmembrane domain (Supplementary Fig. 1), and currently the Universal Protein Resource (UniProt) lists SMIM1 as a type I membrane protein with an N-terminal extracellular domain (residues 1–46) and a C-terminal cytoplasmic domain (residues 68–78). The type I orientation was also favored by Storry et al. [2] who developed rabbit antibodies against peptides from the N-terminal domain of SMIM1 and found these antibodies can detect SMIM1 in western blot analyses but fail to agglutinate Vel-positive red cells.
Given that resolving the orientation of SMIM1 in the plasma membrane is critical to understanding its presentation of the Vel blood group antigen, we conducted a series of mass spectrometric, biochemical, mutational and cytological studies to this end. Here we provide strong evidence that the Vel antigen is harbored on the short, extracellular C-terminus of SMIM1.
2. Materials and Methods
2.1 Protein purification, in-gel digestion, mass spectrometry and data analysis
Biochemical purification of the carrier of the Vel antigen using double SDS-PAGE under non-reducing and reducing conditions; in-gel digestions; mass spectrometry; and mass spectrometry data analysis were all performed as previously described [1] except that during the mass spectrometry analysis searches allowed differential modification of serine, threonine and tyrosine (79.9663 Da for phosphorylation); N-termini (42.0106 Da for acetylation); cysteine (57.0215 Da for carboxyamidomethylation) and methionine (15.9949 Da for oxidation).
2.2 Plasmids and site-directed mutagenesis
The expression construct for SMIM1 with a Flag tag at the N- or C-terminus was obtained by subcloning SMIM1 coding sequence into the p3xFLAG-CMV-10 and p3xFLAG-CMV-14 vectors from Sigma (St. Louis, MO, USA) respectively. The expression constructs for SMIM1 variants were made in the pCEP5 episomal vector as previously described for wild type SMIM1 [1]. The SMIM1 variants S4A (Ser6Ala, Ser17Ala, Ser22Ala and Ser27Ala) and C2S (Cys35 and Cys43), and the KELVEL chimera (Kell (1–47)-SMIM1(44–78)) were synthesized by Eurofins MWG Operon (Ebersberg, Germany). The Val74Met variation that corresponds to the minor A allele of rs373895822 (c.220G>A) was constructed by PCR-amplifying a SnaBI/BamHI fragment of the p3xFLAG-CMV-10-SMIM1 plasmid with a CMVp forward primer (5’- ACTTGGCAGTACATCTACG) and a mutagenic reverse primer (5’- CAGGATCCTTATTTGCACTTGTGCATATAG) and by subcloning it in the pCEP5 vector. All plasmids were sequence-verified by GATC Biotech (Konstanz, Germany).
2.3 293 cell culture, transfection and immunofluorescence
E1A-transformed 293 cells were maintained in Dulbecco’s modified Eagle’s medium (Mediatech, Manassas, VA, USA) supplemented with 5% Fetal Bovine Serum (Hyclone, Logan, UT, USA), 5% Cosmic Calf Serum (Hyclone), 50 units/ml of penicillin and 50 µg/ml of streptomycin (Life Technologies, Carlsbad, CA, USA).
Six cm culture dishes of cells were transfected at 70% of confluence with 0.25 µg of either WT or S4A Flag-tagged SMIM1 using calcium phosphate precipitation. 24 hours after transfection, cells from each dish were then trypsinized and resuspended in 8 mL of growth media, of which 1.5 mL was added onto glass coverslips in 6-well plates which had been pretreated for two minutes with 1 mg/mL poly-L-lysine (Sigma) in phosphate-buffered saline solution (PBS) and then with 3 mg/mL rat tail collagen (Sigma) in PBS until plating (~20 minutes). 24 hours after plating, the cells on each coverslip were fixed in 2 mL of 3.7% paraformaldehyde in PBS for 20 minutes, washed with PBS and then permeabilized in 2 mL of 0.5% Triton X-100 in PBS for 10 minutes. After permeabilization the samples were washed with PBS and then blocked using 1.5% bovine serum albumin (BSA) in PBS for 45 minutes at 4 °C. After a PBS wash the primary antibody incubation occurred while rocking overnight in a humidified container at 4 °C using a 1:5,000 dilution of anti-Flag M2 antibody (Sigma) in 2% BSA in PBS. After a ten minute wash with cold PBS while rocking at 4 °C the secondary antibody incubation occurred while rocking for 1 hour at 4 °C using a 1:10,000 dilution of Goat anti-Mouse Alexa Fluor 488 (Molecular Probes, Life Technologies) in 2% BSA in PBS. The final wash was for ten minutes with cold PBS while rocking at 4 °C. The coverslips were then mounted using hard set DAPI mounting media (Vectashield, Vector Labs, Burlingame, CA, USA), which was allowed to cure overnight at 4 °C.
Imaging was done using a DeltaVision fluorescent microscope (GE Healthcare Bio-Sciences, Pittsburg, PA, USA) with a 100× objective at 1,024×1,024 resolution. Image analysis was done using ImageJ v.1.4.8 (National Institutes of Health, USA), with antibody (green) channels standardized across images based on the mock control. The Find Edges tool of ImageJ was used to generate images with highlighted edges in the antibody channel.
2.3 Cell lysis, immunoprecipitation, phosphatase assay and immunoblotting
E1A-transformed 293 cells transfected with Flag-SMIM1 wild type or Flag-SMIM1 S4A were lysed in 25 mM Tris pH 7.2, 137 mM NaCl, 10% glycerol, 1% NP-40 (Igepal), 25 mM NaF, 10 mM Na4P2O7, 1 mM Na3VO4, 1 mM PMSF, 10 ug/mL leupeptin, and 10 ug/mL pepstatin A. 500 µg of protein extract in 1 mL of lysis buffer was incubated with 20 µL of a 50% slurry of anti-Flag M2 resin (Sigma) that had been washed with lysis buffer. After rocking overnight, the immune complexes were washed thrice in lysis buffer followed by a single wash in PBS. Each immune complex was then resuspended in 500 µL of calf intestinal phosphatase (CIP) buffer (50 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol), which was divided into two tubes each receiving 200 µL. Each tube was then spun, aspirated, and resuspended in 50 µL of CIP buffer. To the plus CIP tubes, 25 U of CIP (New England Biolabs, Beverly, MA, USA) was added. Each tube was incubated for one hour at 37 °C, the supernatant aspirated off, and 40 µL of protein sample buffer (125 mM Tris pH 6.8, 9.4% glycerol, 2% SDS, 5% β-mercaptoethanol) were added. The samples were boiled at 95 °C for 5 minutes, and 10 µL of each sample was loaded onto a 15% SDS-PAGE gel. Following transfer to 0.2 µm pore nitrocellulose membranes (GE Healthcare Bio-Sciences) immunoblotting was done using 10 mL of a 1:1,000 anti-Flag M2 (Sigma) dilution in 1.5% BSA in TBST (Tris-Buffered Saline with 0.05% Tween-20) overnight. This was followed by three TBST washes and then incubation in 20 mL of a 1:5,000 dilution of horse radish peroxidase-conjugated goat anti-mouse IgG (Chemicon, Milipore, Billerica, MA, USA) in TBST for two hours at room temperature. Following three TBST washes the blots were developed using enhanced chemiluminescence reagents (Pierce, Life Technologies, Thermo Scientific) and x-ray film. The CIP time course experiment was conducted in a similar fashion except that all samples received CIP and the shortest time point (0.3 minutes) was done on ice and was never placed at 37 °C.
2.4 K-562 cell culture and transfection
K-562 cell lines stably expressing Flag-SMIM1 or SMIM1-Flag were obtained by electroporation, neomycin selection and limiting dilution as previously described [7]. K-562 cells stably expressing SMIM1 variants were obtained by electroporation and hygromycin selection as previously described [8].
2.5 Flow cytometry analysis
For extracellular staining, native K-562 cells were resuspended in Dulbecco's PBS solution supplemented with 0.15% BSA and then incubated with anti-Flag M2 (1 µg/mL) or anti-Vel SpG213Dc3 (hybridoma supernatant diluted 1:11). The affinity isolated mouse monoclonal anti-Flag M2 was purchased from Sigma while the hybridoma supernatant of human monoclonal anti-Vel SpG213Dc3 was a gift of Franck Vérité (EFS Bretagne, Rennes, France). The monoclonal anti-Vel SpG213Dc3 was recently established from peripheral blood lymphocytes of a Vel-negative patient with serum antibody to Vel and was extensively characterized, especially its strict specificity to the Vel antigen by red cell agglutination tests and flow cytometry analysis (Vérité et al., manuscript in preparation). Labeling with anti-Vel and anti-Flag were revealed with goat F(ab’)2 anti-human IgG(H+L)-PE (1:100, Beckman Coulter, Brea, CA, USA) and goat F(ab’)2 anti-mouse IgG(H+L)-PE (1:100, Beckman Coulter) respectively, and immediately analyzed with a FACSCantoII flow cytometer (BD Bioscience, San Jose, CA) equipped with FACSDiva software (v. 6.1.2) (BD Bioscience). For intracellular staining, K-562 cells were fixed and permeabilized with the Inside Stain Kit from Miltenyi Biotec (Bergisch Gladbach, Germany) and then incubated with anti-Flag (1.25 µg/mL) that was revealed with goat F(ab’)2 anti-mouse IgG(H+L)-FITC (1:100, Beckman Coulter). Ten thousand K-562 cells were collected for each sample and data were post-analyzed with FlowJo software (v. 7.2.5) (TreeStar, Ashland, OR, USA).
3. Results and Discussion
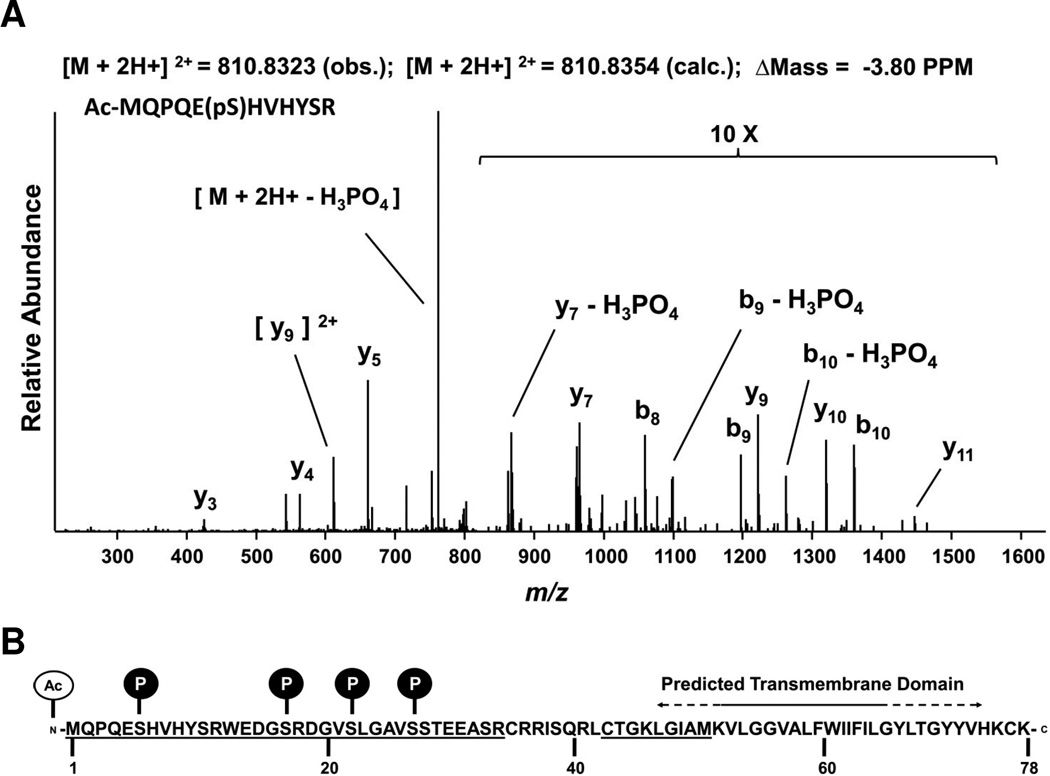
Our original mass spectrometry analyses of the Vel antigen carrier identified chymotryptic peptides that contained the extreme N-terminus of SMIM1. Here we report on a more in-depth mass spectrometry analysis of SMIM1 from red cells during which we identified several posttranslational modifications. These modifications are acetylation of the first methionine and phosphorylation of four serine residues (Ser6, Ser17, Ser22 and Ser27), all N-terminal to the predicted transmembrane domain (Fig. 1, Supplementary Table 1 and Supplementary Figs. 1–3). While a few secreted kinases have been recently documented [9–11], the vast majority of kinases function intracellularly. Therefore, our phosphorylation site identifications supported an orientation for SMIM1 in the membrane with an intracellular N-terminus.
Figure 1. Mass spectrometry-based identification of posttranslation modifications in the N-terminal region of SMIM1.
(A) Low energy CID tandem mass spectrum of a tryptic SMIM1 peptide harboring an acetylated N-terminus (Ac-M) as well as phosphorylation of serine (pS) at position 6. Metrics for the MS analysis include the observed and calculated precursor m/z of the doubly charged ion and the difference in PPM. Intensities of ion in the m/z range of 800–1,600 were raised ten-fold for better visualization. Several ions, including the precursor, show losses of phosphoric acid (H3PO4) characteristic CID spectra of phosphoserine/threonine peptide ion precursors. (B) Visual summary of the SMIM1 posttranslational modifications identified by MS. Underlined are the peptide regions covered by the MS analysis. The predicted transmembrane domain is indicated with a solid line when predicted by all tested bioinformatics sources and with a dashed line when predicted by at least one but not all tested bioinformatics programs. See Supplementary Table 1 and Supplementary Figs. 1–4 for additional information.
Given phosphorylation plays important roles in the trafficking of numerous proteins to the plasma membrane, and given that the expression of SMIM1 at the cell surface is essential for displaying the Vel antigen, we asked if the four identified phosphorylation sites were critical for cell surface expression of SMIM1. We synthetized a SMIM1 construct that harbors the four identified phosphorylation sites mutated to alanine (Ser6Ala, Ser17Ala, Ser22Ala and Ser27Ala, called S4A hereafter) and an N-terminal Flag tag to study its subcellular localization when expressed in human embryonic kidney 293 cells upon transient transfection. We saw no difference in the membrane localization of wild type SMIM1 when compared to the S4A mutant (Fig. 2A–B). Orthogonal to the mass spectrometry results characterizing SMIM1 from red cells, we found that the majority of wild type SMIM1 expressed in 293 cells was phosphorylated on multiple residues as calf intestinal phosphatase (CIP) caused wild type SMIM1 to increase in mobility during SDS-PAGE to the mobility of the S4A mutant (Fig. 2C–D).
Figure 2. Phosphorylation of SMIM1 leads to differential motility in SDS-PAGE but does not affect subcellular localization.
(A) Immunofluorescence-based imaging of Flag-SMIM1 WT shows a tendency for localization to the plasma membrane. Panels show SMIM1 localization (anti-Flag, green channel) with DAPI-stained (blue) nuclei (left panel), SMIM1 localization alone (center panel), or SMIM1 localization with edge detection from ImageJ (right panel). Edge detection (right panel) suggests potential subcellular localization at the membrane as well as in membrane-bound vesicles. Immunofluorescence of Flag-SMIM1 S4A (B) shows that subcellular localization is not dependent on phosphorylation at the four mutated serines. (C) Flag-SMIM1 WT shows distinct banding in SDS-PAGE that increases in motility upon treatment with calf intestinal phosphatase (CIP), while Flag-SMIM1 S4A produces a single band that does not change upon CIP treatment. (D) A time course of CIP treatment shows a progressive shift of higher bands toward the lowest molecular band in WT, while no change is observed in the S4A.
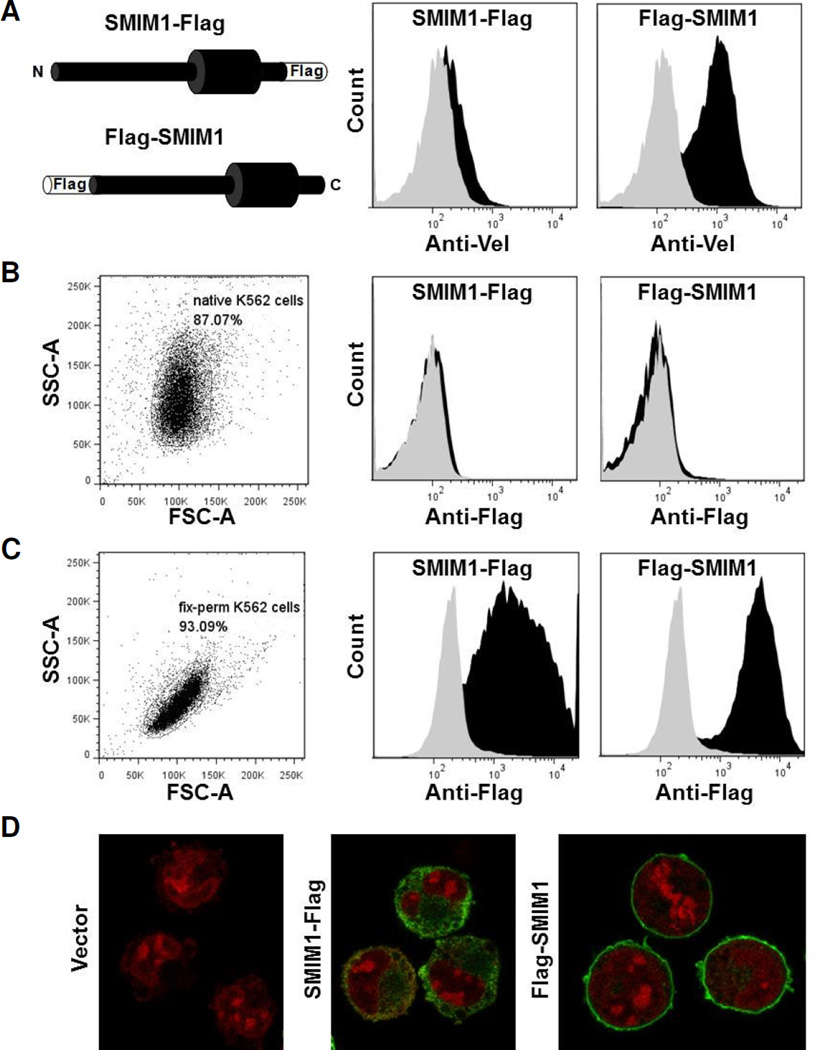
We next set out to determine the orientation of SMIM1 in the plasma membrane. Towards this goal we established human erythroleukemia K-562 cells that stably express SMIM1 with a Flag tag fused to either its N- or C-terminus (Flag-SMIM1 or SMIM1-Flag) and we analyzed them by flow cytometry with anti-Vel (Fig. 3A). Exogenous expression of untagged SMIM1 was previously shown to drive Vel antigen expression at the surface of K-562 cells in a fashion similar to how endogenous SMIM1 drives Vel antigen expression on red cells [1,2]. We found that only the expression of Flag-SMIM1 was associated with strong anti-Vel reactivity by flow cytometry analysis (Fig. 3A, right panel), suggesting that the Flag tag impairs the targeting of SMIM1-Flag to the plasma membrane. Alternatively, SMIM1-Flag might become expressed at the cell surface but the C-terminal tag could hinder the creation of the anti-Vel epitope or the binding of anti-Vel to SMIM1.
Figure 3. Flow cytometry and microscopy analysis of N- and C-terminal Flag-tagged SMIM1 stably expressed in K-562 cells.
(A) Expression constructs for SMIM1 with a Flag tag either at the N- or C-terminus were made as illustrated by the left panel and stably transfected in K-562 cells. Flow cytometry profiles of native cells expressing Flag-SMIM1 or SMIM1-Flag (both in black) and cells stably transfected with the empty vector (grey) that were labeled with anti-Vel (right and middle panels). (B) Flow cytometry profiles of native cells expressing Flag- SMIM1 or SMIM1-Flag (both in black) and cells stably transfected with the empty vector (grey) that were labeled with anti-Flag (right and middle panels). The left panel displays the SSCA/ FSC-A scatter plot of native K-562 cells. (C) Flow cytometry profiles of fixed and permeablized cells expressing Flag-SMIM1 or SMIM1-Flag (both in black) and cells stably transfected with the empty vector (grey) that were labeled with anti-Flag (right and middle panels). The left panel displays the SSC-A/FSC-A scatter plot of fixed and permeabilized K-562 cells. (D) Immunofluorescence microscopy of fixed and permabilized cells expressing Flag-SMIM1 (right), cells expressing SMIM1-Flag (middle) and cells stably transfected with the empty vector (left) that were labeled with anti-Flag (green) and propidium iodide (red).
We therefore analyzed both K-562 stable lines by flow cytometry for the presence of the Flag tag. No anti-Flag reactivity was detected on native K-562 cells expressing SMIM1-Flag (Fig. 3B, middle panel), or on K-562 cells expressing Flag-SMIM1 (Fig. 3B, right panel), suggesting that the Flag tag was intracellular in both cases. This was confirmed by flow cytometry analysis of fixed and permeabilized cells with anti-Flag (Fig. 3C). A similar analysis could not be performed with anti-Vel as it loses its specificity upon fixation and permeabilization of K-562 cells. Consistent with the anti-Flag flow cytometry results of fixed and permeabilized cells, immunofluorescence analysis of such cells with anti-Flag showed that Flag-SMIM1 was robustly expressed at the plasma membrane of stably transfected K-562 cells (Fig. 3D, right panel). In contrast, SMIM1-Flag was found to be cytoplasmic (Fig. 3D, middle panel), consistent with the C-terminal Flag tag impairing the targeting of SMIM1 to the plasma membrane. Together these data suggest that Flag-SMIM1 is properly localized and inserted in the plasma membrane with the N-terminus on the cytoplasmic side, whereas SMIM1-Flag could not reach the plasma membrane. Furthermore, these data support the hypothesis that SMIM1 is a type II membrane protein with an extracellular C-terminus.
With our experimental data indicating that SMIM1 was a type II membrane protein, it follows that the Vel antigen should be carried by the C-terminal domain of SMIM1. To demonstrate this, we fused the SMIM1 transmembrane and C-terminal domain to the N-terminal cytoplasmic domain of Kell which is a known type II membrane protein, most famous for carrying on its C-terminal domain the blood group antigens of the KEL system. Kell is a 732 amino acid glycoprotein and possesses a short N-terminal domain (residues 1–47) upstream of the transmembrane domain, which is typical of type II membrane proteins [12]. Kell and SMIM1 share other hallmarks of type II membrane proteins including the absence of a cleavable signal peptide and the presence of positively charged residues adjacent to the transmembrane domain on the N-terminal side [13]. We therefore created a chimera between Kell (residues 1–47) and SMIM1 (residues 44–78), which we called KELVEL in reference to the KEL and VEL blood group systems. As shown in Fig. 4A–B, the expression of the KELVEL chimera drives the cell surface expression of the Vel antigen in K-562 cells comparable to wild type SMIM1. This unambiguously demonstrates that the Vel antigen is carried by the C-terminal domain of SMIM1 as its N-terminal domain is totally absent from the KELVEL chimera.
Figure 4. Flow cytometry analysis of the Vel antigen in native K-562 cells stably expressing variants of SMIM1.
The variants used are described on the left, and the flow cytometry profiles of native cells expressing those variants and labeled with anti-Vel are shown on the right. (A) KELVEL chimera corresponding to a fusion between the 47 N-terminal residues of Kell and the 35 C-terminal residues of SMIM1. (B) Wild type SMIM1. (C) ΔKCK mutant of SMIM1. (D) S4A mutant of SMIM1. (E) C2S mutant of SMIM1.
Not only did the KELVEL chimera establish that the C-terminal domain of SMIM1 carries the Vel antigen but it also suggested that the N-terminal cytoplasmic domain of SMIM1 per se is not essential for the expression of the Vel antigen on the cell surface. In contrast, its C-terminus appears to be important in expression of the Vel antigen in some manner as a mutant of SMIM1 lacking the final three amino acids (Lys76Ter, called ΔKCK) does not lead to detectable cell-surface expression of the Vel antigen in K-562 cells (Fig. 4C). The mechanism by which the C-terminal amino acids are essential in this regard could be that their loss might destabilize the SMIM1 protein, might prevent the proper membrane targeting of SMIM1, or might ablate the anti-Vel epitope on the C-terminal domain of SMIM1. Consistent with the N-terminal domain being non-essential for the extracellular exposure of the Vel antigen, both S4A and C2S mutants, which harbor four mutated serines (Ser6Ala, Ser17Ala, Ser22Ala and Ser27Ala) and two mutated cysteines (Cys35 and Cys43) respectively in the N-terminal domain, show cell surface expression of the Vel antigen in K-562 cells (Fig. 4D–E) like wild type SMIM1 (Fig. 4B).
As a type II membrane protein, SMIM1 would have only a very short extracellular domain of 3 to 12 residues depending on the prediction program (Fig. 1B, Supplementary Fig. 1). Therefore we decided to study the effect of rs373895822 on the expression of the Vel antigen as the minor A allele of this SNP in SMIM1 (p.Val74Met) is the only non-synonymous SNP that has been reported in publicly available databases to affect the twelve C-terminal residues of SMIM1. The variation p.Val74Met appears to be very rare as it has been found only once in 8,136 “European (Non-Finnish)” individuals according to the Exome Aggregation Consortium (ExAC). If p.Val74Met abolished the expression of the Vel antigen on red cells, it would define a novel Vel-negative allele of SMIM1. Surprisingly, given the side chain differences, we did not detect any significant reduction in anti-Vel reactivity in K-562 cells stably expressing the V74M mutant of SMIM1 (Fig. 5A–B). We hence conclude that implementing rs373895822 in the genotyping of people for VEL is not required as the minor allele of rs373895822 is not another Vel-negative allele of SMIM1.
Figure 5. Analysis of the effect of the SNP rs373895822 (p.Val74Met) in SMIM1 on the expression of Vel in K-562 cells.
(A) Flow cytometry profiles of native cells expressing SMIM1 with Val at position 74 (black) and cells stably transfected with the empty vector (grey) that were labeled with anti-Vel. (B) Flow cytometry profiles of native cells expressing SMIM1 with Met at position 74 (black) and cells stably transfected with the empty vector (grey) that were labeled with anti-Vel.
In conclusion, all our experimental data argue for SMIM1 being a type II membrane protein that displays the Vel antigen on its C-terminus. This is important to not only the field of transfusion medicine, but also to the cellular biochemists studying SMIM proteins and to those studying generally the insertion of proteins into membranes.
Supplementary Material
Highlights.
The Vel blood group system antigen is carried on the C-terminus of SMIM1.
SMIM1 is a type II transmembrane protein with an extracellular C-terminus.
Mass Spectrometry identified SMIM1 as a phosphoprotein with an acetylated N-terminus.
Several SMIM1 point mutants are characterized with respect to their Vel antigen presentation.
Acknowledgements
We thank Carole Saison (INTS) for her help in molecular cloning and Franck Vérité (EFS Bretagne) for his gift of an aliquot of anti-Vel hybridoma supernatant. This work was supported by the Institut National de la Transfusion Sanguine (L.A., V.H., J.-P.C.), the University of Vermont via a REACH award from the Vice President of Research (B.A.B., L.P.K.), the Beckman Scholars Program (B.A.B., L.P.K.), the Vermont Genetics Network through U. S. National Institutes of Health Grant 8P20GM103449 from the INBRE program of the NIGMS (B.A.B., L.P.K.), and U.S. National Institutes of Health Grants 5 P20RR016435 and P20RR021905 (B.A.B., L.P.K.) from the COBRE program of the NIGMS.
Abbreviations
- SMIM
Small Integral Membrane Protein
- SDS-PAGE
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
- PCR
Polymerase Chain Reaction
- PBS
Phosphate-Buffered Saline
- BSA
Bovine Serum Albumin
- TBST
Tris-Buffered Saline Tween-20
- CIP
Calf Intestinal Phosphatase
- SNP
Single Nucleotide Polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
L.A., L.P.K., V.H. and B.A.B. conducted experiments and analyzed data. J.-P.C. provided scientific insights and laboratory oversight. L.A. and B.A.B. designed research and wrote the manuscript. All authors reviewed the manuscript and provided approval for submission.
The authors declare that they have no conflicts of interest, financial or otherwise, regarding the publication of this manuscript.
Supporting information
Supplementary Table 1. SMIM1 peptides identified by mass spectrometry following purification of the carrier of the Vel antigen.
Supplementary Figure 1. Comparisons of bioinformatic predictions of the membrane topology of SMIM1 in the plasma membrane.
Supplementary Figure 2. Low-energy CID tandem mass spectra showing acetylation of the Nterminus of SMIM1 and the phosphorylation of SMIM1 at serine 6.
Supplementary Figure 3. Low-energy CID tandem mass spectra showing phosphorylation of SMIM1 at serine residues 17, 22 and 27.
References
- 1.Ballif BA, et al. Disruption of SMIM1 causes the Vel- blood type. EMBO Mol Med. 2013;5:751–761. doi: 10.1002/emmm.201302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storry JR, Joud M, Christophersen MK, Thuresson B, Akerstrom B, Sojka BN, Nilsson B, Olsson ML. Homozygosity for a null allele of SMIM1 defines the Vel-negative blood group phenotype. Nat Genet. 2013;45:537–541. doi: 10.1038/ng.2600. [DOI] [PubMed] [Google Scholar]
- 3.Cvejic A, et al. SMIM1 underlies the Vel blood group and influences red blood cell traits. Nat Genet. 2013;45:542–545. doi: 10.1038/ng.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sussman LN, Miller EB. New blood factor: Vel. Rev Hematol. 1952;7:368–371. [PubMed] [Google Scholar]
- 5.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 6.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 7.Helias V, et al. ABCB6 is dispensable for erythropoiesis and specifies the new blood group system Langereis. Nat Genet. 2012;44:170–173. doi: 10.1038/ng.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saison C, et al. Null alleles of ABCG2 encoding the breast cancer resistance protein define the new blood group system Junior. Nat Genet. 2012;44:174–177. doi: 10.1038/ng.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagliabracci VS, Pinna LA, Dixon JE. Secreted protein kinases. Trends Biochem Sci. 2013;38:121–130. doi: 10.1016/j.tibs.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bordoli MR, et al. A secreted tyrosine kinase acts in the extracellular environment. Cell. 2014;158:1033–1044. doi: 10.1016/j.cell.2014.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tagliabracci VS, et al. A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell. 2015;161:1619–1632. doi: 10.1016/j.cell.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels G. Human Blood Groups. 3rd edition. Wiley-Blackwell; 2013. Kell and Kx Blood Group Systems; pp. 278–305. [Google Scholar]
- 13.Parks GD, Lamb RA. Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell. 1991;64:777–787. doi: 10.1016/0092-8674(91)90507-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.