Abstract
Ebola virus (EBOV) can lead to severe hemorrhagic fever with a high risk of death in humans and other primates. To guide treatment and prevent spread of the viral infection, a rapid and sensitive detection method is required for clinical samples. Here, we described and evaluated a reverse transcription loop-mediated isothermal amplification (RT-LAMP) method to detect Zaire ebolavirus using the nucleoprotein gene (NP) as a target sequence. Two different techniques were used, a calcein/Mn2+ complex chromogenic method and real-time turbidity monitoring. The RT-LAMP assay detected the NP target sequence with a limit of 4.56 copies/μL within 45 min under 61°C, a similar even or increase in sensitivity than that of real-time reverse transcription-polymerase chain reaction (RT-PCR). Additionally, all pseudoviral particles or non- Zaire EBOV genomes were negative for LAMP detection, indicating that the assay was highly specific for EBOV. To appraise the availability of the RT-LAMP method for use in clinical diagnosis of EBOV, of 417 blood or swab samples collected from patients with clinically suspected infections in Sierra Leone, 307 were identified for RT-LAMP-based surveillance of EBOV. Therefore, the highly specific and sensitive RT-LAMP method allows the rapid detection of EBOV, and is a suitable tool for clinical screening, diagnosis, and primary quarantine purposes.
Keywords: Zaire EBOV, RT-LAMP, sensitivity, specificity, rapid detection, prevalence
Introduction
The 2014 Ebola virus (EBOV) outbreak was the largest to date in West African countries (Frieden et al., 2014; Hampton, 2014). It spread through direct contact with infected individuals, or by touching the blood, organs, bodily secretions and fluids, or the contaminated clothes of such people (MacNeil and Rollin, 2012). Because the initial symptoms of ebolavirus infection can be confused with those of other febrile illnesses such as endemic malaria (Chertow et al., 2014), and because the infection cannot be detected rapidly in patients living in remote areas (World Health Organization [WHO], 2014)1, the numbers of infected people and deaths were the highest yet recorded. To control infection and to prevent further transmission during outbreaks of filoviruses such as EBOV, rapid detection is therefore essential (Grolla et al., 2005).
Current methods for the detection and diagnosis of EBOV infection include virus isolation, electron microscopy, immunohistochemistry (Zaki et al., 1999), enzyme-linked immunosorbent assay testing (Niikura et al., 2001), reverse transcription-polymerase chain reaction (RT-PCR), serologic testing for IgM/IgG virus-specific antibodies (Ksiazek et al., 1999a; Saijo et al., 2001), and point-of-care biosensors (Baca et al., 2015). In general, when the EBOV viral load in the blood gets to a higher case fatality rate, the detection of antigens as a suitable method is used for laboratory diagnosis (Fisher-Hoch et al., 1992; Ksiazek et al., 1999b). Thus, the World Health Organization recommends real-time RT-PCR as the first choice for EBOV diagnosis. However, inhibitors present in crude biological samples can inactivate the Taq DNA polymerase used in PCR-based techniques (de Franchis et al., 1988). Moreover, these methods are relatively complex and require specialized instruments. Thus, to complement PCR-based methods, another rapid, simple, and effective assay is needed.
Loop-mediated isothermal amplification (LAMP) is a one-step nucleic acid detection method developed by Notomi et al. (2000) which relies on autocycling strand displacement DNA synthesis. This novel method is highly specific and sensitive, takes advantage of four or six specific primers to recognize six or eight different sequences of the target gene, and is performed under isothermal conditions in less than 1 h using Bst DNA polymerase. Furthermore, LAMP is less influenced by inhibitors present in complex samples than standard PCR, which is highly beneficial for clinical specimens such as blood components, sputum, feces, or body fluids (Kaneko et al., 2007). LAMP assays have been widely applied to genetic diagnoses, the detection of epidemic bacteria (Hara-Kudo et al., 2005; Song et al., 2005) and viruses (Okafuji et al., 2005), fetal sex identification (Hirayama et al., 2013), and parasite recognition (Chen et al., 2011; Kong et al., 2012).
Kurosaki et al. (2007) developed a simple reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of Zaire ebolavirus, targeting the trailer region of the viral genome. However, this method has yet to be tested in clinical samples. The EBOV genome is approximately 19 kb, and encodes the following seven genes, which are flanked by untranslated regions: nucleoprotein (NP), viral structural protein (VSP)35, VSP40, glycoprotein, VP30, VP24, and RNA-dependent RNA polymerase (Ali and Islam, 2015).
NP is highly conserved among all EBOV species currently known, and plays an important role in intracellular events such as replication and transcription of the viral genome, and nucleocapsid formation (Ali and Islam, 2015). It is therefore recommended by the World Health Organization for use as a target gene for the RT-PCR assay.
In the present work, we developed a point-of-care RT-LAMP assay targeted to NP. Five sets of primers for the detection of EBOV were designed and used in optimization of the RT-LAMP assay. We also evaluated the specificity and sensitivity of the LAMP method. Finally, 417 blood samples collected from patients with clinically suspected infections were analyzed by RT-LAMP and RT-PCR in clinical diagnosis.
Materials and Methods
Viruses, RNA Extraction, and Preparation of Templates
Twenty-six genomes of respiratory pathogens including artificial RNAs of Sudan EBOV (Subtype Sudan, strain Gulu), Zaire EBOV and MARV, SARS coronavirus, influenza A H7N9, H1N1, H2N3, human parainfluenza viruses (PIV) type 1/2/3 and 4, adenoviruses (ADV; serotype 3, serotype 5, and serotype 55), respiratory syncytial virus infection RSVA/RSVB, MERS RNA, human metapneumovirus HMPV, human coronavirus HCoV-229E/ HCoV-OC43/HCoV-NL63, and HCoV-HKU1, bocavirus BoV, as well as three respiratory bacterial pathogens such as Legionella pneumophila 9135, Mycobacterium tuberculosis 005, and Haemophilus influenza ATCC 49247 were used in this study. Total viral RNAs were extracted from 200 μl of each culture using a QIAamp viral RNA mini kit (Qiagen, Hilden, Germany). All infectious materials were handled in biosafety level 3 facilities.
Preparation of Artificial EBOV RNA
Preparation of artificial EBOV RNA was performed as described previously (Watanabe et al., 2004) with modifications. Briefly, 663 kb NP fragments were synthesized (Sangon Biotech Co., Ltd., Shanghai, China) and cloned into vector pGEM-3Zf(+) with inverse orientation of the T7 promoter sequence (Promega). In vitro transcription of artificial EBOV RNA from NP subclones was carried out using 50 U of T7 RNA polymerase (Promega) in a 50-μl reaction volume according to the manufacturer’s instructions. The RNA concentration was determined by measuring the optical density at 260 nm (OD260), and the RNA purity was determined by calculating the OD260/OD280 absorption ratio (ratios were ensured to be >1.8). RNA was then dissolved in 20 μL DEPC-treated water, and stored at -70°C before use.
Primer Design
Based on the NP sequences of strain Mayinga deposited in GenBank (accession no. AF086833), we selected potential target regions and further analyzed the sequence using Primer Explorer V4 software2 by aligning it with other species of EBOV. We designed specific primer sets for the detection of EBO V in RT-LAMP, with each set including an outer forward primer (F3), an outer backward primer (B3), a forward inner primer (FIP), and a backward inner primer (BIP) linked by a four thymidine spacer (TTTT), which can recognize both sense and anti-sense strands. To accelerate the RT-LAMP reaction, an additional loop primer (LB) was designed. All primers were synthesized commercially (Sangon Biotech Co., Ltd.).
RT-LAMP Assays
Reverse transcription loop-mediated isothermal amplification reactions were performed using a Loopamp RNA amplification kit (Eiken Chemical Co., Ltd., Tokyo, Japan) in a volume of 25 μL according to the manufacturer’s protocol. Each reaction included 80 pmol of FIP and BIP, 40 pmol of LB, 10 pmol of F3 and B3, and 2 μL template RNA. The reaction was carried out at 61°C for 60–80 min in dry bath incubators.
Reverse transcription loop-mediated isothermal amplification amplified products were detected by turbidity monitoring as well as visual observation. To assess turbidity, the amount of white magnesium phosphate precipitate produced during the LAMP reaction process was monitored using a Loopamp Real-time Turbidimeter (LA-230; Eiken Chemical Co., Ltd., Tochigi, Japan) recording the reaction curves at 650 nm every 6 s with magnesium ion (Mg2+) in the reaction buffer (Mori et al., 2001). For visual inspection, tubes containing 1 μl of fluorescent calcein were observed by the naked eye and photographed under natural light or UV light at 365 nm. The color changed from orange to green for a positive reaction, while the negative control remained orange.
Real-time RT-PCR Assays
To illustrate RT-LAMP detection sensitivity, we targeted a region of NP using the LiferiverTM EBOV Real Time RT-PCR Kit (Shanghai ZJ BioTech Co., Ltd.) recommended by the World Health Organization. Thermocycler conditions followed the manufacturer’s instructions. During the amplification process, the fluorescence intensity of the reporter dye (FAM) and a quencher dye (TAMRA) was recorded to calculate the normalized reporter signal, which is linked to the amount of product amplified. The standard curve was drawn using the in vitro transcribed RNA standard to calculate the number of RNA copies in viral RNA extracts. The threshold cycle (Ct value) refers to the number of amplification cycles for the fluorescence to reach the threshold.
Clinical Specimens
A total of 417 clinical specimens from whole blood or swabs were collected from patients thought to be infected by EBOV during the outbreak in Sierra Leone, from 2014 to 2015. RT-LAMP assays and real-time RT-PCR were performed simultaneously by the China Mobile Laboratory Testing Team in Freetown, Sierra Leone. Information about the clinical samples is listed in Supplemental Tables S1 and S2. This study was carried out in accordance with the recommendations of the Institute of Disease Control and Prevention, China with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Results
Optimizing the RT-LAMP Assay
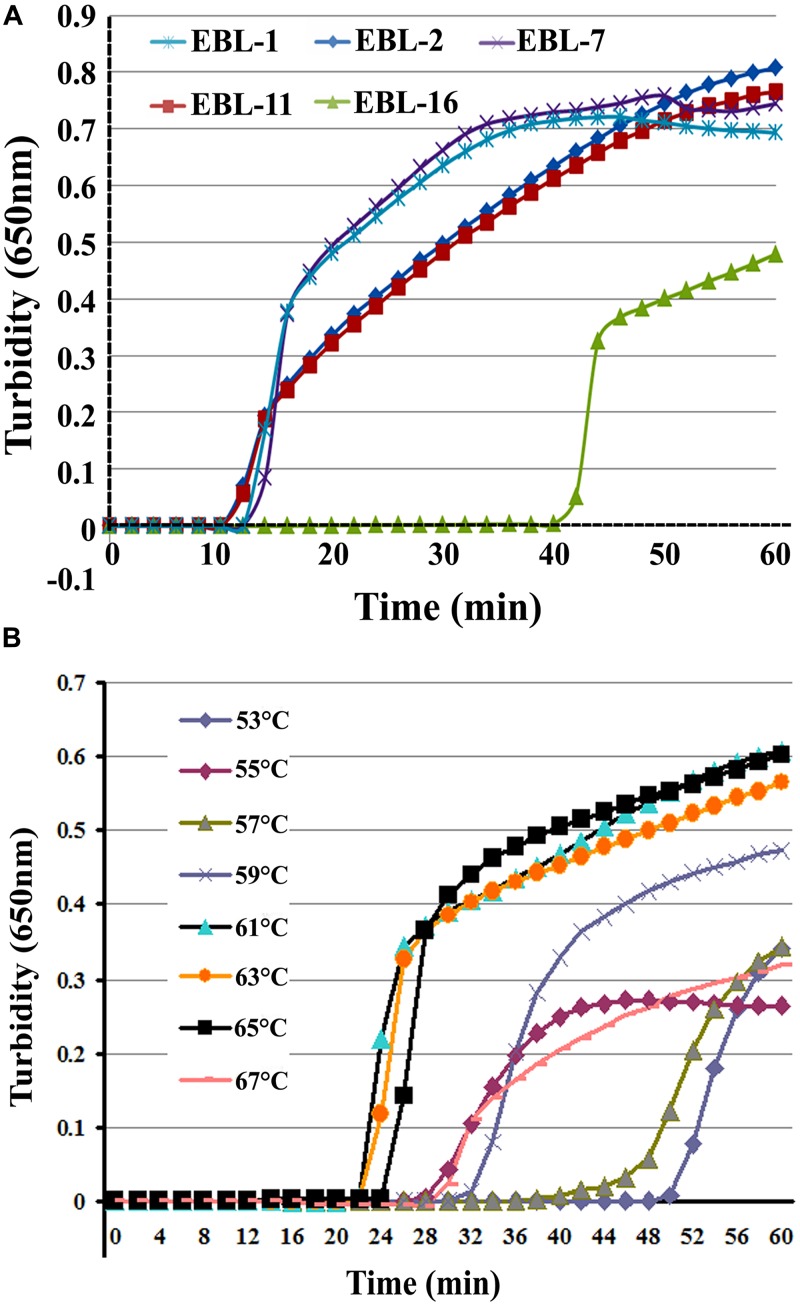
A total of five sets of primers were initially designed to detect artificial EBOV RNA using the Real-time Turbidimeter. As shown in Figure 1A, the EBL-2 primer set amplified NP in the shortest time (∼10 min), so this was chosen as the optimal primer set for RT-LAMP detection of EBOV (Table 1).
FIGURE 1.
The most appropriate primers and reaction temperatures for the reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay. Turbidity was monitored and recorded every 6 s for five sets of primers used to amplify the target gene with a Loopamp real-time turbidimeter at 650 nm. (A) A total of five sets of primers including EBL-1, EBL-2, EBL-7, EBL-11, and EBL-16 were designed to detect artificial EBOV RNA. (B) Reaction temperatures ranged from 53 to 67°C with 2°C intervals.
Table 1.
Sequence of primers used for specific amplification of NP.
| Primer | Type | Sequence(5′–3′) |
|---|---|---|
| EBL-2F3 | Forward outer | GATGGAAGCTACGGCGAAT |
| EBL-2B3 | Backward outer | GTGAGGGCCTGGGACATT |
| EBL-2FIP | Forward inner | AGTGTCCTCGTCGTCCTCGTCTTAGAGTTACTCGGAAAACGGC |
| EBL-2BIP | Backward inner | ATCGACCAAGGGTGGACAACAGTTTGTGTCTGTCTGCCCTCTA |
| EBL-2LB | Loop backward | AAGAACAGTCAAAAGGGCCAGC |
| EBL-2LF | Loop forward | CCAAGTCATCTGGTGCATTCAT |
To further optimize the amplification, we compared reaction temperatures ranging from 53 to 67°C with 2°C intervals. The most suitable reaction temperature range was shown to be 59–65°C (Figure 1B), and 61°C was ultimately chosen as the optimal reaction temperature.
Specificity of NP Detection by RT-LAMP
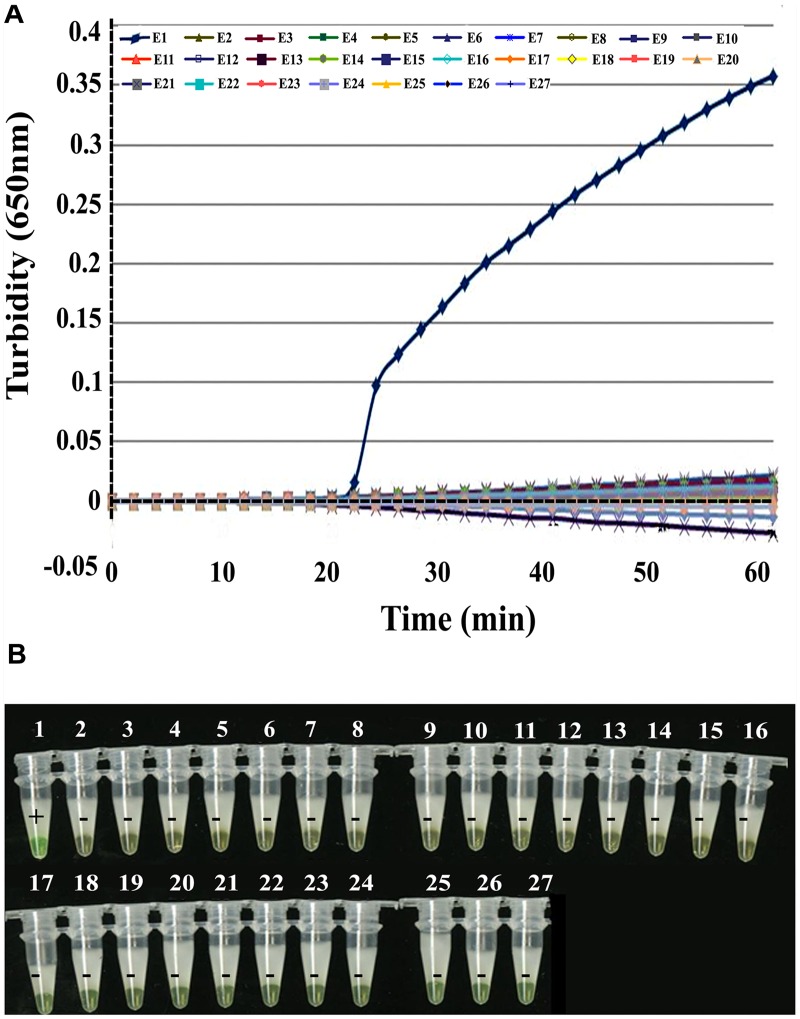
To test the LAMP specificity for NP, we tested 26 non- Zaire EBOV viruses in addition to EBOV itself and in vitro transcribed artificial EBOV RNA as the positive control. Figure 2 shows that EBOV RNA was identified positively by RT-LAMP with the EBL-2 primer set using turbidity monitoring and visual observation. All non- Zaire EBOV strains tested negative, including the blank control, indicating that the RT-LAMP method was specific for EBOV.
FIGURE 2.
Specificity of EBOV NP detection by RT-LAMP. (A) Turbidity was monitored and recorded every 6 s by a Loopamp real-time turbidimeter at 650 nm. (B) Visual detection using a calcein fluorescent detection reagent. Lane 1, positive control (artificial EBOV RNA); lane 2, negative control (double-distilled water); lane 3, Sudan EBOV (artificial Sudan EBOV RNA); lane 4, MARV (artificial MARV RNA); lane 5, SARS coronavirus; lane 6, H7N9; lane 7, H1N1; lane 8, H2N3; lanes 9–12, human parainfluenza viruses (PIV) 1, 2, 3, and 4; lanes 13–15, adenoviruses (ADV; serotypes 3, 5, and 55); lanes 16 and 17, respiratory syncytial virus infection, RSVA, RSVB; lane 18, MERS RNA; lane 19, human metapneumovirus, HMPV; lane 20, bocavirus, BoV; lanes 21–24, human coronavirus, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1; lane 25, Legionella pneumophila 9135; lane 26, Mycobacterium tuberculosis 005; and lane 27, Haemophilus influenza ATCC 49247.
Sensitivity of NP Detection by RT-LAMP
To determine the sensitivity of the RT-LAMP assay for EBOV, a series of dilutions were prepared of artificial EBOV RNA ranging from 4.56 × 104 to 4.56 × 10-2 copies/μL. As shown in Figure 3A, the times of positivity detection ranged from 18 min for 4.56 × 104 copies/μL to 36 min for 4.56 copies/μL of virus RNA by real-time monitoring. Thus, the RT-LAMP detection limit for NP is 4.56 copies/μL of artificial RNA in a 61°C reaction lasting for 60 min. For the visual inspection, all positive reactions changed to green while negative ones remained orange under natural or 365 nm UV light (Figure 3B). These data indicate that the sensitivity of the two detection methods was the same. The detection limit of real-time RT-PCR for NP was 4.56 copies/μL, but this was achieved with a higher Ct value (Ct = 41). Thus, we concluded that the sensitivity of the RT-LAMP assay for EBOV was similar or higher than real-time RT-PCR.
FIGURE 3.
Comparison of RT-LAMP sensitivities in detecting EBOV NP. Artificial EBOV RNA was serially diluted 10-fold from 4.56 × 104 copies/μL to 4.56 × 10-2 copies/μL. (A) Turbidity was monitored with a Loopamp Realtime Turbidimeter at 650 nm every 6 s. (B) The reaction was detected visually using a calcein fluorescent detection reagent. Artificial EBOV RNA concentrations were: tube 1, 4.56 × 104 copies/μL; tube 2, 4.56 × 103 copies/μL; tube 3, 4.56 × 102 copies/μL; tube 4, 4.56 × 101 copies/μL; tube 5, 4.56 copies/μL; tube 6, 4.56 × 10-1 copies/μL; tube 7, 4.56 × 10-2 copies/μL; tube 8, ddH2O.
Clinical Sample Detection
The 417 clinical blood or swab samples were simultaneously analyzed by RT-LAMP and real-time RT-PCR. Of these, 307 patients were confirmed to be infected with EBOV, while 106 tested negative (Figure 4 and Table 2). A higher Ct value (Ct > 36) was recorded for the remaining four samples by RT-PCR, and green fluorescence was observed after ∼45 min by RT-LAMP, indicating a low level of EOBV RNA. We suggested that these patients should be monitored for 2 weeks in hospital.
FIGURE 4.
Visual detection of RT-LAMP findings of 413 patients with clinically suspected infections in Sierra Leone. Sample information is listed in Supplemental Tables S1 and S2. A total of 307 positive (A) and 106 negative (B) samples were tested for EBOV infection by real-time RT-PCR and RT-LAMP. NC, negative control (distilled water); PC, positive control (artificial EBOV RNA).
Table 2.
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) and real-time reverse transcription-polymerase chain reaction (RT-PCR) findings of clinical blood or swab samples.
| Results | RT-LAMP | Real-time RT-PCR |
|---|---|---|
| Positive samples | 307 (Time: 10–30min) | 307 (Ct ≤ 30) |
| Negative samples | 106 (no change in color) | 106 (no peak) |
Discussion
Ebola virus has extremely high morbidity and mortality levels in humans, it reemerged and caused an outbreak in Western Africa where 28,476 suspected, probable, and confirmed cases, and 11,298 deaths were reported in Sierra Leone up until Oct. 21 2015 according to Ebola Situation Report from WHO3). Although several chemical agents, vaccines, and antibodies inhibit the spread of EBOV in humans and animals, effective therapies for clinical treatment are scarce.
To combat the increasing incidence of EBOV infections, we developed an RT-LAMP assay specific for EBOV diagnosis using primers spanning the 663 bp NP sequence of the viral genome. We found that the limit of detection for this technique was 4.56 copies/μL, which compares with 13.4 copies/μL for the Real-Time RT-PCR assay using the LiferiverTM-EBOV Kit according to the manufacturer’s instructions. In comparison, the limit of detection was 10 RNA molecule standards targeting the nucleoprotein gene by RT-PCR reported by Weidmann et al. (2004). In the present work, the RT-LAMP assay showed an equivalent or superior sensitivity to RT-PCR assays, indicating that it is sufficiently sensitive to detect low copy numbers of RNA. Furthermore, the RT-LAMP assay showed a high level of specificity, with no cross-reactivity with other species of EBOV or other viruses. It was also comparable with real-time RT-PCR at confirming cases of EBOV infection in clinical samples.
The RT-LAMP assay can detect the presence of virus in a single step in which reverse transcription and DNA amplification proceed in a single tube at a constant temperature of 61°C. Compared with RT-PCR, the sensitivity of the RT-LAMP assay is far greater in the presence of inhibitors. Moreover, RT-LAMP primers specifically recognize target sequences using five independent target sequence regions, compared with RT-PCR primers that recognize only two independent regions. Therefore, the RT-LAMP assay is more suitable for the rapid detection of NP in clinical samples.
Conclusion
We established a rapid and effective visual RT-LAMP assay targeting EBOV NP, which we showed to be extremely specific and sensitive in the molecular diagnosis of EBOV infections. It is a reliable tool for the identification of EBOV, so could be used as an alternative method of diagnostic testing at clinical laboratories without the need for special apparatus. Moreover, it can provide accurate results within 1 h, so may be of use in the clinical diagnosis of EBOV in developing countries.
Author Contributions
JY and XZ conceived and designed the experiments. XW and WL performed clinical detection in Sierra Leone. HL, PL, and DD performed and developed the RT-LAMP. XY, EL, PL, DD, DZ, LC, and XH performed the experiments. XZ wrote the manuscript. JY and XZ edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of The China Mobile Laboratory Testing Team in Sierra Leone including Xiushan Zhang, Leili Jia, Chuanfu Zhang, Rongzhang Hao, Shuguang Tian, Xiang Zhao, Wei Wu, Lihua Wang, Ziqian Xu, Xitong Yuan, Ruizhong Jia, Rongtao Zhao, Yong Chen, Wenyi Zhang, Guohui Chang, and Zeliang Chen for the clinical detection of EBOV. We are also grateful to Li Fengjing and Xiao Shengli from Beijing Lanpu Bio-tech Co., Ltd. for technical assistance and helpful discussions.
Funding. This work was supported by a grant from the National Natural Science Foundation of China (31370093 and 81201320) to JY and XW, mega-projects of Science and Technology Research of China (Grant 2011ZX10004-001), and a grant from the National High Technology Research and Development Program of China (863 Program; grant no. SS2014AA022210).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01332
References
- Ali M. T., Islam M. O. (2015). A highly conserved GEQYQQLR epitope has been identified in the nucleoprotein of Ebola virus by using an in silico approach. Adv. Bioinform. 2015 278197–278203. 10.1155/2015/278197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca J. T., Severns V., Lovato D., Branch D. W., Larson R. S. (2015). Rapid detection of Ebola virus with a reagent-free, point-of-care biosensor. Sensors (Basel) 15 8605–8614. 10.3390/s150408605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Tong Q., Zhang Y., Lou D., Kong Q., Lv S., et al. (2011). Loop-mediated isothermal amplification: rapid detection of Angiostrongylus cantonensis infection in Pomacea canaliculata. Parasit. Vectors 4 204 10.1186/1756-3305-4-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow D. S., Kleine C., Edwards J. K., Scaini R., Giuliani R., Sprecher A. (2014). Ebola virus disease in West Africa-clinical manifestations and management. N. Engl. J. Med. 371 2054–2057. 10.1056/NEJMp1413084 [DOI] [PubMed] [Google Scholar]
- de Franchis R., Cross N. C., Foulkes N. S., Cox T. M. (1988). A potent inhibitor of Taq polymerase copurifies with human genomic DNA. Nucleic Acids Res. 16 10355 10.1093/nar/16.21.10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher-Hoch S. P., Perez-Oronoz P. I., Jackson E. L., Hermann L. M., Brown B. G. (1992). Filovirus clearance in non-human primates. Lancet 340 451–453. 10.1016/0140-6736(92)91770-9 [DOI] [PubMed] [Google Scholar]
- Frieden T. R., Damon I., Bell B. P., Kenyon T., Nichol S. (2014). Ebola 2014-New challenges, new global response and responsibility. N. Engl. J. Med. 371 1177–1180. 10.1056/NEJMp1409903 [DOI] [PubMed] [Google Scholar]
- Grolla A., Lucht A., Dick D., Strong J. E., Feldmann H. (2005). Laboratory diagnosis of Ebola and Marburg hemorrhagic fever. Bull. Soc. Pathol. Exot. 98 205–209. [PubMed] [Google Scholar]
- Hampton T. (2014). Largest-ever outbreak of Ebola Virus disease thrusts experimental therapies, vaccines into spotlight. JAMA 312 987–989. 10.1001/jama.2014.11170 [DOI] [PubMed] [Google Scholar]
- Hara-Kudo Y., Yoshino M., Kojima T., Ikedo M. (2005). Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253 155–161. 10.1016/j.femsle.2005.09.032 [DOI] [PubMed] [Google Scholar]
- Hirayama H., Kageyama S., Moriyasu S., Sawai K., Minamihashi A. (2013). Embryo sexing and sex chromosomal chimerism analysis by loop-mediated isothermal amplification in cattle and water buffaloes. J. Reprod. Dev. 59 321–326. 10.1262/jrd.2013-028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. (2007). Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70 499–501. 10.1016/j.jbbm.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Kong Q. M., Lu S. H., Tong Q. B., Lou D., Chen R., Zheng B., et al. (2012). Loop-mediated isothermal amplification (LAMP): early detection of Toxoplasma gondii infection in mice. Parasit. Vectors 5 2 10.1186/1756-3305-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T. G., Rollin P. E., Williams A. J., Bressler D. S., Martin M. L., Swanepoel R., et al. (1999a). Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179 (Suppl. 1), S177–S187. 10.1086/514321 [DOI] [PubMed] [Google Scholar]
- Ksiazek T. G., West C. P., Rollin P. E., Jahrling P. E., Peters C. J. (1999b). ELISA for the detection of antibodies to Ebola viruses. J. Infect. Dis. 179 (Suppl. 1), S192–S198. 10.1086/514313 [DOI] [PubMed] [Google Scholar]
- Kurosaki Y., Takada A., Ebihara H., Grolla A., Kamo N., Feldmann H., et al. (2007). Rapid and simple detection of Ebola virus by reverse transcription-loop-mediated isothermal amplification. J. Virol. Methods 141 78–83. 10.1016/j.jviromet.2006.11.031 [DOI] [PubMed] [Google Scholar]
- MacNeil A., Rollin P. E. (2012). Ebola and Marburg hemorrhagic fevers: neglected tropical diseases? PLoS Negl. Trop. Dis. 6:e1546 10.1371/journal.pntd.0001546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289 150–154. 10.1006/bbrc.2001.5921 [DOI] [PubMed] [Google Scholar]
- Niikura M., Ikegami T., Saijo M., Kurane I., Miranda M. E., Morikawa S. (2001). Detection of Ebola viral antigen by enzyme-linked immunosorbent assay using a novel monoclonal antibody to nucleoprotein. J. Clin. Microbiol. 39 3267–3271. 10.1128/JCM.39.9.3267-3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 E63 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okafuji T., Yoshida N., Fujino M., Motegi Y., Ihara T., Ota Y., et al. (2005). Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J. Clin. Microbiol. 43 1625–1631. 10.1128/JCM.43.4.1625-1631.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo M., Niikura M., Morikawa S., Ksiazek T. G., Meyer R. F., Peters C. J., et al. (2001). Enzyme-linked immunosorbent assays for detection of antibodies to Ebola and Marburg viruses using recombinant nucleoproteins. J. Clin. Microbiol. 39 1–7. 10.1128/JCM.39.1.1-7.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Toma C., Nakasone N., Iwanaga M. (2005). Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243 259–263. 10.1016/j.femsle.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Watanabe T., Noda T., Takada A., Feldmann H., Jasenosky L. D., et al. (2004). Production of novel ebola virus-like particles from cDNAs: an alternative to ebola virus generation by reverse genetics. J. Virol. 78 999–1005. 10.1128/JVI.78.2.999-1005.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann M., Muhlberger E., Hufert F. T. (2004). Rapid detection protocol for filoviruses. J. Clin. Virol. 30 94–99. 10.1016/j.jcv.2003.09.004 [DOI] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2014). Urgently Needed: Rapid, Sensitive, Safe and Simple Ebola Diagnostic Tests. Available at: http://www.who.int/mediacentre/news/ebola/18-november-2014-diagnostics/en/ (accessed March 23 2015). [Google Scholar]
- Zaki S. R., Shieh W. J., Greer P. W., Goldsmith C. S., Ferebee T., Katshitshi J., et al. (1999). A novel immunohistochemical assay for the detection of Ebola virus in skin: implications for diagnosis, spread, and surveillance of Ebola hemorrhagic fever. Commission de Lutte contre les Epidemies a Kikwit. J. Infect. Dis. 179 (Suppl. 1), S36–S47. 10.1086/514319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.