Abstract
Cells from a patient with a DNA repair-deficiency disorder are anticipated to bear a large number of somatic mutations. Because such mutations occur independently in each cell, there is a high degree of mosaicism in patients' tissues. While major mutations that have been expanded in many cognate cells are readily detected by sequencing, minor ones are overlaid with a large depth of non-mutated alleles and are not detected. However, cell cloning enables us to observe such cryptic mutations as well as major mutations. In the present study, we focused on a fibroblastic cell line that is derived from a patient diagnosed with xeroderma pigmentosum (XP), which is an autosomal recessive disorder caused by a deficiency in nucleotide excision repair. By making a list of somatic mutations, we can expect to see a characteristic pattern of mutations caused by the hereditary disorder. We cloned a cell by generating an iPS cell line and performed a whole-exome sequencing analysis of the progenitor and its iPS cell lines. Unexpectedly, we failed to find causal mutations in the XP-related genes, but we identified many other mutations including homozygous deletion of GSTM1 and GSTT1. In addition, we found that the long arm of chromosome 9 formed uniparental disomy in the iPS cell line, which was also confirmed by a structural mutation analysis using a SNP array. Type and number of somatic mutations were different from those observed in XP patients. Taken together, we conclude that the patient might be affected by a different type of the disorder and that some of the mutations that we identified here may be responsible for exhibiting the phenotype. Sequencing and SNP-array data have been submitted to SRA and GEO under accession numbers SRP059858 and GSE55520, respectively.
Keywords: Xeroderma pigmentosum, Patient iPS cell, Sun burn, Suntan, Melanoma
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/XP40OS/fibroblast and its iPS cells |
| Sex | Male |
| Sequencer or array type | Illumina HiSeq 1000 and HumanCytoSNP-12 |
| Data format | FASTQ and matrix tables |
| Experimental factors | Fibroblast and its iPS cells |
| Experimental features | Identification of somatic mutations using iPS-cell cloning |
| Consent | Publicly available from NCBI SRA and GEO |
| Sample source location | Japanese Collection of Research Bioresources (JCRB) Catalog No.: JCRB0327 |
1. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/sra/?term=SRP059858
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55520
http://epigenetics.nrichd.ncchd.go.jp/ips/data/XP40OS.vcf.bz2
http://epigenetics.nrichd.ncchd.go.jp/ips/data/XP40OS.tsv.bz2
2. Experimental design, materials and methods
Xeroderma pigmentosum (XP) is caused by a deficiency in base excision repair, resulting in a large number of single-nucleotide mutations, especially in sun-exposed skin [1]. Due to the independent manner of the occurrence of the somatic mutations, the patient tissue, including fibroblasts, may form a high degree of mosaicism. If a mutation had occurred in an early stage of development or in stem cells, it could readily be detected by DNA sequencing. Most of the mutations that have occurred in skin cells would not be expanded much because the greater part of the cells is rather differentiated. Hence, it seems difficult to obtain a genome-wide pattern of XP-derived mutations. However, it may be possible to examine a whole set of mutations of a single cell if the cell could be cloned. This can be achieved by generation of iPS cells owing to the fact that the process is cloning per se [2]. One may suspect that a number of de novo mutations arise during the reprogramming process and cultivation after the establishment of an iPS cell line, but many recent studies have demonstrated that most of the mutations found in iPS cells were preexisting in their progenitor lines, suggesting the high genetic stability of reprogramming and subsequent cultivation [3], [4]. In the present study, we focused on a fibroblast cell line derived from an XP patient and established its iPS cell lines in order to delineate somatic mutations in the exome-target regions.
We obtained the XP40OS cell line, which was derived from a patient diagnosed with XP group C, from the JCRB Cell Bank [5]. This study was approved by the Institutional Review Board at the National Center for Child Health and Development. Employing the same method described in our previous study, we established its iPS cell line and performed whole-exome sequencing and structural alteration analyses for both the progenitor and iPS cell lines [6]. In brief, the iPS cells were generated by reprogramming with Sendai virus infection-mediated expression of the four factors. Extracted DNA samples were treated with Agilent SureSelect Human All Exon V4 + UTRs + lincRNA, whose target size was approximately 80 Mb, to prepare a pair-end library. The library was sequenced on the Illumina platform, yielding 18.8 Gb (93.3 M read pairs) and 18.7 Gb (92.7 M read pairs) data, respectively. After trimming, the data sizes were reduced to 16.9 Gb (84.3 M read pairs) and 16.8 Gb (83.7 M read pairs). For both samples, over 99% bases had q-scores that are more than 15. Our pipeline software included cutadapt-1.7.1, BWA 0.7.12, SAMtools 1.2, Picard 1.130, Genome Analysis Toolkit 3.3, and our own custom programs coded in C or Perl. Reads were mapped to the hs37d5 (GRCh37) reference sequence and PCR duplicate reads were then eliminated. Ratios of PCR duplicates were 27.4% and 26.5%, respectively. Along with in-house control data, multi-sample calling of single-nucleotide and short indel variations was performed. Filtration and identification of somatic mutations were carried out as described in our previous study [6].
First, we searched for possible variants that could affect the XP phenotype. Such variants should be found in both lines. A homozygous one (rs1800975) was found in the 5′ untranslated region of the XPA gene and two heterozygous ones (rs2228000 and rs2228001), which alter amino acid sequences, were found in the protein-coding regions of the XPC gene. Since their allelic frequencies in the population were not rare (50.8%, 32.6%, and 60.6%, respectively, according to the Human Genetic Variation Database, http://www.genome.med.kyoto-u.ac.jp/SnpDB), they cannot be considered as etiologic variants. While we failed to find any other notable variants in XP-related genes, we found homozygous deletions in the BTNL3, GSTM1, GSTT1, and SLC25A24 loci. The GSTM1 and GSTT1 genes lacked the entire coding regions. The list of all possible variants is available from the websites (see the “Direct link to deposited data” section).
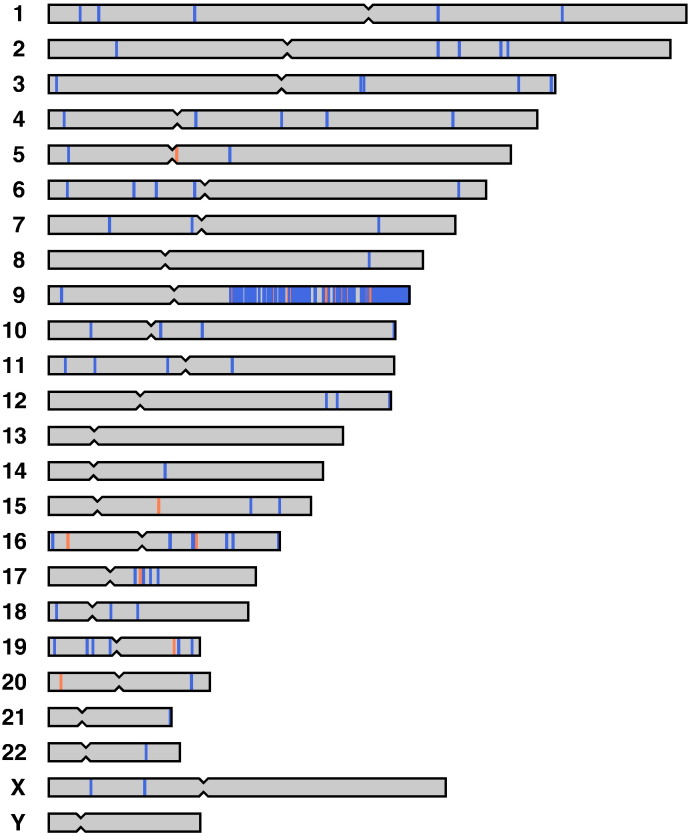
Next, we searched for inconsistent genotypes called with UnifiedGenotyper between the progenitor and its iPS cell lines to identify somatic mutations. Using the criteria described in our previous study [6], 922 single-nucleotide substitutions and 77 indels were identified. Because the numbers were extremely high compared to those of the preceding studies, we examined the positions of all the genotypes that exhibited disagreement between the two lines (Fig. 1). Among these sites, 845 single-nucleotide and 69 indel sites were located on the long arm of chromosome 9. All the called genotypes were homozygous without exception. Using an Illumina HumanCytoSNP-12 v2.1 DNA Analysis BeadChip Kit, we also performed a SNP-genotyping analysis to examine the existence of structural alterations. Copy-neutral loss of heterozygosity was detected in the 9:71,035,938–138,893,874 region (GRCh37) of the iPS cell line, indicating that a large structural mutation formed uniparental disomy. In that case, heterozygous sites in the region became homozygous. The 914 sites in the exome-target regions were considered to be just heterozygous in the progenitor cell, but not individual mutations. The SNP-genotyping analysis also suggested one copy-gain mutation around 16:83,939,438–85,567,156 only in the iPS cell line, but no genotype disagreement was found in this 1.6-Mb region. Among the remaining 77 single-nucleotide substitutions, C-to-T or G-to-A transitions accounted for 29 of the changes.
Fig. 1.
Genome-wide distribution of the somatic mutations identified in the present study.
Blue and red bars indicate single-nucleotide and indel mutations, respectively. Nearly one thousand mutations were detected on the long arm of chromosome 9. The discrepant genotypes between the progenitor and its iPS cell lines were caused by a single structural mutation that formed uniparental disomy in the chromosome arm.
3. Discussion
It is well known that C-to-T or G-to-A transitions in a dipyrimidine context are typical type of UV-specific mutations [7]. A trinucleotide analysis revealed that TCT, TCC, TCA, and their complementary trinucleotides, in which the mutated base is situated in the center, are frequently subject to mutation. Partially due to the relatively low number of mutations, the significance of the mutation type in the sample was not determined. In addition to the fact that we failed to find crucial mutations in XP-related genes in the exome-target regions, the donor of the fibroblast might be affected by another type of XP. It could be possible that some of the mutations that we identified in both of the lines are responsible for the phenotype.
Conflict of interest
The authors declare that there are no conflicts of interests.
Acknowledgements
We would like to express our sincere thanks to Dr. C. Ketcham for English editing and to E. Suzuki and K. Saito for secretarial work. This research is (partially) supported by the Research Project for Practical Applications of Regenerative Medicine (15bk0104028h0003) from Japan Agency for Medical Research and development, AMED. We acknowledge the International High Cited Research Group (IHCRG #14-104), Deanship of Scientific Research, King Saud University, Riyadh, Kingdom of Saudi Arabia. AU thanks King Saud University, Riyadh, Kingdom of Saudi Arabia, for the Visiting Professorship. Computation time was provided by the computer cluster Hitachi HA8000/RS210 at the Center for Regenerative Medicine, National Research Institute for Child Health and Development.
References
- 1.Li L., Bales E.S., Peterson C.A., Legerski R.J. Characterization of molecular defects in xeroderma pigmentosum group C. Nat. Genet. 1993;5(4):413–417. doi: 10.1038/ng1293-413. [DOI] [PubMed] [Google Scholar]
- 2.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460(7251):49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 3.Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E., Lee J.H., Loh Y.H., Manos P.D., Montserrat N., Panopoulos A.D., Ruiz S., Wilbert M.L., Yu J., Kirkness E.F., Izpisua Belmonte J.C., Rossi D.J., Thomson J.A., Eggan K., Daley G.Q., Goldstein L.S., Zhang K. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471(7336):63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young M.A., Larson D.E., Sun C.W., George D.R., Ding L., Miller C.A., Lin L., Pawlik K.M., Chen K., Fan X., Schmidt H., Kalicki-Veizer J., Cook L.L., Swift G.W., Demeter R.T., Wendl M.C., Sands M.S., Mardis E.R., Wilson R.K., Townes T.M., Ley T.J. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10(5):570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato K., Ikenaga M., Sano S. Kinetic analysis of polyethylene glycol-induced cell fusion in cultured human fibroblasts: its application to genetic complementation analysis of xeroderma pigmentosum. Med. J. Osaka Univ. 1982;33(1–2):19–28. [PubMed] [Google Scholar]
- 6.Fukawatase Y., Toyoda M., Okamura K., Nakamura K., Nakabayashi K., Takada S., Yamazaki-Inoue M., Masuda A., Nasu M., Hata K., Hanaoka K., Higuchi A., Takubo K., Umezawa A. Ataxia telangiectasia derived iPS cells show preserved x-ray sensitivity and decreased chromosomal instability. Sci. Rep. 2014;4:5421. doi: 10.1038/srep05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brash D.E., Rudolph J.A., Simon J.A., Lin A., McKenna G.J., Baden H.P., Halperin A.J., Pontén J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. U. S. A. 1991;88(22):10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]