Abstract
Hypoxia causes critical cellular injury both in early human development and in adulthood, leading to cerebral palsy, stroke, and myocardial infarction. Interestingly, a remarkable phenomenon known as hypoxic preconditioning arises when a brief hypoxia exposure protects target organs against subsequent, severe hypoxia. Although hypoxic preconditioning has been demonstrated in several model organisms and tissues including the heart and brain, its molecular mechanisms remain poorly understood. Accordingly, we used embryonic and larval zebrafish to develop a novel vertebrate model for hypoxic preconditioning, and used this model to identify conserved hypoxia-regulated transcripts for further functional study as published in Manchenkov et al. (2015) in G3: Genes | Genomes | Genetics. In this Brief article, we provide extensive annotation for the most strongly hypoxia-regulated genes in zebrafish, including their human orthologs, and describe in detail the methods used to identify, filter, and annotate hypoxia-regulated transcripts for downstream functional and bioinformatic assays using the source data provided in Gene Expression Omnibus Accession GSE68473.
Keywords: Hormesis, Hypoxia–ischemia, Metabolism, Preconditioning, Stress tolerance
| Specifications | |
|---|---|
| Organism/cell line/tissue | Zebrafish (Danio rerio) |
| Sex | Mixed sex |
| Sequencer or array type | NimbleGen Gene Expression Danio Rerio 385k Array [071105_Zv7_EXPR] |
| Data format | RMA Normalized (R/Bioconductor, Limma) |
| Experimental factors | Hypoxia vs. normoxia; shield (gastrula) and 8-somite (segmentation) stages |
| Experimental features | Study of hypoxia-induced differential expression during zebrafish development at gastrulation (shield) and segmentation (8-somite) stages |
| Consent | N/A |
| Sample source location | Boston, Massachusetts, USA |
1. Experimental design and data
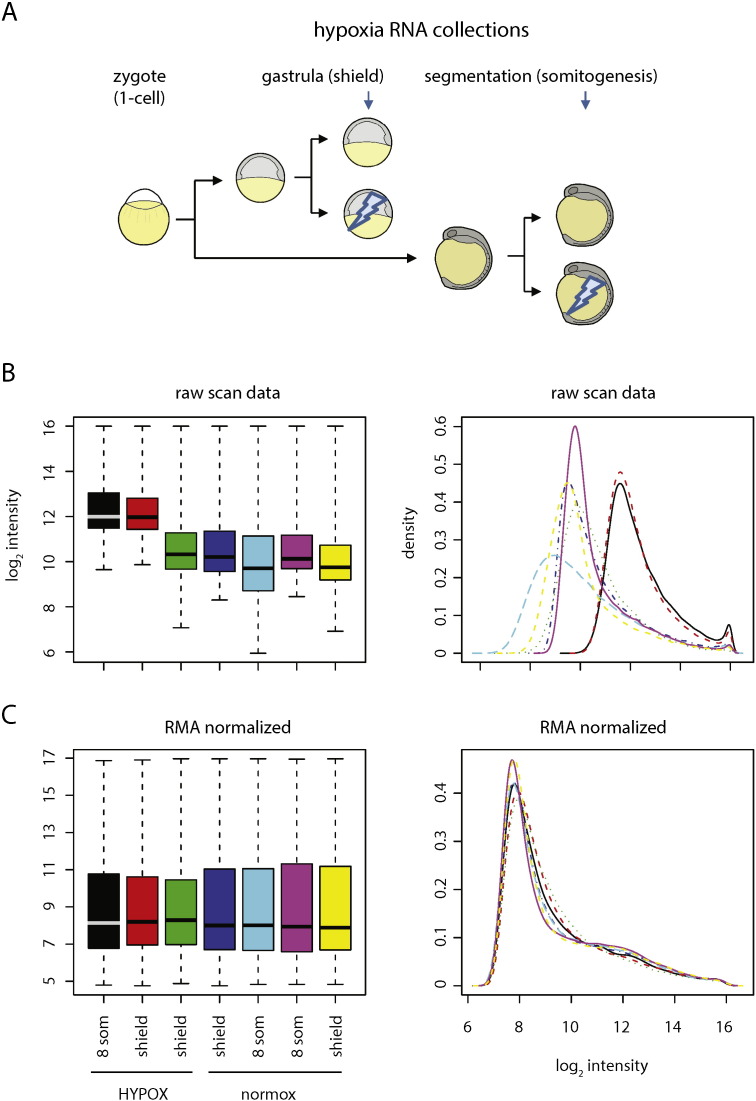
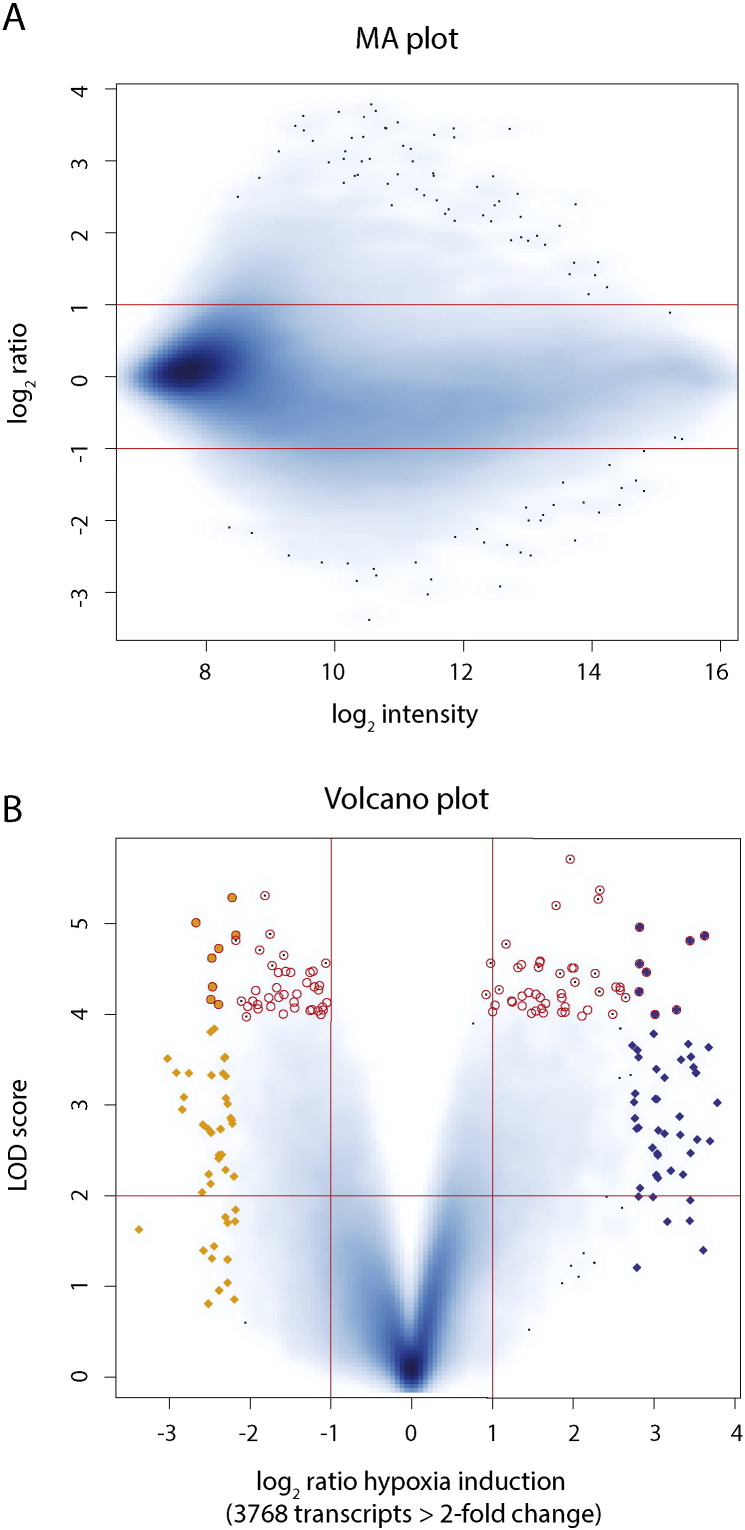
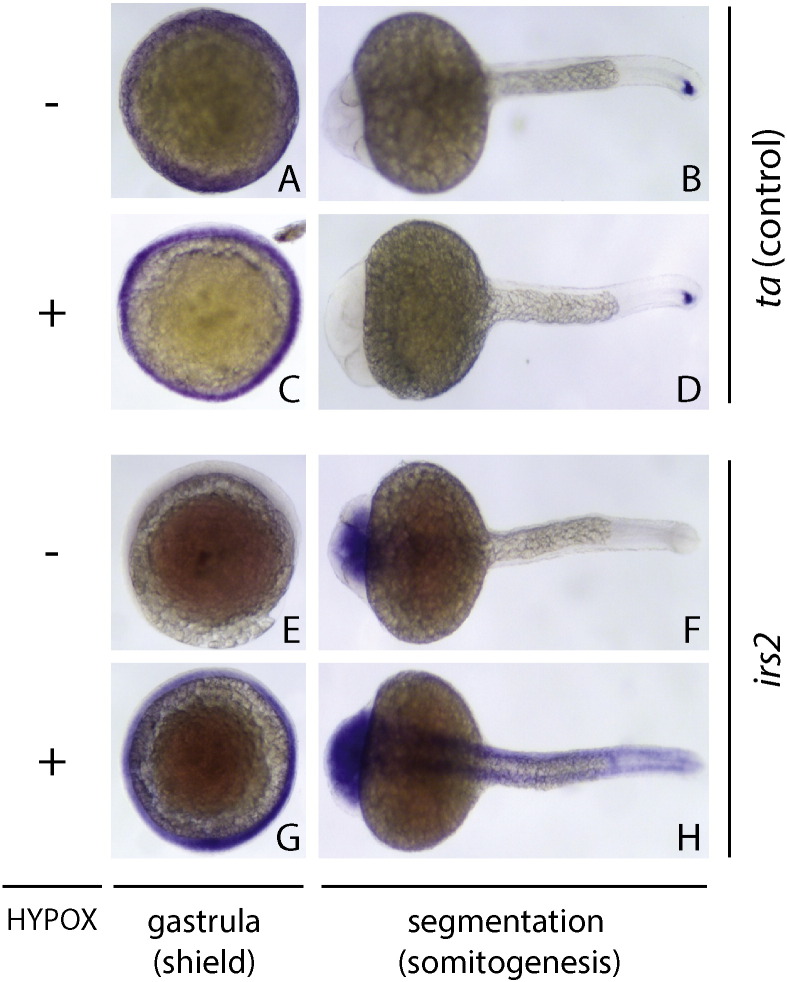
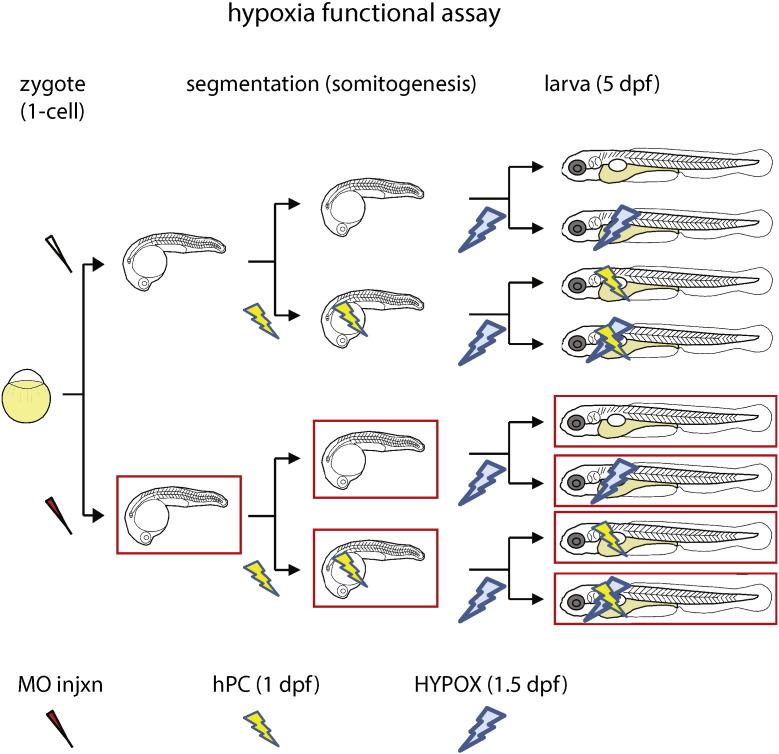
To identify novel hypoxia protective factors, we developed a novel in vivo hypoxic stress assay in zebrafish. First, we optimized hypoxic stress parameters at two different timepoints during zebrafish development and collected control and hypoxia-exposed embryos at gastrula (shield, 6 h postfertilization or hpf) and segmentation (8 somite, 12 hpf) stages to identify shared hypoxia responses at two timepoints (Fig. 1A). We reasoned that hypoxia response genes activated at distinct developmental timepoints were likely to be enriched for evolutionarily conserved hypoxia-protective components relevant for human biology and disease. We performed RNA extraction, labeling, and hybridization to NimbleGen zebrafish microarrays and show both the raw and normalized intensity plots and histograms for these data (Fig. 1B, C). We combined data from normoxic control samples at both gastrula and segmentation in comparison with matched hypoxia-exposed samples and identified 3768 of the 37,157 transcripts measured to be differentially expressed greater than 2-fold under hypoxia (Fig. 2). To further increase the utility of this dataset for the scientific community, we provide additional annotations in this article that includes human homologs and functional properties/domains for all transcripts measured, including the 300 most hypoxia-induced and hypoxia-repressed genes passing statistical significance criteria (Supplementary Table 1, Supplementary Table 2, Supplementary Table 3). From these lists of genes, we validated the expression of a subset of individual hypoxia-induced genes using qPCR and/or in situ hybridization, with irs2 shown as a representative hypoxia-induced gene that exhibited increased and ectopic expression at both gastrula and segmentation (Fig. 3). We further tested individual expression-validated genes for hypoxia-protective function in both acute hypoxic stress buffering and hypoxic preconditioning assays in zebrafish (Fig. 4) and identified several novel hypoxia-induced genes that function to protect against acute hypoxic stress and/or provide hypoxic preconditioning protection [1].
Fig. 1.
Hypoxia sample collection and quality control. (A) RNA collection scheme. Stage-synchronized embryos were collected in duplicate at gastrula (shield, 6 h) and segmentation (8-somite, 12 h) stages for RNA extraction with or without 2 h of severe hypoxia exposure at 0.3% oxygen (blue arrows). (B) Raw data from individual samples is shown both as a log-intensity boxplot and histogram. (C) Data after Robust Multi-array Average (RMA) normalization showing alignment.
Fig. 2.
Genome-wide hypoxia differential expression. (A) log2 hypoxia induction vs. log10 average intensity scatterplot (MA plot) showing the effect of hypoxia on the expression of all 37,157 transcripts measured. Y-axis shows a relatively uniform degree of hypoxia induction for transcripts at all hybridization intensities. Horizontal red lines indicate 2-fold induction and repression. (B) Volcano scatterplot of all transcripts showing statistical significance on Y-axis as determined by empirical Bayesian method and x-axis demonstrating fold hypoxia induction. Orange triangles; 100 most downregulated genes; blue triangles: 100 most upregulated genes; red circles: 100 most statistically significant differentially expressed genes. Vertical red lines indicate 2-fold induction and repression. Horizontal red line indicates p < 0.01. These figure panels are complementary or modified presentations, respectively, of the data presented in Fig. 2A of the primary hypoxia study [1].
Fig. 3.
Candidate hypoxia-regulated gene validation. In situ hybridization images for the hypoxia-induced gene irs2 are shown under normoxia (A, B, E, F) and hypoxia (C, D, G, H) at gastrula (6 h) and segmentation (24 h) stages, dorsal view. Increased intensity and tissue distribution of irs2 expression are evident in the hypoxia-exposed group (G, H). In comparison, a control gene (ta) shows no hypoxia-dependent changes in expression (A–D). These figure panels depict additional embryo views and/or specimens analogous to Fig. 2D,E from the primary hypoxia study [1].
Fig. 4.
In vivo hypoxic stress protection assay. Candidate genes were assessed for a functional role in protection from hypoxic stress through two assays, both of which were performed with targeted morphant (knockdown; red needle and red outline) and embryos injected with control morpholino oligonucleotides. For acute hypoxia, larvae were subjected to hypoxia at 1.5 dpf and evaluated for exacerbation of lethality at 5 dpf (blue bolt). For the hypoxic preconditioning assay, larvae were preconditioned through a milder hypoxia exposure at 1 dpf (yellow bolt) and re-exposed to prolonged severe hypoxia at 1.5 dpf (blue bolt).
2. Materials and methods
2.1. Zebrafish husbandry and strains
Zebrafish from the TL/AB strain were maintained using standard procedures and developmental stages were determined as previously described [2]. Embryos were raised at 28.5 °C in embryo water containing 0.1% Methylene Blue hydrate (Sigma, St. Louis, MO, USA) and 0.03% Instant Ocean sea salt (United Pet Group, Cincinnati, OH, USA). Experiments were performed under UCSD IACUC protocol S13006.
2.2. Hypoxia exposure and RNA isolation for microarrays
For hypoxia treatment of embryos used for RNA isolation and microarray analysis, a closed hypoxia chamber was used with a flow meter attached to a gas source (Billups-Rothenberg, Inc.). Hypoxia chambers were flushed for 4 min at 20 L/min, and repeated 30 min later with nitrogen gas. 50 mm Petri dishes containing 4 mL of embryo water were pre-equilibrated in the hypoxia chamber prior for at least 4 h prior to embryo transfer, as this treatment was determined to be the minimum time necessary to reach < 1% oxygen as measured by a dissolved oxygen meter (DO-5509; Alfa Electronics), and gave similar results when compared with longer pre-equilibration times up to 24 h. A colorimetric resazurin indicator was used to monitor the hypoxic environment during pre-treatment of media and the duration of each experiment (< 1% oxygen if colorless; Bio-Bag, Becton, Dickinson and Company).
Hypoxia was induced by transfer of individual embryos into hypoxic media at the shield or 8 somite stage for 1 to 2 h, as indicated. Embryos from synchronized crosses were dechorionated at < 1 h postfertilization (hpf) and maintained in a normoxic environment. Embryos were examined individually prior to hypoxia exposure and any delayed or abnormal embryos were discarded. Hypoxia exposure was initiated by individual embryo transfer from normoxic to hypoxic media using a glass pipette. Care was taken to minimize chamber opening time and gas flow was turned on immediately prior to and during chamber opening to minimize atmospheric contamination of chamber environment. After embryo addition, the resealed chamber was flushed with gas using the same protocol as for initial pre-equilibration above.
After hypoxic incubation, embryos were removed from the hypoxia chamber and immediately transferred in groups of 10–15 embryos to 1.5 mL tubes for flash freezing in liquid nitrogen and storage at − 80 °C, and samples collected in duplicate. Frozen embryos were subsequently thawed on ice and pestle-homogenized in an initial 250 μL of TRIzol (Life Technologies), to which 500 μL was added once homogenization was complete and the standard extraction protocol completed. RNA was quantitated using a NanoDrop and quality was verified using an Agilent BioAnalyzer prior to further processing.
2.3. Microarray hybridization and analysis
RNA samples verified by Bioanalyzer to show two strong ribosomal bands and minimal degradation were further amplified with the MessageAmp kit (Ambion) and labeled fluorescently with Cy3 or Cy5-dUTP (Amersham) in a reverse transcription reaction suing Superscript (Invitrogen). Labeled cDNA was rinsed and pooled with 20 μg of each of salmon sperm DNA, poly-A RNA, and tRNA, which were then concentrated to small volume in a Microcon YM-30 column. Samples were added to full-genome zebrafish microarrays designed using the Zv7 genome assembly and containing 385,000 probes, at approximately 12 probes for each of 37,157 target transcripts (071105_Zv7_EXPR; NimbleGen, Inc.) [3]. Hybridization and washes were performed per standard protocols, and arrays were stored in a closed, dark slide box until scanning no more than 6 h later (AXON GenePix 4000A Microarray Scanner). Laser intensity was optimized separately for green and red channels and optimized to include a small number of pixels/spots at maximum saturation to ensure adequate dynamic range and sensitivity at the low intensity range. Biological replicates were performed with “dye-flipping”, so that each duplicate sample was labeled with the opposite dye as the first experimental sample. The resulting dataset was filtered and analyzed with R/Bioconductor and Limma to confirm normal distributions of intensities and the absence of significant region-specific artifacts prior to inclusion into the dataset for normalization and downstream analysis. Notably, during this quality control process one sample was discarded due to high background and is not included in the downstream analyses (hypoxia, 8-somite), resulting in 3 hypoxia samples and 4 control normoxia samples as the basis for all downstream bioinformatic analyses (Fig. 1B, C). An established, empirical Bayesian method for differential expression in microarrays (eBayes) was used to determine p-values of individual genes, taking into account multiple hypothesis testing, the overall data distribution for all genes, and the small number of biological replicates typical in genome-wide datasets [4], [5], [6]. Microarray source data is available at NCBI GEO (GSE68473), and the R code used for the analysis can be reviewed in full in the Expanded View Code, supplementary in [1]. Top hypoxia-induced genes were filtered to retain only those genes that (a) had a single, unambiguous probe BLAT match to the later Zv9 genome assembly, (b) for which RNA-seq or EST evidence was available (UCSC genome browser, Z-seq [7], and (c) for which an identifiable human homolog existed in RefSeq and/or Ensembl databases.
2.4. Morpholino injection, developmental hypoxia assay, and phenotypic scoring
Morpholinos (MOs) were designed and obtained from Gene Tools, LLC. Splice-blocking MOs were preferred, but a translation-blocking MO was used for irs2 as no splice-blocking MO was able to be designed that passed QC filters (ENSDART00000053924; personal communication, Gene Tools). Standard Control and irs2 MOs were solubilized in water at a stock concentration of 1 mM (~ 8 mg/mL). The resulting stock solution was diluted 1:4 (~ 2 mg/mL) in water containing a phenol red tracer. MO's used: Standard Control Oligo (CCTCTTACCTCAGTTACAATTTATA), irs2 (CCCCTTTAAGAGGCGGACTTGCCAT).
As consistency of phenotype and survival of MO-injected embryos was observed to be higher when chorionated, we used chorionated embryos for MO microinjection and subsequent hypoxia exposure for the developmental stress assay. Wild-type embryos at the 1 to 4-cell stages were injected with MOs into the yolk near the embryo interface at a volume of 1 to 2 nL (2 to 4 ng) and both Standard Control and irs2 MOs were injected at 2 ng.
For hypoxia treatment of morphant embryos, an adjustable, self-regulating hypoxia chamber was used at 0.3% oxygen in a 28.5 °C humidified incubator (Biospherix C-chamber). Embryos were individually transferred with a pipette into 4 mL of pre-equilibrated hypoxic media in 6-well plates for hPC for 5 h at 1 dpf and/or prolonged sH for 38 h at 1.5 dpf, both at 0.3% oxygen. To end hypoxia exposure, plates were removed from the hypoxia chamber, 2 mL of normoxic embryo water was added to each well, and wells were kept open to the ambient air for 10 min prior to replacement of lids and return to the normoxic 28.5 °C incubator. During normoxic recovery, embryos were checked daily for lethality and provided fresh water.
2.5. In situ expression analysis
Digoxigenin (DIG)-labeled antisense RNA probes to desired gene transcripts were in vitro transcribed from linearized plasmid templates containing the clone of interest with T7 or SP6 RNA polymerase as appropriate (Promega). Primers used to amplify and clone probe regions for irs2 are: forward-CTTCAGTCAGCCCCACTAAC, reverse-CCTGCTTTACAACAACCGCC, and for ta are as described [8]. In situ hybridization of zebrafish embryos was performed according to standard procedures [9], using immunohistochemical detection of the DIG-labeled RNA–RNA hybrids by an anti-DIG Alkaline-Phosphatase coupled antibody and nonfluorescent, colorimetric detection with BCIP/NBT. Digital images were obtained on a Zeiss Axio-Imager microscope and scaled, cropped, and contrast-adjusted with Photoshop.
3. General methods
For embryo injection and/or stress experiments, embryos were examined immediately prior to initiation of stress and were excluded if they exhibited developmental delay and/or malformations. This criterion was pre-established. Embryos with normal development and morphology were then assigned randomly to control or experimental protocols, and the order of injection of control and experimental embryos was alternated for successive experiments. After exposure to stress, embryo samples were labeled numerically and survival from each well was quantitated prior to assignment of identity. Each well contained a minimum of eight and a maximum of 12 embryos in order to prevent crowding, which can cause abnormal, delayed and/or asynchronous embryos. Biological duplicates were placed on opposite corners of plates in order to minimize edge or location effects in the hypoxia chamber.
3.1. Zebrafish gene annotation
Microarray probes were assigned to unique zebrafish cDNAs via BLAST search of publicly available databases (RefSeq, VEGA, Ensembl, ZGI, EST). Results were filtered to report only matches at > 99% identity and then prioritized in the above order such that manually curated and full-length sequences were favored when available. Zebrafish cDNAs were then assigned putative human matches via BLAST searches against the RefSeq database of human proteins, and results were filtered to obtain the top matching human protein. From each top match, full-length sequences were obtained and run through prediction algorithms (http://www.cbs.dtu.dk), including a TMHMM algorithm to predict transmembrane proteins (score = number of predicted transmembrane segments), as well as a SignalP algorithm to predict secreted proteins (score = number of algorithms out of 2 total predicting a signal peptide). In addition, links to the OMIM database were generated for each human protein when available. Genesets were downloaded from the Molecular Signatures Database (molsigdb version 5, http://www.broadinstitute.org/gsea/msigdb). Three sets were tested: (a) the complete database of 10,348 genesets, (b) 615 sets of transcription factor target motifs, and (c) 1454 sets of gene ontology (GO) terms. These genesets and the human proteins matching the top and bottom 300 array hits were tested for overlap, and sets with significant enrichment were identified using a hypergeometric distribution, and corrected for multiple tests via the Benjamini–Hochberg technique. The set of full annotation information for all 37,157 transcripts is provided in Supplemental Table 3.
4. Discussion
What happens when an organ or organism is stressed? The assumption in the field of molecular biology and developmental genetics has been that individual genes that are important for a particular biological process are differentially expressed under conditions relevant for that process. This “just in time” phenomenon of gene regulation has been validated at the systems level via genome-wide studies of mitosis, meiosis, and cellular stress in single-celled and multicellular animals using measures of both transcription and translation [10], [11], [12], [13], [14].
Given our interest in cellular mechanisms of protection against hypoxic stress and the success of the above genome-wide strategies, we performed a transcriptional study of responses to hypoxia during development in zebrafish to identify and functionally validate novel hypoxia protective factors and successfully identified irs2, crtc3, camk2g2, brt01, and ncam2 as novel hypoxia-protective genes [1]. Of these five genes, only irs2 has been demonstrated to be a target of the established hypoxia-inducible factor (HIF) pathway [15], [16], raising the interesting possibility that these new genes may be HIF-independent hypoxia protective factors. Future studies are indicated to determine whether these novel hypoxia-protective genes function via the established HIF pathway or via a HIF-independent mechanism, and whether other identified hypoxia-regulated genes that have not yet been functionally tested in our assay may also play hypoxia-protective roles.
Here we present additional details for sample preparation, sample processing, data analysis, and functional in vivo testing in the zebrafish developmental hypoxia model. In addition, we provide extensive annotation for each of the 37,157 transcripts measured in the microarray study, including homology to human and mouse proteins, human disease genes, and predicted functions.
The following is the supplementary data related to this article.
Top 300 Hypoxia-Upregulated Genes.
Top 300 Hypoxia-Downregulated Genes.
Full Annotation Table for 37,157 Transcripts on Zebrafish Microarray.
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Manchenkov T., Pasillas M.P., Haddad G.G., Imam F.B. Novel genes critical for hypoxic preconditioning in zebrafish are regulators of insulin and glucose metabolism. G3 Genes Genom. Genet. 2015 doi: 10.1534/g3.115.018010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. (PubMed PMID: 8589427) [DOI] [PubMed] [Google Scholar]
- 3.Bai X., Kim J., Yang Z., Jurynec M.J., Akie T.E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., Zhou Y., Grunwald D.J., Lin S., Cantor A.B., Orkin S.H., Zon L.I. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142(1):133–143. doi: 10.1016/j.cell.2010.05.028. (PubMed PMID: 20603019; PubMed Central PMCID: PMC3072682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Efron B., Tibshirani R., Storey J.D., Tusher V. Empirical Bayes analysis of a microarray experiment. J. Am. Stat. Assoc. 2001 [Google Scholar]
- 5.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. (Article3. PubMed PMID: 16646809) [DOI] [PubMed] [Google Scholar]
- 6.Smyth G.K. Limma: linear models for microarray data. In: Gentleman R., Carey V., Huber W., Irizarry R., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 7.Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A., Schier A.F. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–591. doi: 10.1101/gr.133009.111. (PubMed PMID: 22110045; PubMed Central PMCID: PMC3290793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulte-Merker S., Ho R.K., Herrmann B.G., Nüsslein-Volhard C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development. 1992;116(4):1021–1032. doi: 10.1242/dev.116.4.1021. [DOI] [PubMed] [Google Scholar]
- 9.Thisse C., Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. (PubMed PMID: 18193022) [DOI] [PubMed] [Google Scholar]
- 10.DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278(5338):680–686. doi: 10.1126/science.278.5338.680. (PubMed PMID: 9381177) [DOI] [PubMed] [Google Scholar]
- 11.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. (PubMed PMID: 9843569; PubMed Central PMCID: PMC25624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laub M.T., McAdams H.H., Feldblyum T., Fraser C.M., Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290(5499):2144–2148. doi: 10.1126/science.290.5499.2144. (PubMed PMID: 11118148) [DOI] [PubMed] [Google Scholar]
- 13.Arbeitman M.N., Furlong E.E., Imam F., Johnson E., Null B.H., Baker B.S., Krasnow M.A., Scott M.P., Davis R.W., White K.P. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297(5590):2270–2275. doi: 10.1126/science.1072152. (PubMed PMID: 12351791) [DOI] [PubMed] [Google Scholar]
- 14.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. (PubMed PMID: 19213877; PubMed Central PMCID: PMC2746483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi C.M., Finger E.C., Krieg A.J., Wu C., Diep A.N., LaGory E.L., Wei K., McGinnis L.M., Yuan J., Kuo C.J., Giaccia A.J. Cross-talk between hypoxia and insulin signaling through Phd3 regulates hepatic glucose and lipid metabolism and ameliorates diabetes. Nat. Med. 2013;19(10):1325–1330. doi: 10.1038/nm.3294. (PubMed PMID: 24037093; PubMed Central PMCID: PMC4089950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei K., Piecewicz S.M., McGinnis L.M., Taniguchi C.M., Wiegand S.J., Anderson K., Chan C.W., Mulligan K.X., Kuo D., Yuan J., Vallon M., Morton L.C., Lefai E., Simon M.C., Maher J.J., Mithieux G., Rajas F., Annes J.P., McGuinness O.P., Thurston G., Giaccia A.J., Kuo C.J. A liver Hif-2alpha-Irs2 pathway sensitizes hepatic insulin signaling and is modulated by Vegf inhibition. Nat. Med. 2013;19(10):1331–1337. doi: 10.1038/nm.3295. (PubMed PMID: 24037094; PubMed Central PMCID: PMC3795838) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top 300 Hypoxia-Upregulated Genes.
Top 300 Hypoxia-Downregulated Genes.
Full Annotation Table for 37,157 Transcripts on Zebrafish Microarray.