Abstract
We report draft genome sequence of Morganella sp. Strain SA36, isolated from water spring in Aljouf region, Saudi Arabia. The draft genome size is 2,564,439 bp with a G + C content of 51.1% and contains 6 rRNA sequence (single copies of 5S, 16S & 23S rRNA). The genome sequence can be accessed at DDBJ/EMBL/GenBank under the accession no. LDNQ00000000.
Keywords: Water springs, Morganella, Secondary metabolites, Heavy metals resistance, Whole genome sequencing
| Specifications | |
|---|---|
| Organism/cell line/tissue | Morganella sp. |
| Strain(s) | SA36 |
| Sequencer or array type | Sequencer; Roche 454 |
| Data format | Processed |
| Experimental factors | Microbial strains |
| Experimental features | Draft genome sequence of Morganella sp. SA36 |
| Assembly and annotation | |
| Consent | N/A |
| Sample source location | Water spring in Aljouf, Saudi Arabia |
1. Direct link to deposited data
2. Experimental design, materials and methods
Morganella sp. a motile gram-negative rod belonging to the family Enterobacteriaceae, has low pathogenicity, but compromised patients can develop diarrhoea, wound infections, urinary tract infections, bacteraemia, and sepsis due to Morganella morganii [1], [2]. Genomic DNA was extracted from pure culture of bacterial strain and subsequently sequenced using Roche 454 GS (FLX Titanium) pyrosequencing. All of the reads were assembled using GS De Novo Assembler version 2.8 (454 life science), which generated 1355 contigs with N50 2419 bp. The G + C content was calculated using the draft genome sequence. The G + C content for the draft genome is 51.1%. The genome contains 34 tRNA genes and 6 rRNA genes (5S-23S-16S) predicted by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP).
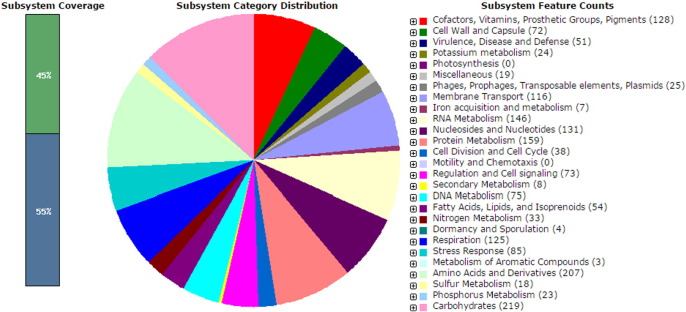
A total of 3229 protein coding sequences in 322 subsystems were functionally annotated by Rapid Annotation using the Subsystems Technology (RAST) [3] server (Fig. 1). Genome analysis revealed that the genome of Morganella sp. strain SA36 contains various gene clusters for biosynthesis of secondary metabolites and antimicrobial peptides. The draft genome was annotated using an automated annotation pipeline based on the Prodigal gene prediction algorithm [4], which predicted 2552 candidate protein-encoding gene models for strain SA36. The predicted CDSs were used to search the NCBI non-redundant database using BLAST tools [5]. Genome analysis revealed that the genome of Morganella sp. SA36 contains various gene clusters encoding putative virulence genes and heavy metal resistance proteins (cobalt, molybdenum, manganese and copper).
Fig. 1.
Subsystem distribution of Morganella strain SA36 (based on RAST annotation server).
Functional comparison of genome sequences in the RAST server revealed the closest neighbours of Providencia rustigianii DSM 4541 (score 549) followed by Providencia stuartii ATCC 25827 (score 276), Providencia alcalifaciens DSM 30120 (score 251), Providencia stuartii MRSN 2154 (score 245) and M. morganii subsp. morganii KT (score 150). On the other hand, analysis of the complete 1355 contig sequence in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch) under default settings (with matches only against cultured strains) M. morganii subsp. morganii KT. Overall the various in silico results confirmed that the present environmental isolate is a member of the genus Morganella, though further characterization work is required to determine its species.
3. Nucleotide sequence accession number
The Morganella sp. SA36 whole genome shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession no LDNQ00000000.
Conflict of interest
The authors declare that there is no conflict of interests on the work published in this paper.
Acknowledgements
This work was funded by the Deanship of Scientific Research (DSR), Aljouf University, Aljouf, KSA, under grant no. (34/155, 2014).The authors, therefore, acknowledge with thanks DSR technical and financial support. We also would like to acknowledge ArrayGen Technologies, Pune, India for contributing to data analysis and bioinformatics support.
References
- 1.Chang H.Y., Wang S.M., Chiu N.C., Chung H.Y., Wang H.K. Neonatal Morganella morganii sepsis: a case report and review of the literature. Pediatr. Int. 2011;53:121–123. doi: 10.1111/j.1442-200X.2010.03241.x. [DOI] [PubMed] [Google Scholar]
- 2.Tucci V., Isenberg H.D. Hospital cluster epidemic with Morganella morganii. J. Clin. Microbiol. 1981;14:563–566. doi: 10.1128/jcm.14.5.563-566.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]