Abstract
We present the annotation of the draft genome sequence of Serratia sp. strain TEL (GenBank accession number KP711410). This organism was isolated from entomopathogenic nematode Oscheius sp. strain TEL (GenBank accession number KM492926) collected from grassland soil and has a genome size of 5,000,541 bp and 542 subsystems. The genome sequence can be accessed at DDBJ/EMBL/GenBank under the accession number LDEG00000000.
Keywords: Serratia, Entomopathogenic nematodes, Whole-genome sequencing, Annotation
| Specifications | |
|---|---|
| Organism | Serratia sp. |
| Strain | TEL |
| Sequencer or array type | Sequencer; Illumina MiSeq |
| Data format | Processed |
| Experimental factors | Microbial strains |
| Experimental features | Draft genome sequence of Serratia sp. TEL, assembly and annotation |
| Consent | N/A |
| Sample source location | Grassland in Suikerbosrand Nature Reserve near Johannesburg in South Africa |
1. Direct link to deposited data
2. Experimental design, materials and methods
Entomopathogenic nematodes (EPNs) are soil dwelling insect-killing microscopic worms which have been studied as potential insect pest biological control agents across the world [1]. The non-feeding infective juvenile (IJ) stage functions as vectors for insect pathogenic symbiotic enterobacteria. After the IJs have infected the insect host the bacteria are regurgitated from the alimentary into the insect's haemolymph. The bacteria release antimicrobials that suppress opportunistic colonialization by microbial saprophytes and lethal insecticidal toxins which kill the insect within 48–72 h after IJ ingress into the haemolymph [2]. EPNs belonging to Heterorhabditis, Steinernema, and Oscheius genera have evolved symbiotic associations with Photorhabdus, Xenorhabdus, and an insect pathogenic species of Serratia, respectively [3], [1], [4]. Not all species or strains of bacteria in the genus Serratia are pathogens of insects [5]. An insect pathogenic strain of Serratia marcescens has also been found to be associated with Caenorhabditis briggsae and the IJs of this nematode are able to infect larvae of Galleria mellonella [6]. Similarly, in other studies, insect pathogenic bacteria belonging to the genus Serratia have been found to be symbiotically associated with Oscheius carolinensis and this nematode is also capable of acting as a vector for the bacteria by infecting insect hosts [1].
In this paper we discuss Serratia sp. TEL (GenBank accession KP711410) and several functional features identified from the annotation results. Serratia sp. TEL (Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Serratia) is a Gram-negative, non-spore forming bacterium. This bacterium is symbiotically associated as an insect pathogen with a novel entomopathogenic nematode species Oscheius species TEL-2014 (GenBank accession number KM492926).
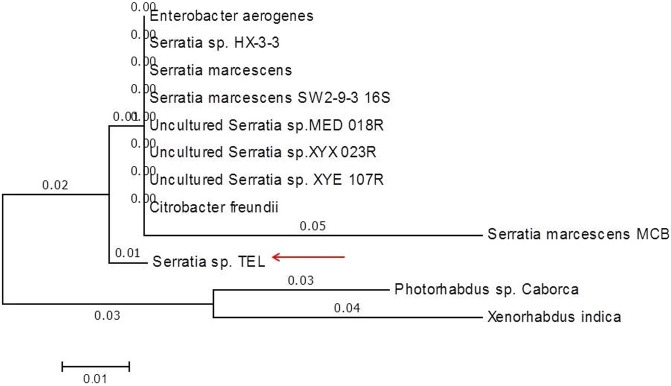
To isolate the bacterial pathogen, non-feeding infective juveniles were surface sterilized with 0.1% sodium hypochlorite for 4 h and rinsed with autoclaved distilled water. Sterile IJs were carefully suspended in 1 ml of sterile nutrient broth in a microtube and homogenized with a small sterile plastic pestle. The homogenate was streaked onto NBTA and McConkey agar plates and incubated at 25 °C for 24 h in the dark. DNA was extracted from the bright green colonies and pink colonies, pure cultures obtained from the selective media and genomic DNA was extracted using the ZR bacterial DNA miniprep kit (Zymo Research). A polymerase chain reaction was employed to amplify the 16S rRNA gene sequence of the isolated bacterial DNA using EUB968 forward primer 5′-ACGGGCGGTGTGTC-3′ Tm (°C) = 62 and UNIV1382 reverse primer 5′-AACGCGAAGAACCTTAC-3′ Tm (°C) = 66. The same primers were used for the sequencing of this gene. The sequence obtained was subjected to NCBI BLAST under the default settings for highly similar alignments. The analysis revealed that among all the matches for the 16S rRNA gene sequences, the unknown sequence had a high affinity to a novel Serratia species which was then assigned the name Serratia sp. strain TEL. Other high scoring bacteria 16S rRNA sequences were downloaded from GenBank to be used for phylogenetic analysis. The evolutionary relatedness of Serratia sp. strain TEL with several Serratia species was based on the 16S rRNA ITS regions using the Maximum Likelihood method based on the Tamura–Nei model using MEGA6. The bootstrap consensus tree was inferred from 1000 replications and the tree was drawn to scale, with branch lengths measuring the number of substitutions per site (next to the branches) as shown in Fig. 1. Bacteria belonging to the well-known genera Photorhabdus and Xenorhabdus were used as possible out-groups to root the tree and see if there is any significant relationship they might have with the identified Serratia species.
Fig. 1.
The evolutionary history of Serratia sp. TEL (pointed with a red arrow) inferred by using the Maximum Likelihood method based on the Tamura–Nei model on MEGA6 software. The bootstrap consensus tree inferred from 1000 replications.
The whole genome was shotgun sequenced on the Illumina Miseq, following sequence quality control using Fastq-mcf toolkit and de novo assembly using SPADES were performed as described in [7]. A draft annotated genome of this organism was constructed using the best-placed reference protein set; GeneMarkS + annotation method set on the NCBI Prokaryotic Genome Automatic Annotation Pipeline and has been reported in [7]. Furthermore, the functional annotation was carried out by RAST (Rapid Annotation using Subsystem Technology) hosted by Fellowship for Interpretation.
RAST revealed the closest neighbours of Serratia sp. TEL as S. marcescens Db11 (reference genome id 615.1), followed by Serratia proteamaculans 568 (399741.3) and Serratia plymuthica A30 (1206776.3).
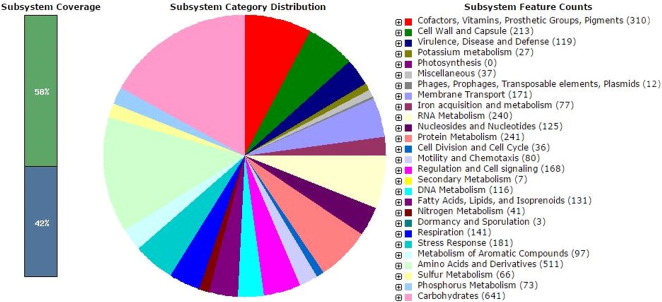
RAST identified a total of 4618 coding regions and 103 RNAs. Fig. 2 shows that the annotated genome has 119 genes potentially involved in virulence, disease and defence including 88 genes for resistance to antibiotics and toxic compounds such lysozyme inhibitors, multiple antibiotic resistance (MAR) locus, beta lactamase, copper homeostasis genes, streptothricin resistance genes, multidrug resistance efflux pumps, multidrug resistance tripartite systems found in Gram negative bacteria and resistance to chromium compounds. An example of a gene involved in stress response, virulence and defence is 16 kDa heat shock protein A and will be briefly discussed.
Fig. 2.
The subsystem distribution of Serratia sp. TEL based on RAST annotation.
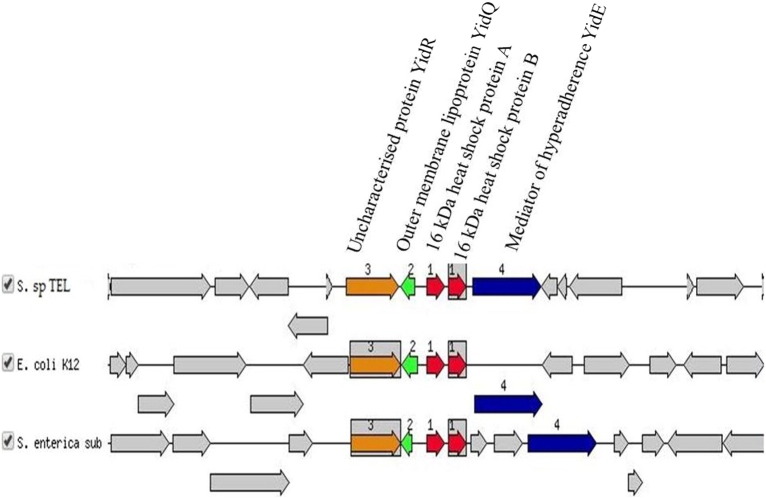
The focus gene coding for a 16 kDa heat shock protein A is involved in stress response, virulence and defence was compared with two similar organisms. The graphic is centred on the focus gene, which is red and numbered 1. Sets of genes with similar sequence are grouped with the same number and colour. Genes whose relative position is conserved in at least two other species are functionally coupled and share grey background boxes. The focus gene always points to the right, even if it is located on the minus strand.
It was found that some of the genes involved in virulence, disease and defence as shown in Fig. 3 such as the 16 kDa heat shock protein B and the outer membrane lipoprotein YidQ which were closely associated with genomic regions containing 16 kDa heat shock protein A and were also similarly arranged together in Serratia sp. TEL, Escherichia coli K12 and S. enterica subsp. enterica serovar Choleraesuis strain SC-B67. Additionally, the annotated genome has 80 genes involved in motility and chemotaxis in which 24 of these genes encode for bacterial chemotaxis and 56 of them encode for flagellum motility.
Fig. 3.
Comparative analysis of genomic regions containing 16 kDa heat shock protein A found in Serratia sp. TEL to Escherichia coli K12 and Salmonella enterica subsp. enterica serovar Choleraesuis strain SC-B67.
Genetic subsystems discussed were highlighted as important because their presence contributed to the insect pathogenicity of the bacterial species for which the nematode acted as the vector for the location and infection of potential insect hosts.
3. Nucleotide sequence accession numbers
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LDEG00000000.
Conflict of interest
The authors declare that there is no conflict of interest on any work published in this paper.
Acknowledgements
Thanks to Gauteng Department of Agriculture and Rural Development (GDARD) (GDARO 12 TS) for funding the research project. Thanks also for the Innovation Doctoral Scholarship from the NRF National Research Foundation (NRF) with the grant number [SFH1208147793] and the Wits Postgraduate Merit Award (PMA) from the University of the Witwatersrand which supported the first author. Thanks to the Agricultural Research Council (ARC) Biotechnology platform for Illumina technology sequencing services.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2015.08.010.
Appendix A. Supplementary data
The following is the supplementary data to this article.
Supplementary figure.
References
- 1.Torres-Barragan A., Suazo A., Buhler W.G., Cardoza Y.J. Studies on the entomopathogenicity and bacterial associates of the nematode Oscheius carolinensis. Biol. Control. 2011;59:123–129. [Google Scholar]
- 2.Malan A.P., Nguyen K.B., Addison M.F. Entomopathogenic nematodes (Steinernematidae and Heterorhabditidae) from the southwestern parts of South Africa. Afr. Entomol. 2006;12:65–69. [Google Scholar]
- 3.Stuart R.J., Barbercheck M.E., Grewal P.S., Taylor R.J., Hoy W.C. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. Microbiololgy. 2006;121:303–309. [Google Scholar]
- 4.Zhang C., Liu J., Xu M., Sun J., Yang S., An X., Gao G., Lin M., Lai R., He Z., Wu Y., Zhang K. Heterorhabditidoides chongmingensis gen. nov., sp. nov. (Rhabditida: Rhabditidae), a novel member of the entomopathogenic nematodes. J. Invertebr. Pathol. 2008;98:153–168. doi: 10.1016/j.jip.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Kwak Y., Khan A.R., Shin J. Genome sequence of Serratia nematodiphila DSM 21420T, a symbiotic bacterium from entomopathogenic nematode. J. Biotechnol. 2015;193:1–2. doi: 10.1016/j.jbiotec.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Abede E., Jumba M., Bonner K., Gray V.M., Morris K., Thomas W.K. An entomopathogenic Caenorhabditis briggsae. J. Exp. Biol. 2010;213:3223–3229. doi: 10.1242/jeb.043109. [DOI] [PubMed] [Google Scholar]
- 7.Lephoto T.E., Featherston J., Gray V.M. Draft whole-genome sequence of Serratia sp. strain TEL, associated with Oscheius sp. TEL-2014 (Nematoda: Rhabditidae) isolated from a grassland in South Africa. Genome Announc. 2015;3(4):e00747-15. doi: 10.1128/genomeA.00747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure.