Abstract
Autism spectrum disorder (ASD) is a clinically complex and heterogeneous disorder. It is characterized by impaired social abilities, disordered language, isolated areas of interest, and repetitive behaviors. Evidence suggested that the neuropathology of ASD is widely distributed, involving epigenetic regulation in the brain. MiRNAs are a group of endogenous non-coding RNAs that play a critical role in neurodevelopment, neuroplasticity, and other fundamental neurobiological processes. To study miRNA profiling in Autism spectrum disorder in China, we performed miRNA microarray followed quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Here, we describe detailed methods and analysis on these microarray data which has been deposited in Gene Expression Omnibus (GEO): GSE67979.
Keywords: MiRNA, MiRNA microarray, qRT-PCR
1. Direct link to deposited data
The deposited data can be found at: http://www.ncbi.nlm.nih.gov/pubmed/?term=GSE67979.
2. Experimental design, materials and methods
2.1. Sample collection and RNA isolation
We totally recruited 20 patients and 20 controls. Peripheral blood samples were obtained from patients and controls at Xiangya Hospital [1]. Total RNA was harvested by the TRIzol (Life Technologies) according to the manufacturer's instructions [2]. The quality and quantity of RNA were examined using an Agilent Bioanalyzer (Santa Clara) and K5500 micro-spectrophotometer (Kaiao). The total RNA was saved at − 80 °C until use.
2.2. miRNA expression profiling by microarray
RiboArray™ miDETECT™ Human Array microarrays (RIBOBIO), which contained 2578 assay probes corresponding to the entire set of primate miRNAs, were used to screen the miRNA expression. Microarray was incubated 10 min in 65 °C followed by 1 h in 37 °C for prehybridization. 2.5 μg total RNA of each sample was labeled by Cy5. The Cy5-labeled RNA samples could be readily visualized with comparable intensity under UV. Labeling efficiency (1.0 to 3.6 was accepted for good microarray result) can be calculated by the concentration of CyDye and RNA measured by K5500 micro-spectrophotometer. Cy5-labeled RNA was denaturated 3 min in hybridization solution, incubated 20 s on ice, and hybridized to microarray 16 h in 37 °C. All microarrays were successively washed by 6 × SSPET, 3 × SSPET, 0.5 × SSPET, and 0.5 × SSPET. Then, developer and coverslip were plus for scan (GenePix 4000B laser scanner), followed by bioinformatic analysis (digitized using the R software). Fig. 1 showed part of the signal collection. Fig. 2 showed the log2 scale of the expression signal values that were plotted, including control probes.
Fig. 1.
Part of the signal collection.
Fig 2.
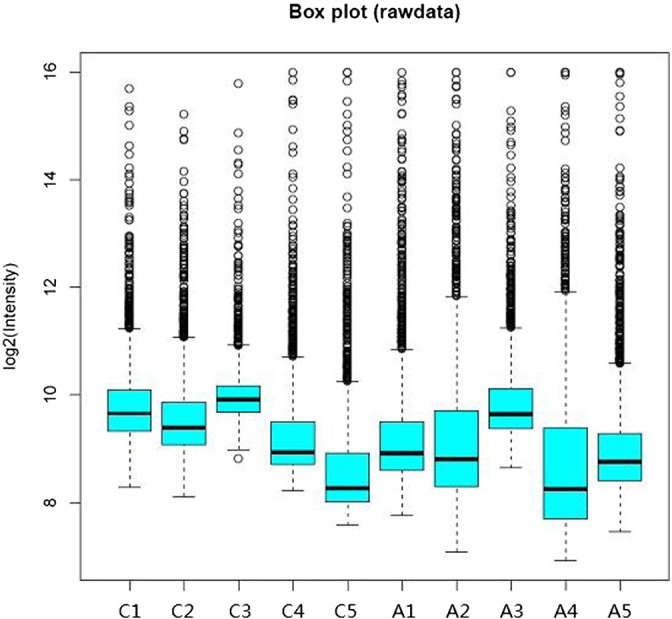
Box plot of raw data.
Note: C1-5 means controls, A1-5 means patients.
2.3. miRNA qRT-PCR analysis
Reverse transcription of total RNA to cDNA was carried out in a 20 μL reaction volume using RevertAid First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions. Quantitative PCR assays (CFX96, Bio-Rad) was carried out in a 20 μL reaction volume containing 1 μL cDNA in the 15 autism patients and 15 health controls. It was programmed for an initial denaturation step (95 °C, 3 min) followed by 40 amplification cycles (95 °C, 10 s; 60 °C, 20 s). The miRNA qRT-PCR primers were provided by RiboBio (Guangzhou, China) [3]. Has-miR-16-5p was used as an internal reference in the qRT-PCR experiments [4], [5]. Differences between control and experimental samples were calculated using the 2 − ΔΔCt method.
3. Discussion
We have studied miRNA profiling of ASD by using RiboArray™ miDETECT™ Human Array (A10101-1-12-19, 1 × 12K). Here, we describe the detailed steps of the microarray analysis. Part of the signal collection and box plot of raw data were provided. As a result, several aberrantly expressed miRNAs related to ASD were discovered. This study confirmed that using miRNA array to study the easily accessible peripheral blood is valid and repeatable [6], [7], [8].
Conflict of interest
The authors declare no conflict of interests.
Acknowledgements
This study was supported by the National Basic Research Program (973 Program) (Nos. 2012CB944601, 2012CB517902 and 2011CB510002 to Hong Jiang), the National Natural Science Foundation of China (Nos. 81410308019, 81471156, 81271260, 30971585 to Hong Jiang; No. 31401135 to Rong Qui), Hunan Funds for Distinguished Young Scientist (No. 14JJ1008 to Hong Jiang), Xinjiang Natural Science Foundation (No. 201318101-4 to Hong Jiang), Natural Science Foundation of Hunan Province (No. 2012SK3194 to Wei Zhuang), the Undergraduate Innovation Project of Central South University (No. YB13028, 201410533324 to Hong Jiang), Graduate Innovation Project of Central South University (No. 2014zzts078 to Zhao Chen), Research project of the First People's Hospital of Chenzhou (No. N2014-017, to Fengzhen Huang) and High-level medical personel of Hunan province “225” Project.
References
- 1.Shi Y., Huang F., Tang B., Li J., Wang J., Shen L., Xia K., Jiang H. MicroRNA profiling in the serums of SCA3/MJD patients. Int. J. Neurosci. 2014;124(2):97–101. doi: 10.3109/00207454.2013.827679. [DOI] [PubMed] [Google Scholar]
- 2.Huang F., Zhang L., Long Z., Chen Z., Hou X., Wang C., Peng H., Wang J., Li J., Duan R., Xia K., Chuang D.M., Tang B., Jiang H. miR-25 alleviates polyQ-mediated cytotoxicity by silencing ATXN3. FEBS Lett. 2014;588(24):4791–4798. doi: 10.1016/j.febslet.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y.Q., Zhang M.F., Wen H.Y., Hu C.L., Liu R., Wei H.Y., Ai C.M., Wang G., Liao X.X., Li X. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics. 2013;68(1):75–80. doi: 10.6061/clinics/2013(01)OA12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroh E.M., Parkin R.K., Mitchell P.S., Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song J., Bai Z., Han W., Zhang J., Meng H., Bi J., Ma X., Han S., Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012;57(4):897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 6.Abu-Elneel K., Liu T., Gazzaniga F.S., Nishimura Y., Wall D.P., Geschwind D.H., Lao K., Kosik K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9(3):153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 7.Ghahramani Seno M.M., Hu P., Gwadry F.G., Pinto D., Marshall C.R., Casallo G., Scherer S.W. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380:85–97. doi: 10.1016/j.brainres.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 8.Goyal D.K., Miyan J.A. Neuro-immune abnormalities in autism and their relationship with the environment: a variable insult model for autism. Front. Endocrinol. 2014;5:29. doi: 10.3389/fendo.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]