Abstract
In order to analyze the production of small RNA (sRNA) by viroids upon infecting the plants, the tomato plants (Solanum lycopersicum cultivar Rutgers) were inoculated with the variants of Potato spindle tuber viroid (PSTVd). After 21-days of postinoculation, total RNA was extracted and subjected for deep-sequencing using Illumina HiSeq platform. The primers were trimmed and only 21- to 24-nt long sRNAs were filtered after quality check of the raw data. The filtered sRNA population was then mapped against both the genomic (+) and antigenomic (−) strands of the respective PSTVd variants using standard pattern-matching algorithm. The profiling of viroid derived sRNA (vd-sRNA) revealed that the viroids are susceptible to host RNA silencing mechanism. High-throughput sequence data linked to this project have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE69225.
Keywords: Viroids, Potato spindle tuber viroid, Viroid derived small RNA, RNA silencing
| Specifications | |
|---|---|
| Organism | Solanum lycopersicum cultivar Rutgers (tomato, cv. Rutgers) |
| Tissue | Tomato plant leaf |
| Sequencer or array type | Illumina HiSeq |
| Data format | Processed data in .txt |
| Experimental factors | Control vs PSTVd infected |
| Experimental features | Comparison of viroid derived small RNA |
| Sample source location | Hirosaki, Japan |
1. Direct link to deposited data
2. Experimental design, materials and methods
Viroids are the non-coding, single stranded, circular RNAs molecules with sizes range of 246 to 401 nucleotides (nt). They can infect certain plants. Recent findings, however, show that viroid infection is associated with the appearance of viroid-derived small RNA (vd-sRNA). These vd-sRNAs have sizes similar to endogenous small interfering RNA and microRNA (miRNA) indicating the possibility of connection of vd-sRNA to the induced symptoms. Interestingly the symptoms vary dramatically, depending on both the plant cultivar and the viroid strain [1].
In order to verify the production of small RNA (sRNA) by the PSTVd variants: PSTVd-mild (PSTVd-M, synonym PSTVd-Dah; GenBank Acc. No. AB623143) and PSTVd-intermediate (PSTVd-I; GenBankAcce. No. AY937179) upon infection, the tomato plants were inoculated with the respective viroid variants as described previously [2]. Mock inoculated plants were used as control. At 21-days postinoculation (dpi) leaf samples were collected and subjected to total RNA extraction using the mirVana™ miRNA isolation kit (Ambion, Austin, TX, USA), with slight modifications. Briefly, 3 to 4 leaf discs were homogenized with 400 μL of lysis/binding buffer. Then, 60 μL of miRNA homogenate additive was added. The resulting solution was purified using acid phenol:chloroform (5:1) followed by DNase I (Promega, Madison, USA) treatment. The RNA was precipitated by adding 2.5 vol of absolute alcohol. RNA integrity was examined in a 2100 Bioanalyzer (Agilent Technologies, California, USA). The total RNA obtained was subjected for northern blot analysis in order to confirm the accumulation of PSTVd-specific sRNA as described previously [2]. By referring to the RNA size marker, a gel slice containing the sRNAs of 15 to 50 nucleotides were excised and eluted from gel using 1 mL gel elution buffer (0.5 M ammonium acetate, 1 mM EDTA [pH 8.0], 0.1% (w/v) SDS) as described previously [3]. All the recovered sRNAs were sent to Hokkaido System Science Co. (Sapporo, Japan) for deep-sequencing using a Genetic Analyzer IIx platform (Illumina, San Diego, CA, USA).The samples were quantified in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and were processed simultaneously in the Illumina system using an index-sequence. The adapter sequences were trimmed from the ends of the resulting raw data. Filtered sequences where re-grouped after eliminating the sequence reads less that 21 nt and longer that 24 nt [3]. Approximately 4.3 million reads of sRNA were obtained from the leaf samples of mock inoculated (GSM1695655), PSTVd-M (GSM1695656) and PSTVd-I (GSM1695657) inoculated plants.
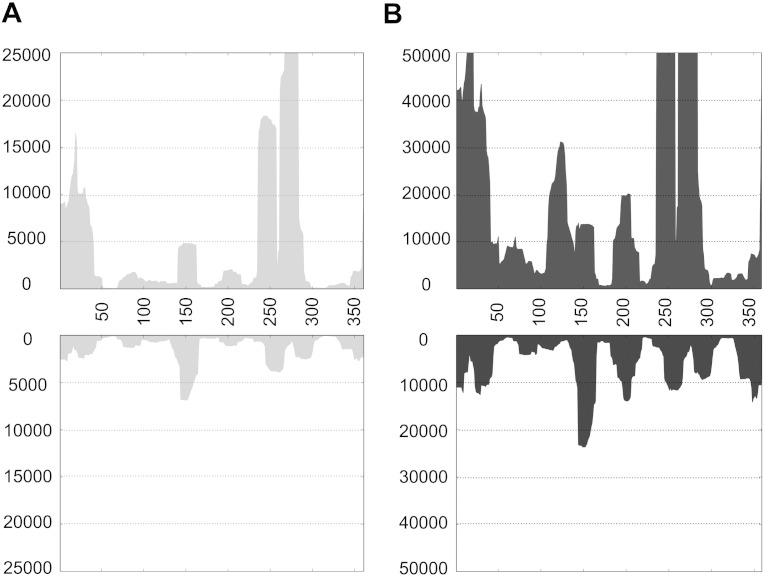
Sequence analysis of over 4.28 million sRNAs obtained from mock inoculated plants identified about 108 and 106 viroid specific small RNAs (vd-sRNA) of PSTVd-M and PSTVd-I type, respectively, matching the genomes of PSTVd-M and PSTVd-I. Analysis of 4.54 million sRNA reads obtained from the PSTVd-M inoculated plants against both the (+) and (−) strands of PSTVd-M revealed 103,933 vd-sRNAs. Similarly, analysis of 4.88 million reads obtained from the PSTVd-I inoculated plants against both the (+) and (−) strands of PSTVd-I underscored 488,146 vd-sRNAs. That said, PSTVd-I inoculated plants showed more vd-sRNA (11.3%) than PSTVd-M (2.3%) inoculated plants. This difference in the vd-sRNA recovery can be attributed to the lower accumulation of PSTVd-M compared to PSTVd-I, as described previously [2]. All 21- to 24-nt long vd-sRNAs were profiled on both polarity strands of the respective PSTVd variants using the standard pattern-matching algorithm in order to understand the production of the vd-sRNAs (Fig. 1). Although PSTVd-M and PSTVd-I produced different amount of sRNAs, both showed similar vd-sRNA profile. Further, profiling of vd-sRNA on the PSTVd genome revealed that certain region of PSTVd produced more sRNA than others, indicating that these regions are more susceptible to RNA silencing.
Fig. 1.
Sequence profiles of the genomic (+) and the antigenomic (−) PSTVd-sRNA populations recovered from the leaf tissues of infected tomato plants. (A) and (B) represent the profiles of the sRNAs derived from the (+) and the (−) strands of the PSTVd-M and PSTVd-I, respectively. Please note that different scales are used so as to compensate for the lower numbers of sRNA sequences recovered for the PSTVd-M infected plants.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC, grant number 155219-12) to JPP. The RNA group is supported by grants from the Université de Sherbrooke. JPP holds the Research Chair of Université de Sherbrooke in RNA Structure and Genomics and is a member of the Centre de Recherche du CHUS. TS received a grant from the Japan Society for the Promotion of Science KAKENHI (Grant No. 24380026 and 15H04455-0001). The funders had no role in study design, data collection and analysis, decision to publish or in the preparation of the manuscript.
References
- 1.Hammann C., Steger G. Viroid-specific small RNA in plant disease. RNA Biol. 2012;9:809–819. doi: 10.4161/rna.19810. [DOI] [PubMed] [Google Scholar]
- 2.Tsushima D., Adkar-Purushothama C.R., Taneda A., Sano T. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild isolates of potato spindle tuber viroid. J. Gen. Plant Pathol. 2015;81:49–62. [Google Scholar]
- 3.Adkar-Purushothama C.R., Zhang Z., Li S., Sano T. Analysis and application of viroid-specific small RNAs generated by viroid-inducing RNA silencing. Methods Mol. Biol. 2015;1236:135–170. doi: 10.1007/978-1-4939-1743-3_12. [DOI] [PubMed] [Google Scholar]