Abstract
In response to muscle damage the muscle adult stem cells are activated and differentiate into myoblasts that regenerate the damaged tissue. We have recently showed that following myopathic damage the level of the Runx1 transcription factor (TF) is elevated and that during muscle regeneration this TF regulates the balance between myoblast proliferation and differentiation (Umansky et al.). We employed Runx1-dependent gene expression, Chromatin Immunoprecipitation sequencing (ChIP-seq), Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) and histone H3K4me1/H3K27ac modification analyses to identify a subset of Runx1-regulated genes that are co-occupied by the TFs MyoD and c-Jun and are involved in muscle regeneration (Umansky et al.). The data is available at the GEO database under the superseries accession number GSE56131.
Keywords: Runx1 transcription factor, Runx1-mediated transcription program in muscle regeneration, Genome-wide expression profile
| Specifications | |
|---|---|
| Organism/cell line/tissue |
In vitro experiments: Microarray, ChIP and ATAC-sequencing: primary myoblasts (PMs) In vivo experiments: RNA-sequencing: Soleus muscles |
| Sex | PM and Soleus muscle samples were derived from female and male mice, respectably. |
| Sequencer or array type | Illumina HiSeq 2500; Affymetrix Mouse Gene 1.0 ST microarrays |
| Data format | Microarray: Raw: CEL files ChIP and ATAC-sequencing: Raw: SRA files Analyzed: bed files (peak files) RNA-sequencing: Raw: SRA files Analyzed: csv file (DESeq2 differential expression) |
| Experimental factors | Transcriptome using microarrays: 4 cultures of Runx1L/L[1] and Runx1f/f derived PMs were used for RNA purification and transcriptome analysis. ChIP- and ATAC-sequencing: 2 wild type PM cultures were used for specific antibodies (Abs) and relevant controls (non-immune serum or specie-specific IgG). RNA-sequencing: 4 Soleus muscle samples were extracted from P60 mdx/Runx1L/L and mdx/Runx1f/f mice [1]. |
| Experimental features | Transcriptome analysis was performed comparing Runx1f/f PMs to Runx1L/L control in vitro, and mdx/Runx1f/fvs. mdx/Runx1L/Lin vivo; TF (Runx1, MyoD and c-Jun) binding and enhancer markers were examined in WT PMs. |
| Consent | The experiments were conducted in strict accordance with the recommendations of the US National Institutes of Health Guide for the Care and Use of Laboratory Animals [1]. |
| Sample source location | N.A. |
1. Direct link to deposited data
The data is available at the GEO database under:
2. Experimental design, materials and methods
2.1. Experimental groups and design
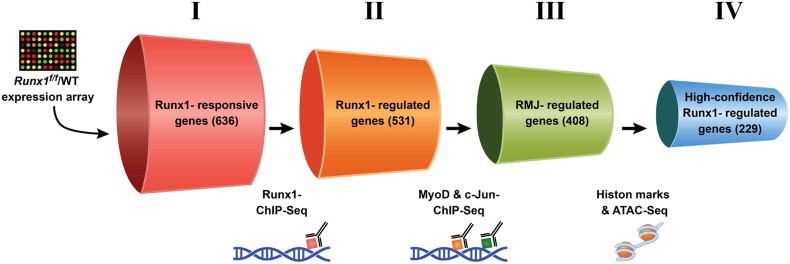
To elucidate the Runx1-mediated myoblast transcriptional program during muscle regeneration, we employed genetically modified Runx1f/f and mdx/Runx1f/f mice [1]. The experimental design is schematically described in Fig. 1: First, we derived the Runx1-responsive genes by comparing the Runx1f/f PM transcriptome to that of Runx1L/L PMs. Next, we defined the Runx1-regulated gene subset by cross-analyzing the Runx1-responsive gene subset with genome-wide Runx1 ChIP-seq data in wild type PMs. To further characterize this Runx1-regulated gene subset we singled out Runx1-bound gene loci that were co-bound by Runx1 transcriptional collaborators MyoD and c-Jun (Fig. 1). Finally, we characterized the open/active chromatin by genome-wide mapping of active enhancer markers (H3K4me1, H3K27Ac) and ATAC-seq analyses. The combination of this comprehensive analysis generated a list of high-confidence Runx1-regulated genes. The expression profile of this high-confidence was validated in vivo using RNA-seq of RNA derived from muscles of mdx/Runx1f/f vs. mdx/Runx1L/L mice [1].
Fig. 1.
Experimental design.
Schematic representation of the selection procedures used to identify high-confidence Runx1-regulated genes (adapted from Umansky et al. [1]). Each cylinder represents a gene subset, with the gene number given in brackets. I — Runx1-responsive genes were derived from Runx1L/Lvs. Runx1f/f PM microarray expression data [1]. II — Runx1-regulated genes were derived by cross-analysis of the Runx1-responsive gene dataset and Runx1 ChIP-seq data. This gene subset represents Runx1-responsive genes that are also occupied by Runx1. III — RMJ-regulated genes are Runx1-responsive genes that are co-occupied by Runx1, MyoD and c-Jun. IV — High-confidence Runx1-regulated gene subset is RMJ-regulated genes that were also marked as having adjacent active regulatory elements by both anti-histone modifications (H3K4me1 & H3K27ac) ChIP-seq and ATAC-seq analysis.
2.2. RNA purification
PM cultures were established as previously described [1], [2]. For transcriptome analysis, 1e6–5e6 PMs were collected after three stages of myoblast enrichment using pre-plating, a total of 6 days post-muscle extraction. To avoid RNA degradation, the cells were washed twice with cold PBS and then subjected to flash-freezing in liquid nitrogen. RNA was isolated by the PerfectPure RNA tissue kit (# 2302410, 5 PRIME, Germany) according to the manufacturer's instructions (cell culture protocol), using a rotor–stator as the disruption method (Omni-TH 02, Omni international, USA). For RNA-seq, Soleus muscles were harvested from 2 month old mice, and RNA was isolated as described above (tissue protocol, Proteinase K added).
2.3. Transcriptome analysis
For microarray analysis, purified total RNA was reverse-transcribed, amplified, and labeled with the Affymetrix GeneChip whole transcript sense target labeling kit. Labeled cDNA was analyzed using Affymetrix Mouse Gene 1.0 ST microarrays, according to the manufacturer's instructions. Microarray data were analyzed using the Partek Genomic Suite software. CEL files (containing raw expression measurements) were imported and data was preprocessed and normalized using the Robust Multichip Average (RMA) algorithm [3]. To identify differentially expressed genes ANOVA was applied and gene fold-changes were calculated. For RNA-seq analysis, purified total RNA was subjected to Illumina TruSeq®. RNA Sample Preparation v2 was used according to the manufacturer's instructions. Indexed samples were sequenced in an Illumina HiSeq 2500 machine in a single read mode. The obtained reads, 50 bp long, were mapped to the mm9 mouse genome assembly using TopHat2 [4] version 2.0.12.0.10 with default options. Expression at the gene level was quantified by HTSeq (version 0.6.1) [5], and using the known genes from the UCSC browser in General Feature Format (GFT) as annotation. Differential expression was calculated utilizing the DESeq2 software (version 1.2.10) [6].
2.4. ChIP-seq analysis
For ChIP, we used WT PM cultures similar to those described above in Section 2.2. Crosse-linked chromatin from 1.2e8 cells (Runx1 ChIP), 6e7 cells (MyoD and c-Jun ChIP) and 1e7 cells (H3K4me1 and H3K27Ac ChIP) was prepared and fragmented to an average size of approximately 200 bp by 20–35 cycles of sonication (30 s each) in 15-ml tubes using the Bioruptor UCD-200 sonicator (Diagenode, USA). Relevant antibodies and controls are described in the Materials and methods section of [1]. DNA was purified using QIAquick spin columns (QIAGEN) and sequencing was performed using Illumina HiSeq 2500. Two biological repeats were conducted and separately sequenced for each ChIP-seq experiment. For ChIP-seq analysis, the reads were aligned to the mouse genome (mm9) allowing one mismatch and using the Bowtie aligner [7]. Reads with a unique best alignment were retained for further processing. Immunoprecipitated samples were compared against the negative control to find binding sites using the MACS2 software with the callpeak function and default parameters. The broad peak setting was used only for the histone marks [8].
2.5. ATAC-seq analysis
ATAC was performed as previously described [9]. Briefly, 5 × 104 PMs were harvested, and underwent the recommended transposition protocol without the lysis stage. The resulting transposed DNA was enhanced using 12 cycles of PCR, as described. The resulting libraries were sequenced using Illumina HiSeq 2500. For ATAC-seq analysis, we obtained paired-end reads of 50 bp length. Adapters were trimmed from the reads, Bowtie2 was run as previously described for ChIP-seq analysis, duplicated reads were removed using the Picard MarkDuplicates function module, and MACS2 was applied as detailed above for ChIP-seq analysis (Section 2.4).
2.6. Functional analysis
The Ingenuity Pathway Analysis tool (https://apps.ingenuity.com/) was used for pathway and bio-function annotation of Runx1-regulated genes and the GREAT software [10] was used for ChIP-seq peak Gene Ontology (GO) analysis. The GSEA software [11] was used to perform gene set enrichment analysis on groups of genes.
3. Results & discussion
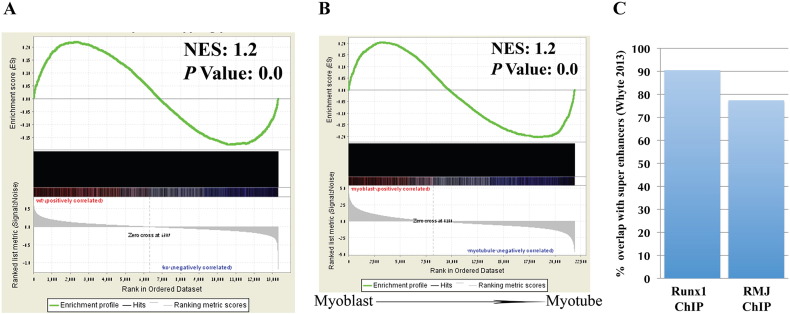
Skeletal muscle is a highly regenerative tissue. Upon muscle injury, the adult stem cells the satellite cells (SCs) are activated and differentiate and fuse to create myofibers that regenerate the tissue. While not expressed in healthy muscle tissue, expression of TF Runx1 significantly increases in response to various types of muscle damage including neuropathic damage [12], [13] and myopathic damage [1]. Specifically, we have recently shown that Runx1 prevents premature differentiation of SC-derived myoblasts during muscle regeneration [1]. In the myoblastic cell line C2C12 Runx1 was co-bound with the major myogenic factor, MyoD [14]. Using in vivo loss- and gain-of-function models and genome-wide transcriptional analyses we identify a subset of Runx1-regulateed genes that participate in muscle regeneration. Significantly, Runx1 transcriptome and ChIP-seq analysis show good correlation (Fig. 2A). Our ChIP-seq data also showed correlation with myoblast-specific transcriptional-profiling data. For example, we found that the proximity of myoblast-specific expressed genes [15] was highly enriched for co-binding of Runx1, MyoD and c-Jun (RMJ) (Fig. 2B). In addition, analysis of myoblast-specific Super-enhancers [16], revealed a strong overlap with our ChIP-seq data in sites regulating myoblast fate (Fig. 2C). Interestingly, Runx1 is involved in cell fate decision of additional adult stem cells populations, including hair follicle stem cells [17], and differentiation of mesenchymal stem cells into myofibroblasts [18]. It is conceivable that common transcription regulatory mechanisms are shared by the three systems. In summary, our data suggests that Runx1 regulates the core transcriptional program that determines myoblastic cell fate and facilitates muscle regeneration by preventing premature differentiation of proliferating myoblasts. The comprehensive data set provides a useful recourse for deciphering the molecular mechanisms underlying muscle regeneration.
Fig. 2.
Correlation with previously-acquired data sets.
(A, B) GSEA analysis of genes bound by all three TFs in comparison to Runx1-responsive genes in proliferating PMs (A) or gene expression data of WT myoblasts vs. differentiated myotubes (B). (C) The Runx1- and RMJ-bound loci in PM ChIP-seq were compared to published C2C12 cell line derived MyoD bound “Super enhancers” [16] loci. Percent overlapping loci of the relevant dataset are presented. C2C12 MyoD bound “Super-enhancers” compared to Runx1 and RMJ ChIP-seq. Overlap presented as percent of Super-enhancers.
Acknowledgments
We thank Dr. Daniella Amann Zalcennstein and Dr. Shlomit Gilad from the Israel National Center for Personalized Medicine (INCPM) for help with Illumina sequencing, Dr. Shirely Horn Saban of Biological Services for help in gene expression data acquisition, Dr. Gilgi Friedlander for help in bioinformatics analysis, Dr. Inbal Biton for help with body composition assays and Rafael Saka and Sharon Ovadia for animal husbandry. We thank Dr. Nancy Speck for providing the Runx1L/L mice, Dr. Benny Shilo for the Myf5::Cre mice, and Dr. Itamar Harel, Dr. Ditsa Levanon and Dr. Joseph Lotem for helpful comments throughout the work.
References
- 1.Umansky K.B., Gruenbaum-Cohen Y., Tsoory M., Feldmesser E., Goldenberg D. Runx1 transcription factor is required for myoblasts proliferation during muscle regeneration. PLoS Genet. 2015;11:e1005457. doi: 10.1371/journal.pgen.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenbaum-Cohen Y., Harel I., Umansky K.B., Tzahor E., Snapper S.B. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11211–11216. doi: 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 4.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015 Jan 15;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love M.I.H.W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. bioRxiv. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Blagden C., Fan J., Nowak S.J., Taniuchi I. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005;19:1715–1722. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X., Yeadon J.E., Burden S.J. AML1 is expressed in skeletal muscle and is regulated by innervation. Mol. Cell. Biol. 1994;14:8051–8057. doi: 10.1128/mcb.14.12.8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blum R., Vethantham V., Bowman C., Rudnicki M., Dynlacht B.D. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh M.S., Parker M.H., Scime A., Parks R., Rudnicki M.A. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoi C.S., Lee S.E., Lu S.Y., McDermitt D.J., Osorio K.M. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol. Cell. Biol. 2010;30:2518–2536. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim W., Barron D.A., San Martin R., Chan K.S., Tran L.L. RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16389–16394. doi: 10.1073/pnas.1407097111. [DOI] [PMC free article] [PubMed] [Google Scholar]