Abstract
The formation of foamy macrophages by sequestering extracellular modified lipids is a key event in atherosclerosis. However, there is controversy about the effects of lipid loading on macrophage phenotype, with in vitro evidence suggesting either pro- or anti-inflammatory consequences. To investigate this in vivo we compared the transcriptomes of foamy and non-foamy macrophages that accumulate in experimental subcutaneous granulomas in fat-fed ApoE null mice or normal chow-fed wild-type mice, respectively. Consistent with previous studies in peritoneal macrophages from LDL receptor null mice (Spann et al., 2012 [1]), we found that anti-inflammatory LXR/RXR pathway genes were over-represented in the foamy macrophages, but there was no change in M1 or M2 phenotypic markers. Quite unexpectedly, however, we found that genes related to the induction of fibrosis had also been up-regulated (Thomas et al., 2015 [2]). The progression of the foamy macrophages along anti-inflammatory and pro-fibrotic pathways was confirmed using immunohistochemistry (described fully in our primary research article (Thomas et al., 2015 [2]). Here we provide additional details on production of the macrophages and their transcriptomic comparison, with the raw and processed microarray data deposited in GEO (accession number GSE70126). Our observations on these cells are indeed paradoxical, because foamy macrophages have long been implicated in promoting inflammation, extracellular matrix degradation and atherosclerotic plaque rupture, which must be provoked by additional local mediators. Our findings probably explain how very early macrophage-rich lesions maintain their structural integrity.
Keywords: Foam cell macrophage, Atherosclerosis, Inflammation, Fibrosis, Non-foamy macrophage
Specifications
| Organism/cell line/tissue | Mus musculus (C57BL background)/NA/sponge granuloma macrophages, atherosclerotic plaque |
| Sex | Male |
| Sequencer or array type | MouseRef8 v2.0 Expression BeadChips, Illumina |
| Data format | Raw and analysed |
| Experimental factors | Sponge granuloma macrophages from fat-fed ApoE−/− mice (FCM: foamy cell macrophages) vs control (ApoE+/+) mice fed a normal chow diet (NFM: non-foamy macrophages). |
| Experimental features | Sponges were surgically placed s.c. in fat-fed ApoE−/− mice or control mice fed a normal chow diet, to produce FCM or NFM, respectively. Four weeks later the sponges were retrieved and the macrophages isolated and purified using differential adherence, with (FCM) or without (NFM) buoyant density centrifugation. RNA of high quality was isolated and compared using Illumina beadchips. Results were obtained from Ingenuity Pathway Analysis and GO annotation and clustering, and were confirmed using RT-qPCR, immunocytochemistry (on macrophages isolated from the sponges) and immunohistochemistry (on sections taken through sponges or diseased arteries from ApoE−/− mice fed a high-fat diet for 12 weeks). |
| Consent | The housing and care of all animals and procedures used in these studies was in accordance with and under licence of the Animals (Scientific Procedures) Act 1986 (London, United Kingdom), which transposes EU Directive 2010/63/EU. |
| Sample source location | NA |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Experimental groups and conditions
We generated FCM and NFM in mice in vivo, in order to understand how the macrophages found in atherosclerotic plaques differ from non-foamy macrophages. Consistent with previous studies in peritoneal macrophages from LDL receptor null mice [1], we found that anti-inflammatory LXR/RXR pathway genes were over-represented in the foamy macrophages, but there was no change in M1 or M2 phenotypic markers. Quite unexpectedly, however, we found that genes related to the induction of fibrosis had also been up-regulated [2].
2.1.1. Animals
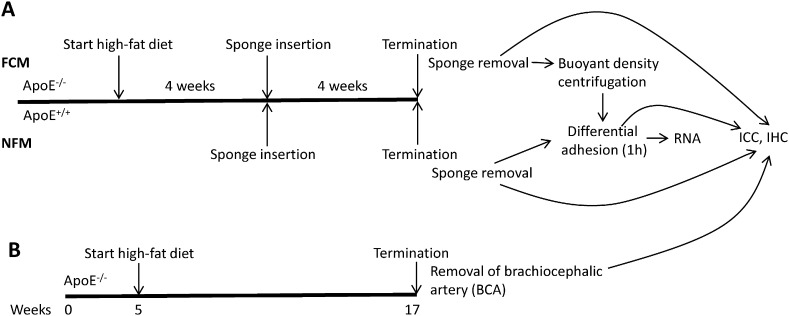
The housing and care of all animals and procedures used in these studies was in accordance with and under licence of the Animals (Scientific Procedures) Act 1986 (London, United Kingdom), which transposes EU Directive 2010/63/EU. We obtained homozygous ApoE−/− and ApoE+/+ mice on a C57/BL background from University of Bristol colonies. Animals were housed conventionally, and were given food and water ad libitum throughout the experiments. Animals were treated as outlined in Fig. 1.
Fig. 1.
Representation of macrophage production in mice. Foam cell macrophages (FCM) or non-foamy macrophages (NFM) were obtained from subcutaneous sponges inserted into mice. RNA was harvested from isolated macrophages and further samples were taken for examination using immunocytochemistry (ICC) or immunohistochemistry (IHC).
2.1.2. Surgical sponge implantation and harvest
Polyurethane sponges (Truclean, Merck) were prepared as follows: 0.5 cm3 pieces of sponge were boiled for 2 h in ethanol, washed in dH2O and autoclaved. Shortly before implantation the sponges were injected with ~ 50 μl Matrigel (BD Biosciences, UK) and soaked in saline (all under sterile conditions). Adult male ApoE−/− mice were placed onto a high-fat diet containing 23% fat from lard, supplemented with 0.15% (w/w) cholesterol (821424, Special Diets Services, UK). Age-matched control mice remained on a normal chow diet (3.5% fat, EURodent Diet 22%, LabDiet, USA). Three weeks later the animals were anaesthetised using halothane, and the sponges inserted subcutaneously. The animals were shaved on their lower back. Using aseptic technique, and with buprenorphine analgesia, a 1-cm long incision was made in the skin. Six pockets were made under the skin, and a 0.5 cm3 piece of sterile sponge placed into each pocket and the incision closed. When fully recovered the animals were returned to their original cages, and the diet regimen continued for a further four weeks.
2.1.3. Macrophage isolation and purification from sponges
The mice were terminated with a lethal dose of anaesthetic and the sponges removed using the aseptic technique. All further manipulations occurred within a laminar flow cabinet. The sponges were cleaned and incubated in Dispase (BD Biosciences) to partially degrade the Matrigel, and cellular exudates squeezed out of the sponges by twisting. NFM, from control mice fed a normal chow diet, were obtained by differential adherence in RPMI 1640 media (Life Technologies) without foetal calf serum, after evaluation by Trypan blue exclusion as described previously [3], [4]. After 1 h the non-adherent cells were removed and fresh medium added. 0.5 h later, the adherent cells were washed in fresh medium and RNA harvested. FCM were obtained from ApoE−/− mice on the high-fat diet. Cellular exudates were obtained as outlined above, and FCM obtained by density centrifugation over a metrizamide gradient (1.3507 refractive index, OPTI-prep, Sigma, USA). Only foam cells float, because of the relatively low buoyant density of lipid. Differential adhesion steps were subsequently performed as above and RNA harvested. Some isolated FCM and NFM were placed onto coverslips for assessment of purity.
2.1.4. Assessment of macrophage purity
Cells that had been adhered onto coverslips were used to assess their lipid content (as indicated by Oil-Red-O staining (2% Oil-red-O in isopropanol; Sigma) and identification of cell type (using immunocytochemistry). All cells from the preparations from sponges from ApoE null mice contained foamy inclusions, whereas no cells in the preparations from control mice staining positively for lipid. 96 ± 5.8% of cells from sponges obtained from the ApoE null mice were monocyte/macrophages (MOMA2, Biosource International, USA) and 0.86 ± 1.2% were smooth muscle cells (α/γ-SM actin, HHF35, Dako, UK), indicating that these preparations were highly pure. Similarly, 87 ± 10% of cells from control mice were MOMA2 positive, and 1.4 ± 1.8% were α/γ-SM actin positive. We confirmed that the preparations had little smooth muscle cell contamination using RT-qPCR (less than 2.5 copies of SM myosin/ng RNA). Hence cells isolated from the fat-fed ApoE null mice were indeed foam cell macrophages (FCM), whereas those from the control mice were non-foamy macrophages (NFM).
2.1.5. RNA isolation, use and quality control assessment
RNA lysates were collected using RLT solution (Qiagen Ltd., UK) with β-mercaptoethanol (Sigma) and the total RNA extracted using the Qiagen RNeasy kit (Qiagen Ltd) according to the manufacturer's instructions. RNA concentration was determined using NanoDrop spectrometry (NanoDrop Technologies, USA), with the samples having RNA Integrity Numbers (RINs) between 8.0 and 9.7. RNA samples of high quality (A260/280 > 2) (n = 4) were compared using Illumina beadchips (MouseRef8 v2.0 Expression BeadChips, Illumina, USA) at 1 μg/sample. Samples were labelled and hybridized as described in the Illumina “Whole-Genome Gene Expression Direct Hybridization Assay Guide (11322355 A)”. Samples were labelled using the Ambion Illumina TotalPrep RNA Amplification Kit (Ambion, USA). The amplification of RNA is based on the Eberwine method, and results in biotinylated, amplified cRNA. The signal was developed with streptavidin-Cy3 (Fisher Scientific, USA) and the BeadChip scanned using the Illumina iScan system. Quality control was performed using Illumina BeadStudio software.
2.2. Comparison of FCM and NFM
2.2.1. Functional annotation
All genes from the data set that met the unadjusted P-value cut-off of 0.01 were uploaded to the Ingenuity Pathways Analysis system (Ingenuity Systems, www.ingenuity.com) and included in the analysis. Each identifier was mapped to its corresponding object in the Ingenuity Knowledge Base. Functional analysis identified the biological functions and canonical pathway analysis identified the pathways that were most significant to the data set. Network maps were also generated within Ingenuity Pathway Analysis. Differentially expressed genes were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base and networks of these molecules algorithmically generated based on their connectivity. Differentially expressed genes were also submitted to GO annotation and clustering using DAVID Bioinformatics Resources (National Institute of Allergy and Infectious Diseases (NIAID) 2008, NIH, http://david.abcc.ncifcrf.gov).
2.2.2. Real time quantitative PCR (RT-qPCR) validation
The array results were validated and expanded on RNA samples prepared in the same way using RT-qPCR (n = 5–7). For reverse transcription, 100 ng of total RNA was used to make cDNA, using a Qiagen Quantitect Reverse Transcriptase Kit (Qiagen Ltd., UK) according to the manufacturer's instructions. cDNA was diluted 1:1 in 10 mM Tris·HCl, pH 8.0. RT-qPCR was performed in a Roche Light Cycler 1.5 (Roche, UK) to quantify the steady-state concentration of RNA, using the QuantiTect SYBR Green PCR Kit (Qiagen Ltd). Each reaction contained 2.5–7 ng RNA and 0.5 μM primers. Initial denaturation (15 min at 95 °C) was followed by 55 cycles of denaturation (15 s at 95 °C), annealing (20 s at 60–66 °C) and extension (25 s at 72 °C). Copy numbers of gene transcripts per total ng RNA input were calculated using standard curves constructed as recommended by from purified amplicon (Bioline, USA). Sequences of the PCR primer pairs used to amplify the respective cDNAs were designed using Ensembl and Primer3, and the specificity of the sequence confirmed using Nucleotide blast (NCBI).
2.3. Statistical analysis
Statistical analysis of the microarrays was performed using the ‘R’ Bioconductor Lumi and Limma packages, applying the linear models and empirical Bayes methods included in the package [5]. Variance stabilizing transformation was performed using the Robust Spline Normalization (RSN) algorithm [6] with unsupervised analysis methods such as Principal Component Analysis and Hierarchical Clustering used for initial data exploration. Statistical analysis of differential expression consisted of a univariate model to detect individual genes that are significantly different in abundance between the conditions [5]. P-values were adjusted for multiple comparisons, using the False Discovery Rate (FDR) Benjamini–Hochberg method. Differential expression was classified as significant (P < 0.01 after FDR correction for multiple testing) or suggestive (P < 0.01 unadjusted). The significance of the association between the data set and the canonical pathway was measured by either a ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway, or by using a right-tailed Fisher's exact test to calculate a P-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone. For the RT-qPCR, data differences between the two groups were tested for significance with Student's t-test, using a logarithmic transformation or the Mann–Whitney U-test if the data were not normally distributed (GraphPad Instat, USA). P-values of less than 0.05 were considered statistically significant.
Third-party financial support
This work was supported by the British Heart Foundation (ACT and ACN). The funder had no role in study design, data collection, analysis and interpretation, decision to publish or preparation of the manuscript.
References
- 1.Spann N.J., Garmire L.X., McDonald J.G., Myers D.S., Milne S.B., Shibata N. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas A.C., Eijgelaar W.J., Daemen M.J.A.P., Newby A.C. Foam cell formation in vivo converts macrophages to a pro-fibrotic phenotype. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chase A.J., Bond M., Crook M.F., Newby A.C. Role of nuclear factor-κB activation in metalloproteinase-1, -3, and -9 secretion by human macrophages in vitro and rabbit foam cells produced in vivo. Arterioscler. Thromb. Vasc. Biol. 2002;22:765–771. doi: 10.1161/01.atv.0000015078.09208.92. [DOI] [PubMed] [Google Scholar]
- 4.Thomas A.C., Sala-Newby G.B., Ismail Y., Johnson J.L., Pasterkamp G., Newby A.C. Genomics of foam cells and nonfoamy macrophages from rabbits identifies arginase-I as a differential regulator of nitric oxide production. Arterioscler. Thromb. Vasc. Biol. 2007;27:571–577. doi: 10.1161/01.ATV.0000256470.23842.94. [DOI] [PubMed] [Google Scholar]
- 5.Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 6.Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]