Abstract
Filamentous growth is an important virulence trait of the human pathogenic fungi within the genus Candida, and the greater propensity of C. albicans to form hyphae has been proposed to account for the greater virulence of this species relative to the less pathogenic species C. dubliniensis. In this meta-analysis, we compare the transcriptional response of C. dubliniensis and C. albicans to the individual environmental stimuli that shape the gene expression profiles during filamentation in 10% serum, namely alkaline pH, 37 °C and reduced cell density. We could identify conserved core temperature and pH responses, however many signature Efg1-regulated, hypha-induced transcripts (e.g. ECE1, HWP1) exhibited reduced or lack of induction in C. dubliniensis. Comparison of the activity of the HWP1 and ECE1 promoters in both species using GFP fusions showed a lag in serum induced fluorescence in C. dubliniensis relative to C. albicans and nutrient depletion was required for maximal expression of these Efg1-regulated transcripts in C. dubliniensis.
Keywords: Candida albicans, Candida dubliniensis, Transcription, Efg1
1. Introduction
Candida dubliniensis is an opportunistic fungal pathogen that was first identified as a common cause of oral candidosis in HIV-infected patients [1]. C. dubliniensis is closely related to Candida albicans, the major fungal pathogen of humans [2]. However, C. albicans is far more prevalent as a pathogen in the human population, particularly in the case of systemic fungal infection where C. dubliniensis is responsible for fewer than approximately 2% of infections [3]. Virulence studies have associated the reduced capacity of C. dubliniensis to establish infection with a reduced ability to undergo the yeast to hypha transition [4], [5], [6]. Analysis of C. dubliniensis cells in the stomach and kidney of infected mice revealed that they grow predominantly in the yeast phase, whereas C. albicans could be recovered in both the yeast and hyphal phases [5]. Models of in vitro infection support this finding; C. dubliniensis remains in the yeast phase when inocluated on reconstituted human epithelium (RHE) and filaments less efficiently than C. albicans following phagocytosis by murine macrophages [6], [7]. Coupled with this reduced filamentation, comparative genomics has revealed that the C. dubliniensis genome does not contain orthologues of the C. albicans hypha-specific virulence factors ALS3, HYR1, SAP4 and SAP5 [2], [8].
Primary regulation of filamentation in vivo and in liquid media in vitro (e.g. serum) is via the cAMP–PKA pathway and the cognate transcription factor Efg1 [9]. Recent studies have shown that a variety of additional stimuli can modulate the activity of this pathway such as temperature (via interaction with Hsp90), farnesol, CO2 and bacterial peptidoglycan [10], [11], [12], [13]. In addition, the activity of Efg1 is regulated by interactions with other transcription factors [14]. Filamentation is also stimulated by additional alkaline pH regulated signals mediated via the Rim101 pathway and the recently described pH regulator Mds3 [15]. The net result of this stimulation is activation of a transcriptional response mediated by the transcription factors Efg1 and Ume6 and activation of the hypha-specific cyclin Hgc1 [16], [17]. Incubation of C. dubliniensis in 10% serum at 37 °C results in abundant production of true hyphae and activation of a conserved transcriptional response [18]. However, true hypha production in serum is nutrient sensitive in C. dubliniensis and addition of nutrients, in particular peptone, to alkaline media greatly inhibits induction of UME6 transcription and filamentation. This nutrient repression may involve the activity of Tor1 kinase, as preliminary data suggests that rapamycin inhibition of Tor1 enhances filamentous growth in nutrient rich media [19]. In this meta-analysis, we compare the transcriptional response of C. dubliniensis and C. albicans to the individual environmental stimuli that shape the gene expression profiles during filamentation in 10% serum, namely alkaline pH, 37 °C and reduced cell density. We could identify conserved core temperature and pH responses with C. albicans, however many signature Efg1-regulated, hypha-induced transcripts (e.g. ECE1, HWP1) exhibited reduced or lack of induction under these conditions relative to C. albicans. In subsequent experiments comparing the activity of HWP1 and ECE1 promoters in both species using GFP fusions, a lag in serum induced fluorescence was observed in C. dubliniensis relative to C. albicans and nutrient depletion was required for maximal expression of these Efg1-regulated transcripts in C. dubliniensis.
2. Methods
2.1. Strains and culture conditions
All C. albicans and C. dubliniensis strains and derivatives used in this study are listed in Table 1. Candida strains were routinely maintained on Yeast extract peptone dextrose (YPD) agar medium. Liquid culture was also carried out in YPD broth and a nutrient depleted YPD broth (i.e. YPD diluted to 10% [v/v] of the standard concentration). Buffering of YPD to pH 5.0 or pH 7.2 was achieved with 0.1 M potassium phosphate buffer. Liquid culture was also carried out in the liquid medium of Lee et al. [20]. Lee's medium was adjusted to pH 4.5 or 7.2 as required. Hyphal induction was carried out in sterile Milli-Q H2O supplemented with 10% (v/v) foetal calf serum with shaking at 200 r.p.m. at 37 °C. The proportion of germ-tubes or hyphae in each culture was assessed at intervals by microscopic examination of an aliquot of culture with a Nikon Eclipse 600 microscope (Nikon U.K., Surrey, U.K.).
Table 1.
Strains used in this study and their genotypes.
2.2. Transcript profiling of C. dubliniensis
C. dubliniensis microarrays representing 5999 orfs from the CD36 genome were used as described by O'Connor et al. [18]. To examine the effects of cell density changes, nutrient depletion, a shift to 37 °C and a shift to alkaline pH, 18 h (early stationary phase) Lee's medium cultures (pH 4.5, 30 °C) were washed and inoculated at 2 × 106 cells/ml in (i) fresh Lee's medium (pH 4.5) at 30 °C, (ii) 10% (v/v) Lee's medium (pH 4.5) at 30 °C, (iii) Lee's medium (pH 4.5) at 37 °C and (iv) Lee's medium (pH 7.2) at 30 °C, respectively. RNA was extracted from these cultures following 3 h incubation under each condition. Poly A mRNA isolation, Cy3/Cy5-labelling and array processing we carried out as described by O'Connor et al. [18]. For each experiment, four biological replicate experiments were performed, including two dye swap experiments. Data was normalized in GeneSpring GX11 using Loess normalization. A t test was performed on each data set using the variance derived from replicate spots. Those genes with a p value ≤ 0.05 were selected for analysis and genes exhibiting at least a 2-fold difference in expression are described here. Categorization of gene ontology (GO) terms was carried out using the “GO term finder” tool at the Candida Genome Database (http://www.candidagenome.org/). Results from all microarrays have been submitted to the GEO archive (Accession: GSE20537).
2.3. Creation of ECE1 and HWP1 promoter-GFP fusions
In order to create strains harbouring PECE1-GFP or PHWP1-GFP fusions, we used the integrating vector pCDRI [6]. A derivative of this plasmid was created by inserting yEGFP fused to the actin terminator on a HindIII/MluI fragment to create pGM175. A HWP1 promoter fragment from bases − 1 to − 1535 to was amplified with primers HWP1AF (GGCGGGCCCGTAAACAAACTCCCACAACCAATCG) and HWP1XR (CTAGCTCGAGTATTGACGAAACTAAAAGCGAG) which included the ‘HCR’ regulatory region described by Kim et al. and cloned upstream of yEGFP following digestion of both fragments with ApaI and XhoI [21]. Similarly, an ECE1 promoter fragment from bases − 1 to − 921 was amplified from C. albicans SC5314 with primers ECEAF (GTACGGGCCCAAGAGTCTCATTCAGATAACG) and EXEXR (GCATCTCGAGTTTAACGAATGGAAAATAGTTG) and cloned upstream of yEGFP in an identical fashion. The resulting plasmids (pHWPGFP and pECEGFP) were linearised within the CDR1 region and used to transform C. albicans SC5314 and C. dubliniensis Wü284 to create the strains CaHWPGFP and CaEGFP and WüHWPGFP and WüEGFP, respectively (Table 1). Ectopic integration in the CDR1 gene was confirmed by Southern hybridization. Fluorescence was examined using a Nikon Eclipse 600 microscope (Nikon U.K., Surrey, U.K.) fitted with a GFP filter. Fluorescent micrographs were taken with a Nikon Coolpix digital camera using a 2 s exposure time. Micrographs in each panel were taken contemporaneously.
For temporal studies of GFP-induction during serum induction, flow cytometry was used. Hyphal induction was carried out in sterile Milli-Q H2O supplemented with 10% (v/v) foetal calf serum with shaking at 200 r.p.m. at 37 °C. An aliquot of each sample (1 ml) was removed at each time point, cells were collected by centrifugation and washed × 3 in PBS at 4 °C. Each sample was fixed with a solution of formaldehyde (4%) and methanol (10%) before analysis by flow cytometry using a Beckman Coulter EPICS XL machine. Mean fluorescence intensities of 10,000 events (cells) were calculated for each time point.
3. Results and discussion
3.1. Morphological response of C. dubliniensis to changes in environment
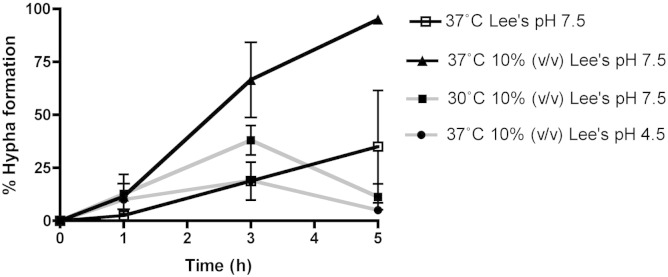
In C. albicans, a shift to alkaline pH and 37 °C is sufficient to induce hypha formation in vitro in nutrient rich culture media [22]. Using Lee's medium as a defined basal medium, we independently examined the role of temperature, pH and nutrient depletion in true hypha formation in C. dubliniensis. Following a shift from Lee's medium pH 4.5 at 30 °C to Lee's medium pH 7.2 at 37 °C, less than 50% of C. dubliniensis cells had formed germ-tubes following 5 h incubation. In contrast, in nutrient depleted (10% v/v) Lee's pH 7.2 at 37 °C, > 95% of cells formed germ-tubes by 5 h. Shifts to 37 °C or pH 7.2 alone in nutrient depleted (10% v/v) Lee's resulted in transient germ-tube formation by < 50% of C. dubliniensis yeast cells (Fig. 1). Sustained hypha formation up to 5 h required the combination of a temperature and pH shift in nutrient depleted medium (Fig. 1).
Fig. 1.
Hypha formation by C. dubliniensis Wü284 in Lee's medium. Cells grown overnight in YPD broth at 30 °C were inoculated in 2 ml Lee's medium (1 × 106 cells) in the well of a 6-well tissue culture dish. The proportion of germ-tube forming cells was assessed hourly using an inverted microscope. Lee's medium was used at standard concentration or diluted to 10% (v/v) standard concentration to induce nutrient depletion.
3.2. C. dubliniensis response to alkaline pH is highly conserved
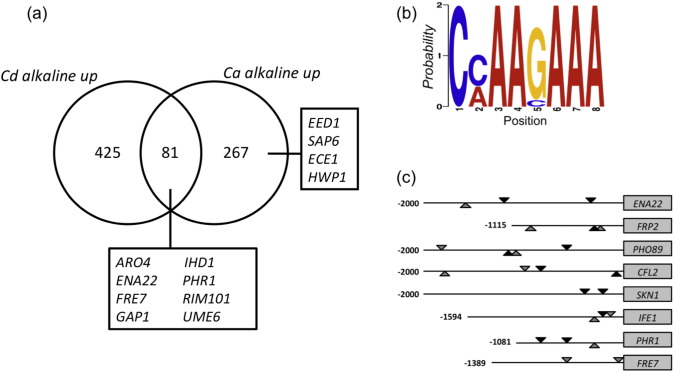
As the above data indicated that the alkaline pH shift plays an important role in inducing filamentation in C. dubliniensis, we analysed the transcriptional response of C. dubliniensis following a transition from acidic to alkaline pH. A total of 425 genes exhibited 2-fold up-regulation during a shift from pH 4.5 to 7.2 in Lee's medium (t-test p < 0.05). This group included genes associated with the cell surface (orthologues of PHR1, EAP1, IFF11, SUN41), metal ion transport (orthologues of CFL11, CFL4, ENA22, PHO84, PHO89) filamentous growth (orthologues of SFL1, UME6, RAS1, RFG1, TEC1) and amino acid metabolism (orthologues of ARO4, HIS3, SAM4). We observed strong conservation between the pH responses of C. albicans and C. dubliniensis, with 81 of the C. albicans alkaline up regulated genes described by Bensen et al. up regulated at least 2-fold in C. dubliniensis (t-test p < 0.05) at pH 7.2. (Fig. 2a) [23]. This group included many of the signature pH responsive genes such as PHR1, ENA22, RIM101 and several ferric reductases (Table S1). However, unlike C. albicans, significant (i.e. > 2-fold) induction of the orthologues of the Efg1-regulated genes EED1, SAP6, ECE1 and HWP1 was not observed at alkaline pH in C. dubliniensis. C. albicans also exhibited approximately 10-fold higher levels of induction of the Efg1-regulated transcripts CSA1 and RBT1.
Fig. 2.
(a) Venn diagram showing the similarity of the alkaline induced response (≥ 2-fold) in C. dubliniensis and C. albicans from Bensen et al. [3]. (b) Position-specific probability matrix generated with MEME showing the motif identified in the promoters of pH-regulated genes in C. dubliniensis. The probability of each nucleotide appearing at each position corresponds to the height of each individual letter at that position multiplied by the total height of the ‘stack’ at that position. (c) Cartoon showing the relative position and frequency of the motif in the promoters of pH-regulated genes identified by ANOVA (see text). The line represents the promoter length (up to − 2000 bases or to the next chromosomal feature) relative to the respective ORF (not to scale). Black triangles represent to location of motifs with a 5′ ‘CC’ and grey triangles those with a 5′ ‘CA’. Triangles on top of the line indicate motifs on the sense strand, lower symbols indicate antisense motifs. Exact positions of motifs are given in Table S1.
In order to identify genes in C. dubliniensis that were exclusively alkaline induced, we used ANOVA to identify a group of 8 genes upregulated by alkaline pH, but not affected by a shift in temperature or cell density in C. dubliniensis. We used the motif finding tool MEME to identify conserved elements in the promoters of these alkaline regulated genes (Table S2). We could identify a conserved C[C/A]AAGA motif within the promoters of each of these genes, similar to the predicted Rim101 binding site described in C. albicans (GCCAAG) suggesting that the C. dubliniensis RIM101 orthologue is important for the alkaline pH response (Fig. 2b). Two elements were present in the SKN1 promoter and three elements in the PHR1 and ENA22 promoters, respectively (Fig. 2c) Conversely, a search for the CCAAGA motif in the promoter regions of the top 25 NRG1-regulated genes in C. dubliniensis revealed that only 6 genes contained this motif (data not shown).
Genes repressed by alkaline pH in both species included RIM8, several heat shock proteins (HSP30, HSP70, HSP78 and HSP104) and several hexose transporters (HXT5, HGT17, HGT19; Table S3).
3.3. C. albicans has a unique response to cell density and temperature shifts in vitro
Analysis of the data set of Kadosh and Johnson showed that following a switch to 37 °C, C. albicans mounted a transcriptional response consisting of 419 genes whose expression was regulated > 2-fold (t-test p < 0.05). The list of temperature-induced genes (223 genes) was enriched for genes involved in ribosome biogenesis (39 genes), amino acid biosynthesis (15 genes) and lipid biosynthesis (18 genes). When the effects of cell density and temperature shifts on global gene expression were investigated in C. dubliniensis, it was revealed that this response was largely conserved in and is likely a response to inoculation in fresh nutrient rich media (Fig. 3). However, our analysis identified a specific response in C. albicans that included several genes normally associated with filamentation at 37 °C. As previously reported, temperature shifts to 37 °C induced expression of ECE1 and HWP1 in C. albicans [24], [25]. Indeed, even in the absence of a temperature shift, a change in cell density was sufficient to induce expression of ECE1 and HWP1 in C. albicans [24]. In C. dubliniensis, the orthologues of ECE1 and HWP1 exhibited weak (< 2-fold) or no induction in response to 37 °C, respectively (Fig. 3a). An additional cluster of C. albicans highly induced genes exhibited down-regulation in C. dubliniensis, including the C. dubliniensis orthologue of EED1, an activator of UME6 expression (Fig. 3b). Additional genes encoding factors with roles in the cell cycle (CCN1, GIN4) cytoskeleton organisation (CDC12, ARF3, CCT3, TCP2), maintenance of hyphal growth (CLA4) and DNA replication (POL30, CDC47) that were induced > 2-fold in C. albicans at 37 °C exhibited no induction in C. dubliniensis (Fig. 3c). Like ECE1 and HWP1, the majority of these temperature (37 °C) induced genes in C. albicans also exhibited induction following culture dilution at 30 °C (e.g. EED1, CLA4, RNR1; Fig. 3). The similarity of the species-specific response of C. albicans to culture dilution and the shift to 37 °C is likely due to the similar affects of these stimuli on the cAMP-PKA pathway. Culture dilution is thought to remove farnesol-mediated repression of the cAMP-PKA pathway, perhaps at the level of adenylate cyclase [24]. Temperature shifts are thought to act by relieving Hsp90 mediated repression of the cAMP-PKA pathway [12]. However, the level of induction was generally highest following the temperature shift, indicating that either this stimulus has a stronger effect on the cAMP-PKA pathway or that the effects of dilution and temperature are cumulative.
Fig. 3.
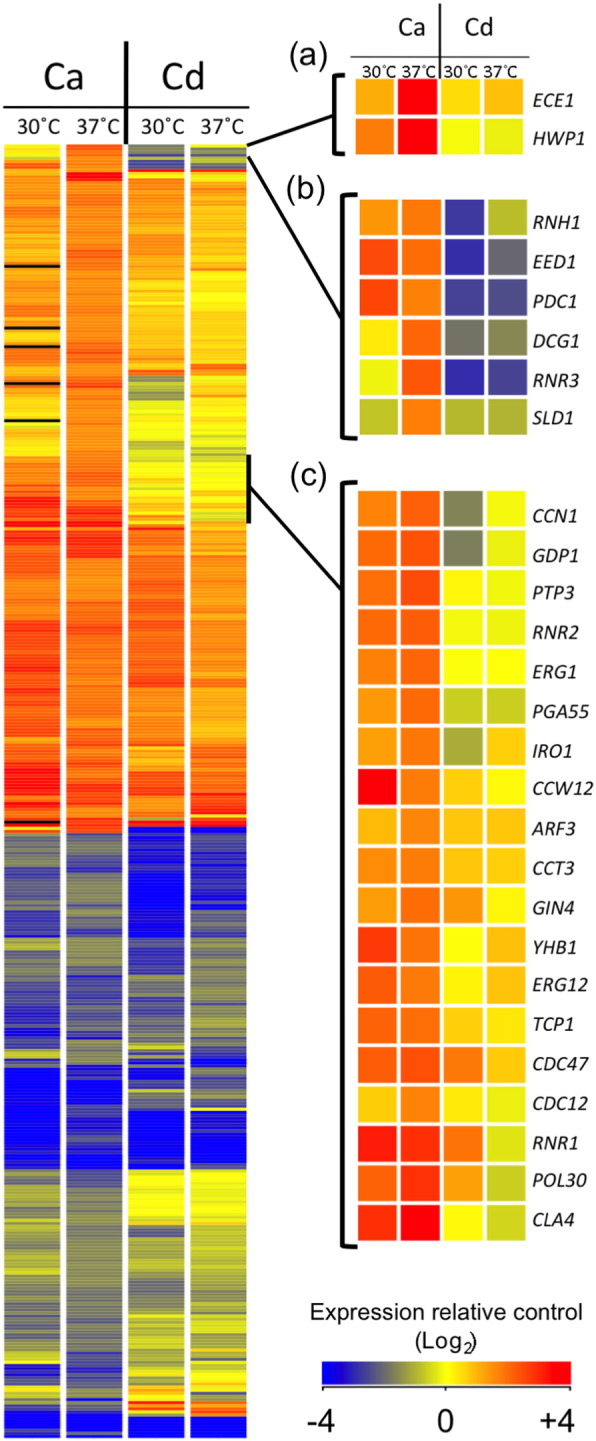
A comparison of the transcriptional responses of C. albicans (Ca) and C. dubliniensis (Cd) to dilution at 30 °C and 37 °C. The ‘heat-map’ on the left shows the expression patterns for 419 genes identified as > 2-fold regulated (t-test p < 0.05) in C. albicans following a switch to 37 °C (from (10). Expression following dilution to medium at 30 °C is also shown. On the right, the expression of the orthologous gene in C. dubliniensis under similar conditions is shown. Expression refers to fold-change relative to the preculture conditions (30 °C) and is denoted by the colour according to the legend. Certain sections are expanded in panels (a), (b) and (c) for clarity, as detailed in the text.
Despite the specific nature of the response to growth at 37 °C in C. albicans, a conserved temperature response could be identified between the two species (Table S4). This consisted of 115 genes induced > 2.0-fold in both species and was enriched for genes involved in ribosome biogenesis (33 genes) and amino metabolism (15 genes). Few genes associated with filamentous growth could be identified from this group, however induction of IHD1 was observed in both C. albicans and C. dubliniensis (6.5 and 8-fold respectively). A common core of 116 genes down regulated > 2.0-fold in both species could also be identified, including cell wall proteins (PIR1, ALS2, ALS4, RBT5, RBT6, RBT8, RBE1, RBR1) and genes involved in carbohydrate and lipid metabolism (Table S5). The specific response of C. dubliniensis to 37 °C included many additional genes involved in amino acid metabolism and transport (16/79 genes) and the transcription factors CPH1 (induced 3.3-fold) and UME6 (induced 4-fold; Table S6). CPH1 is required for filamentous growth on solid media and mating, and its induction here is unexpected as neither process would be expected to occur under the current conditions. Induction of UME6 implicates this transcription factor in filamentation at 37 °C. Unexpectedly, UME6 was induced in the absence of significant induction of other Efg1-regulated transcripts, suggesting an alternative mechanism of induction.
3.4. Nutrient depletion activates a starvation response and UME6 in C. dubliniesis
As nutrient depletion has a strong effect on filamentous growth in C. dubliniensis, we examined the transcript profile of strain Wü284 following dilution to 10% (v/v) Lee's medium (pH 4.5). Genes that exhibited a significant change in expression relative to dilution alone were examined (Tables S7 and S8). This group of genes was found to be enriched for those involved in carbohydrate metabolism, particularly mobilisation of glycogen stores (GLC1, GSY3) and phosphorylation of glucose for entry to glycolysis (GLK1, GLK4, PGM2). Catabolism of alternative sugars was also induced (ARA1, GRE3, XYL1, XYL2). Nitrogen starvation was suggested by increased expression of genes involved in amino acid degradation (CAR1, PNG2), protein degradation (PRC2, LAP3) and scavenging of nitrogen from the environment (DAL7, GAP2, DUR3). These data provide a link between the response to nutrient depletion and filamentation as a 4.3-fold induction of UME6 transcription was observed. Previously, we have observed induction of UME6 in nutrient poor, alkaline, hypha inducing conditions (water plus 10% serum), and this induction was repressed by the addition of peptone [18]. The data presented here demonstrate that the nutrient depleted media, in the absence of temperature or pH shifts, can contribute to UME6 induction. Previous microarray and QRT-PCR studies have shown that the level of UME6 induction in C. dubliniensis is of the order of ~ 100-fold following inoculation in water plus 10% serum (Fig. 4a). The microarray data presented here indicate that this level of induction may involve the integration of several different stimuli including temperature, nutrient depletion and alkaline pH (Fig. 4a). The level of UME6 induction has previously been shown to dictate the level of hyphal elongation and these data provide further evidence that efficient filamentation in C. dubliniensis requires multiple environmental stimuli, including nutrient depletion [26].
Fig. 4.
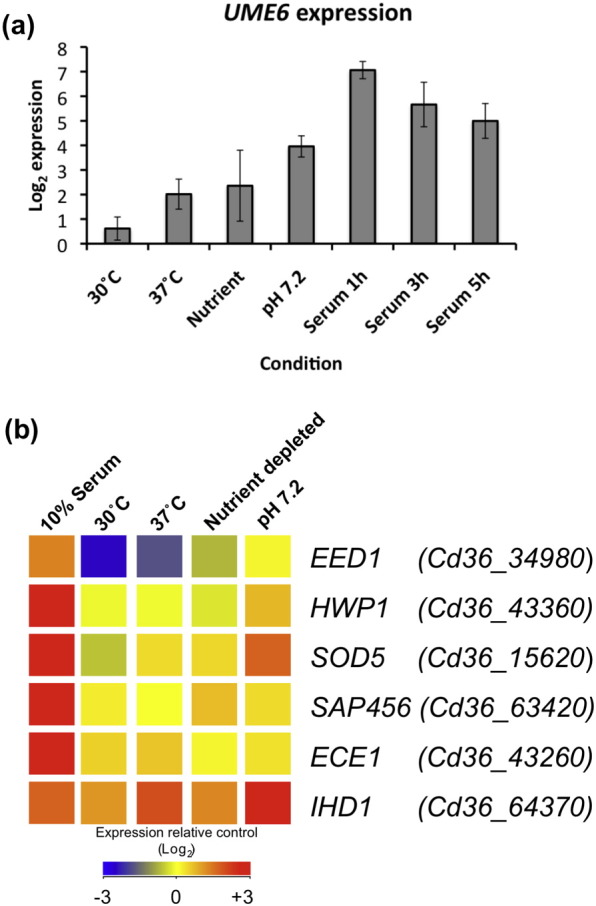
Expression of selected genes in C. dubliniensis from microarray experiments. (a) Expression of UME6 following inoculation of cells from overnight Lee's medium pH 4.5 cultures to fresh medium at 30 °C, 37 °C, nutrient depleted Lee's medium or Lee's buffered to pH 7.2, respectively. Data from previously reported experiments where Lee's grown cells were inoculated to 10% (v/v) bovine serum and grown for 1 h, 3 h and 5 h are also shown for comparison. (b) Heat map showing the expression patterns of selected Efg1-regulated and hypha-induced transcripts in C. dubliniensis.
3.5. Analysis of the intensity and kinetics of hypha-specific gene expression
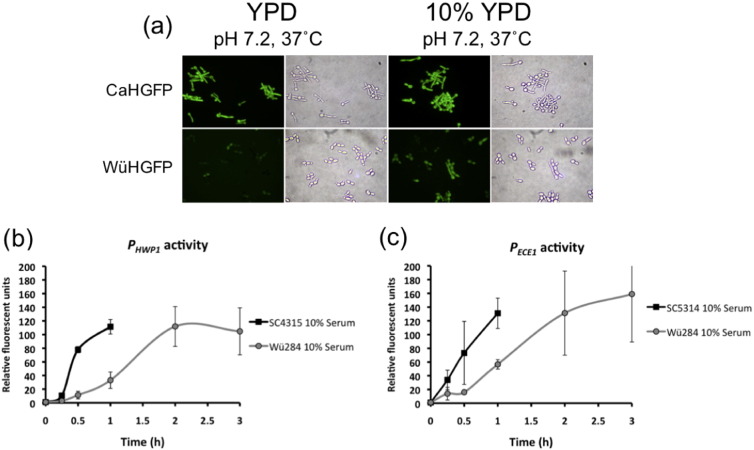
The data presented here indicate that pH and temperature shifts alone have little effect on the induction of several hypha-specific transcripts in C. dubliniensis. In order to characterise this in more detail, we carried out qualitative and quantitative analysis of an Efg1-regulated promoter in C. albicans and C. dubliniensis using a GFP fusion. Induction of GFP expression from the HWP1 promoter (PHWP1-GFP) was observed in C. albicans in response to a shift in both temperature (37 °C) and pH 7.2 (Fig. 5a). A shift in temperature or pH alone was also sufficient to induce visible fluorescence in C. albicans (data not shown). In C. dubliniensis, temperature or pH shifts in YPD medium induced lower levels of fluorescence from the HWP1 promoter relative to that observed in C. albicans, which concurs with the expression data generated by microarray analysis (Fig. 5a). However, nutrient depletion (10% v/v YPD) increased the fluorescence intensity at pH 7.2 and 37 °C (Fig. 5a). Even in the presence of combined temperature and pH shifts nutrient depletion was required to visualise fluorescence and for the formation of filaments, mainly pseudohyphae.
Fig. 5.
Analysis of fluorescence in C. albicans SC5314 (CaPHWP1-GFP) and C. dubliniensis Wü284 (CdPHWP1-GFP) harbouring a PHWP1-GFP fusion construct. (a) Photomicrographs showing fluorescent intensity in CaPHWP1-GFP and CdPHWP1-GFP during growth in YPD buffered to pH 7.2 at 37 °C and 10% (v/v) YPD pH 7.2 at 37 °C. (b & c) Temporal analysis of fluorescence in (a) CaPHWP1-GFP and CdPHWP1-GFP fusion strains and (b) CaPECE1-GFP and CdPECE1-GFP fusion strains. Fluorescence was measured by flow cytometry and mean fluorescence per 10,000 cells was measured and expressed in arbritrary fluorescent units.
We also examined the kinetics hypha-specific gene expression in C. albicans and C. dubliniensis by flow cytometry using the PHWP1-GFP and PECE1-GFP fusions (Fig. 5b & c). In human serum (10% v/v) induction of fluorescence from the PHWP1-GFP and PECE1-GFP fusions was more rapid in C. albicans, occurring within 15 min of induction (Fig. 5b & c). Induced fluorescence was not detected in C. dubliniensis until 60 min, which approximates to the slower rate of germ-tube formation in this species. By 60 min, C. albicans hyphae had clumped to such an extent that measurements could no longer be taken. Maximal fluorescence in C. dubliniensis at 2–3 h was similar to that observed in C. albicans at 1 h (Fig. 5b & c). These data support previous findings that C. dubliniensis expresses hypha-specific genes under nutrient depleted conditions, but indicate that even under permissive conditions, there appears to be a significant lag in expression relative to C. albicans.
4. Conclusions
The combination of microarray analysis and promoter fusions examined here demonstrate that many Efg1-regulated, hypha-specific genes of C. dubliniensis are poorly expressed in response to elevated temperature and alkaline pH relative to the orthologous genes in C. albicans. Although C. dubliniensis exhibited a robust Rim101 mediated response to alkaline pH, activation of Efg1-regulated genes in response to pH was weak. Similarly, growth at 37 °C in YPD medium did not yield significant induction of ECE1 or HWP1. The overall weakness of pH and temperature induction of Efg1-regulated transcripts in C. dubliniensis suggested that additional stimulation was required for full activation of Efg1-regulated transcripts. This may be due to either lower activity of the cAMP-PKA pathway in C. dubliniensis or tight repression of Efg1 activity. Alternatively, repression of these transcripts may be due to another factor such as Nrg1. Transcript profiling has confirmed that the C. dubliniensis orthologue of Nrg1 represses ECE1 and HWP1 and we have previously noted differential expression of this transcriptional repressor under hypha-inducing conditions in C. dubliniensis [6], [27]. Maximal fluorescence from the PHWP1-GFP and PECE1-GFP promoters fusions in C. dubliniensis required nutrient depletion. It is apparent from these studies that maximal induction of these transcripts and hypha formation in C. dubliniensis requires a combination of stimuli and in this regard nutrient depletion appears to be as important as temperature shifts and alkaline pH. This concurs with the study of Grumaz et al. who examined the transcript profile of C. dubliniensis using RNA-seq in YEPD plus serum [28]. Grumaz and colleagues noted weak induction of many hypha-induced genes in these conditions, likely due to the nutrient rich medium used to examine their induction [28]. Previous studies demonstrated that UME6 expression in C. dubliniensis was directly repressed by peptone [18]. These findings suggest that the increased levels of filamentation and virulence exhibited by C. albicans are linked to the ability of this species to form hyphae and express hypha-associated virulence genes at alkaline pH irrespective of local nutrient availability. This is manifested in C. albicans by increased levels of UME6 expression and enhanced expression of virulence factors such as HWP1, SOD5 and SAP6. Recent studies have implicated the large TLO gene family of C. albicans in the regulation of filamentous growth and studies are underway to determine whether this gene family plays a role in conferring increased morphological flexibility in C. albicans [29].
Acknowledgements
This work for supported by funding from Science Foundation Ireland (grant no. 11/RFP.1/GEN/3044) and the Irish Health Research Board (research grant RP/2004/235).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2015.08.026.
Appendix A. Supplementary data
Supplementary tables.
References
- 1.Sullivan D.J., Westerneng T.J., Haynes K.A., Bennett D.E., Coleman D.C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141(Pt 7):1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 2.Jackson A.P., Gamble J.A., Veomans T., Moran G.P., Saunders D., Harris D., Aslett M., Barrell J.F., Butler G., Citiulo F., Coleman D.C., de Groot P.W.J., Goodwin T.J., Quail M.A., McQuillan J., Munro C.A., Pain A.A., Poulter R.T., Rajandream M.-A.A., Renauld H., Spiering M.J., Tivey A.A., Gow N.A.R., Barrell B., Sullivan D.J., Berriman M. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 2009;19:2231–2244. doi: 10.1101/gr.097501.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kibbler C.C., Seaton S., Barnes R.A., Gransden W.R., Holliman R.E., Johnson E.M., Perry J.D., Sullivan D.J., Wilson J.A. Management and outcome of bloodstream infections due to Candida species in England and Wales. J. Hosp. Infect. 2003;54:18–24. doi: 10.1016/s0195-6701(03)00085-9. [DOI] [PubMed] [Google Scholar]
- 4.Asmundsdóttir L.R., Erlendsdóttir H., Agnarsson B.A., Gottfredsson M. The importance of strain variation in virulence of Candida dubliniensis and Candida albicans: results of a blinded histopathological study of invasive candidiasis. Clin. Microbiol. Infect. 2009;15:576–585. doi: 10.1111/j.1469-0691.2009.02840.x. [DOI] [PubMed] [Google Scholar]
- 5.Stokes C., Moran G.P., Spiering M.J., Cole G.T., Coleman D.C., Sullivan D.J. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet. Biol. 2007;44:920–931. doi: 10.1016/j.fgb.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Moran G.P., MacCallum D.M., Spiering M.J., Coleman D.C., Sullivan D.J. Differential regulation of the transcriptional repressor NRG1 accounts for altered host–cell interactions in Candida albicans and Candida dubliniensis. Mol. Microbiol. 2007;66:915–929. doi: 10.1111/j.1365-2958.2007.05965.x. [DOI] [PubMed] [Google Scholar]
- 7.Spiering M.J., Moran G.P., Chauvel M., MacCallum D.M., Higgins J., Hokamp K., Veomans T., d'Enfert C., Coleman D.C., Sullivan D.J. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot. Cell. 2010;9:251–265. doi: 10.1128/EC.00291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran G., Stokes C., Thewes S., Hube B., Coleman D.C., Sullivan D. Comparative genomics using Candida albicans DNA microarrays reveals absence and divergence of virulence-associated genes in Candida dubliniensis. Microbiology. 2004;150:3363–3382. doi: 10.1099/mic.0.27221-0. [DOI] [PubMed] [Google Scholar]
- 9.Biswas S., Van Dijck P., Datta A.A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornby J.M., Jensen E.C., Lisec A.D., Tasto J.J., Jahnke B., Shoemaker R., Dussault P., Nickerson K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001;67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klengel T., Liang W.-J., Chaloupka J., Ruoff C., Schröppel K., Naglik J.R., Eckert S.E., Mogensen E.G., Haynes K., Tuite M.F., Levin L.R., Buck J., Mühlschlegel F.A. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro R., Uppuluri P., Zaas A.A., Collins C., senn H., Perfect J., Heitman J., Cowen L. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 2009 doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X.-L., Lee R.T.H., Fang H.-M., Wang Y.-M., Li R., Zou H., Zhu Y., Wang Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4:28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Noffz C.S., Liedschulte V., Lengeler K., Ernst J.F. Functional mapping of the Candida albicans Efg1 regulator. Eukaryot. Cell. 2008;7:881–893. doi: 10.1128/EC.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zacchi L.F., Gomez-Raja J., Davis D.A. Mds3 regulates morphogenesis in Candida albicans through the TOR pathway. Mol. Cell. Biol. 2010 doi: 10.1128/MCB.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlisle P.L., kadosh D. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot. Cell. 2010;9:1320–1328. doi: 10.1128/EC.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoldt V.R., Sonneborn A.A., Leuker C.E., Ernst J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor L., Caplice N., Coleman D.C., Sullivan D.J., Moran G.P. Differential filamentation of Candida albicans and Candida dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryot. Cell. 2010;9:1383–1397. doi: 10.1128/EC.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan D.J., Moran G.P. Differential virulence of Candida albicans and C. dubliniensis: a role for Tor1 kinase? Virulence. 2011;2:77–81. doi: 10.4161/viru.2.1.15002. [DOI] [PubMed] [Google Scholar]
- 20.Lee K.L., Buckley H.R., Campbell C.C. An amino acid liquid synthetic medium for the development of mycellal and yeast forms of Candida albicans. Med. Mycol. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 21.Kim S., Wolyniak M.J., Staab J.F., Sundstrom P. A 368-base-pair cis-acting HWP1 promoter region, HCR, of Candida albicans confers hypha-specific gene regulation and binds architectural transcription factors Nhp6 and Gcf1p. Eukaryot. Cell. 2007;6:693–709. doi: 10.1128/EC.00341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadosh D., Johnson A.D. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bensen E.S., Martin S.J., Li M., Berman J., Davis D.A. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 24.Enjalbert B., Whiteway M. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell. 2005;4:1203–1210. doi: 10.1128/EC.4.7.1203-1210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadosh D., Johnson A.D. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee M., Thompson D.S., Lazzell A., Carlisle P.L., Pierce C., Monteagudo C., López-Ribot J.L., Kadosh D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran G.P. Transcript profiling reveals rewiring of iron assimilation gene expression in Candida albicans and C. dubliniensis. FEMS Yeast Res. 2012;12:918–923. doi: 10.1111/j.1567-1364.2012.00841.x. [DOI] [PubMed] [Google Scholar]
- 28.Grumaz C., Lorenz S., Stevens P., Lindemann E., Schöck U., Retey J., Rupp S., Sohn K. Species and condition specific adaptation of the transcriptional landscapes in Candida albicans and Candida dubliniensis. BMC Genomics. 2013;14:1-1. doi: 10.1186/1471-2164-14-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haran J., Boyle H., Hokamp K., Yeomans T., Liu Z., Church M., Fleming A.B., Anderson M.Z., Berman J., Myers L.C., Sullivan D.J., Moran G.P. Telomeric ORFs (TLOs) in Candida spp. Encode mediator subunits that regulate distinct virulence traits. PLoS Genet. 2014;10:e1004658. doi: 10.1371/journal.pgen.1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillum A.M., Tsay E.Y., Kirsch D.R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 31.Morschhäuser J., Ruhnke M., Michel S., Hacker J. Identification of CARE-2-negative Candida albicans isolates as Candida dubliniensis. Mycoses. 1999;42:29–32. doi: 10.1046/j.1439-0507.1999.00259.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.