Abstract
Mitochondrial metabolic dysfunction is often seen in cancers. This paper shows that the defect in a mitochondrial electron transport component, the cytochrome c oxidase (CcO), leads to increased glycolysis and carcinogenesis. Using whole genome microarray expression analysis we show that genetic silencing of the CcO subunit Cox4i1 in mouse C2C12 myoblasts resulted in metabolic shift to glycolysis, activated a retrograde stress signaling, and induced carcinogenesis. In the knockdown cells, the expression of Cox4i1 was less than 5% of the control and the expression of the irreversible glycolytic enzymes (Hk1, Pfkm and Pkm) increased two folds, facilitating metabolic shift to glycolysis. The expression of Ca2+ sensitive Calcineurin (Ppp3ca) and the expression of PI3-kinase (Pik3r4 and Pik3cb) increased by two folds. This Ca2+/Calcineurin/PI3K retrograde stress signaling induced the up-regulation of many nuclear genes involved in tumor progression. Overall, we found 1047 genes with 2-folds expression change (with p-value less than 0.01) between the knockdown and the control, among which were 35 up-regulated genes in pathways in cancer (enrichment p-value less than 10− 5). Functional analysis revealed that the up-regulated genes in pathways in cancer were dominated by genes in signal transduction, regulation of transcription and PI3K signaling pathway. These results suggest that a defect in CcO complex initiates a retrograde signaling which can induce tumor progression. Physiological studies of these cells and esophageal tumors from human patients support these results. GEO accession number = GSE68525.
Keywords: Genome expression, Functional analysis, Tumor progression, Mitochondrial metabolic dysfunction, Cytochrome c oxidase defects
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus/C2C12 myoblasts |
| Sequencer or array type | Affymetrix GeneChip Mouse Gene 2.0ST Arrays scanned by Affymetrix GeneChip Scanner 3000 |
| Data format | Raw CEL files and RMA summary |
| Experimental factors | Tumorigenic vs. control cells |
| Experimental features | Genetic silencing of Cox4i1 by shRNA induced a tumorigenic phenotype in C2C12 myoblasts, the mRNA expression of which was compared to the control with scrambled shRNA |
1. Direct link to deposited data
2. Direct link to deposited source codes
3. Experimental Design, Materials and Methods
3.1. Sample preparation
Cells were grown in Dulbecco's Modified Eagle medium supplemented with 10% fetal bovine serum and 0.1% penicillin/streptomycin. In case of Cox4i1 silenced (C4KD) cells, the medium was further supplemented with 1 mM Pyruvate and 50 μg/ml Uridine. RNA was prepared using Qiagen Rneasy kit following manufacturer's instructions. 5.5 μg single-stranded cDNA was fragmented and labeled using the Affymetrix GeneChip WT Terminal Labeling kit (PN: 900671). Labeled cDNA (3.25 μg) was hybridized for 17 h at 45°C to Affymetrix GeneChip Mouse Gene 2.0ST Arrays (PN: 902118).
3.2. Microarray analysis
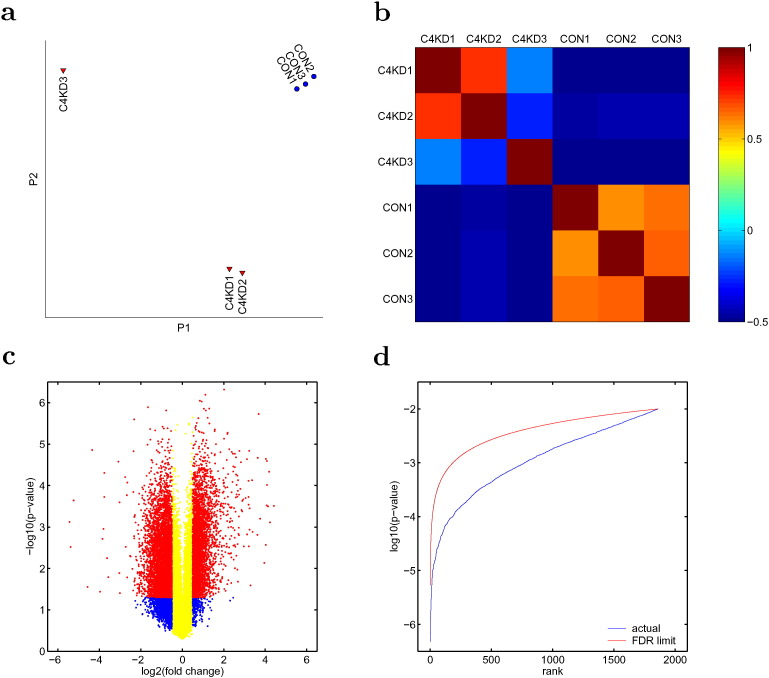
The data were processed by Affymetrix Expression Console using Probeset-Summarize-Engine with default setting of the robust multi-array average (RMA, [1]) method to calculate the expression level for each probeset of the array (in log2 unit, online file: rma-gene-full.summary.txt). For quality control, we performed principal component analysis (PCA) based on RMA expression levels of the samples. The projection to the most dominant two components is shown in Fig. 1a. The control samples formed a single cluster separable from the knockdown. However, the knockdown samples are further branched into two groups. The grouping is also clearly shown (Fig. 1b) by the correlation coefficient value (with each probe mean over all samples removed) between two samples: if the two samples come from the same group, the value is high (above 0.6); if they come from different groups, the value is low or negative. Further investigation of the biological samples showed that C4KD1 and C4KD2 cells contained nearly 90% reduced Cox4i1 mRNA and protein as compared to control cells [2], while in C4KD3 cells the reduction was not as much (probably due to a less effective population selection by antibiotic resulting a mixture of knockdown and non-knockdown cells). This is consistent with microarray results: for both C4KD1 and C4KD2 the reduction were ~ 20 folds, for C4KD3 only ~ 2 folds. In the following analysis, C4KD3 was excluded.
Fig. 1.
Quality control for data analysis. The array expression data (from all 41,345 probes) of each sample are projected to the first two components of PCA, P1 and P2 in a, showing the separation of 6 samples into three groups. This separation is also shown in the 6 × 6 matrix of the correlation coefficients between samples (b). The plot in (c) shows the selection criteria for significant fold change in individual gene expression: the absolute fold change has to be bigger than 20.5 (red and blue dots), and the p-value has to be smaller than 0.05. For functional analysis, the selected genes are ranked by p-value and are further selected by Benjamini-Hocheberg method to limit FDR (d).
3.3. Statistical analysis
Fold changes are presented as the mean ± s.e.m. (standard error of the mean) and p-values were determined by one-tailed Welch's t-test, which is used to test the hypothesis that two populations have equal means and is more reliable than Student's t-test when the samples from two populations have unequal variances and unequal sample sizes [3]. So for a statistic t and a degrees of freedom v, the p-value is calculated by
| (1) |
where I is the regularized incomplete beta function. The statistic t is defined by the following formula:
| (2) |
where m1 and s1 are the mean and the s.e.m of the 1st sample, and m2 and s2 of the 2nd sample. The degrees of freedom v is approximated by
| (3) |
where v1 and v2 are the 1st and 2nd sample size, respectively. All calculations are performed using RMA expression levels, i.e., in log2 unit. For a given gene, a fold change > 20.5 with a p-value < 0.05 was considered statistically significant. Fig. 1b shows the distribution of fold change versus p-value for the full probeset of the Affymetrix GeneChip Mouse Gene 2.0ST Arrays. Out of 41,345 probes, 6049 had statistically significant fold change (red dots in Fig. 1c). Among those are 4908 uniquely identified genes (out of 26,515 Entrez genes of the array). Each individual gene expression presented in the paper is among those.
3.4. Pathway analysis
The list of genes with significant fold change was uploaded to KEGG Mapper to search pathways (http://www.genome.jp/kegg/tool/map_pathway1.html, [4]). To further narrow down the list of pathways, we calculated the p-value of pathway enrichment. With H the hits, i.e., the number of genes from our list present in a pathway of size S, N the number of genes of the list, and T the total number of genes of the array, the enrichement p-value was determined by
| (4) |
in which F(h; t, s, n) is the hypergeometric cumulative distribution function
| (5) |
We found N = 4908, used T = 26515 and retrieved S from the KEGG database (July, 2015). Table 1 shows the top 64 most enriched pathways—excluding the ones in organ functions and specific diseases.
Table 1.
Top 64 most enriched pathways in the C4KD cells compared to the control cells.
The top pathways were selected with the number of hits ≥ 20 and the p-value < 0.15.
| Hits | Size | % | p-value | Kegg id | Name of pathway |
|---|---|---|---|---|---|
| 330 | 1271 | 26 | 1.3e-11 | 01100 | Metabolic pathways |
| 127 | 397 | 32 | 6.4e-11 | 05200 | Pathways in cancer |
| 95 | 352 | 27 | 5.3-e05 | 04151 | PI3K-Akt signaling pathway |
| 74 | 232 | 32 | 6.3e-07 | 05203 | Viral carcinogenesis |
| 73 | 206 | 35 | 6.1e-09 | 05205 | Proteoglycans in cancer |
| 71 | 276 | 26 | 1.8e-03 | 05206 | MicroRNAs in cancer |
| 68 | 254 | 27 | 7.2e-04 | 04010 | MAPK signaling pathway |
| 66 | 208 | 32 | 3.0e-06 | 04510 | Focal adhesion |
| 64 | 170 | 38 | 3.4e-09 | 03013 | RNA transport |
| 64 | 218 | 29 | 6.3e-05 | 01130 | Biosynthesis of antibiotics |
| 64 | 235 | 27 | 6.3e-04 | 04144 | Endocytosis |
| 60 | 145 | 41 | 1.1e-10 | 03010 | Ribosome |
| 58 | 230 | 25 | 6.9e-03 | 04014 | Ras signaling pathway |
| 54 | 214 | 25 | 8.7e-03 | 04015 | Rap1 signaling pathway |
| 50 | 133 | 38 | 1.8e-07 | 03040 | Spliceosome |
| 49 | 118 | 42 | 5.5e-09 | 04919 | Thyroid hormone signaling pathway |
| 48 | 177 | 27 | 3.0e-03 | 05202 | Transcriptional misregulation in cancer |
| 45 | 168 | 27 | 5.1e-03 | 04141 | Protein processing in endoplasmic reticulum |
| 45 | 172 | 26 | 8.1e-03 | 04022 | cGMP-PKG signaling pathway |
| 45 | 179 | 25 | 1.7e-02 | 00230 | Purine metabolism |
| 44 | 198 | 22 | 1.1e-01 | 04062 | Chemokine signaling pathway |
| 43 | 139 | 31 | 2.7e-04 | 00190 | Oxidative phosphorylation |
| 43 | 143 | 30 | 5.4e-04 | 04120 | Ubiquitin mediated proteolysis |
| 41 | 142 | 29 | 1.7e-03 | 04910 | Insulin signaling pathway |
| 41 | 129 | 32 | 2.0e-04 | 04152 | AMPK signaling pathway |
| 41 | 174 | 24 | 5.5e-02 | 04145 | Phagosome |
| 40 | 181 | 22 | 1.3e-01 | 04020 | Calcium signaling pathway |
| 40 | 143 | 28 | 3.6e-03 | 04310 | Wnt signaling pathway |
| 39 | 159 | 25 | 3.5e-02 | 04921 | Oxytocin signaling pathway |
| 39 | 125 | 31 | 4.3e-04 | 04110 | Cell cycle |
| 38 | 154 | 25 | 3.4e-02 | 04390 | Hippo signaling pathway |
| 37 | 104 | 36 | 2.8e-05 | 00240 | Pyrimidine metabolism |
| 37 | 124 | 30 | 1.5e-03 | 04142 | Lysosome |
| 37 | 103 | 36 | 2.2e-05 | 04922 | Glucagon signaling pathway |
| 36 | 134 | 27 | 1.1e-02 | 04068 | FoxO signaling pathway |
| 35 | 122 | 29 | 4.0e-03 | 04722 | Neurotrophin signaling pathway |
| 35 | 115 | 30 | 1.3e-03 | 04114 | Oocyte meiosis |
| 34 | 140 | 24 | 5.2e-02 | 04530 | Tight junction |
| 33 | 140 | 24 | 7.8e-02 | 04550 | Signaling pathways regulating pluripotency of stem cells |
| 31 | 94 | 33 | 5.6e-04 | 00564 | Glycerophospholipid metabolism |
| 30 | 109 | 28 | 1.3e-02 | 04066 | HIF-1 signaling pathway |
| 29 | 101 | 29 | 8.1e-03 | 05231 | Choline metabolism in cancer29 |
| 29 | 117 | 25 | 5.5e-02 | 01200 | Carbon metabolism |
| 28 | 124 | 23 | 1.5e-01 | 04071 | Sphingolipid signaling pathway |
| 27 | 87 | 31 | 3.3e-03 | 04540 | Gap junction |
| 26 | 88 | 30 | 7.9e-03 | 04512 | ECM-receptor interaction |
| 26 | 96 | 27 | 2.5e-02 | 03015 | mRNA surveillance pathway |
| 25 | 83 | 30 | 7.0e-03 | 03008 | Ribosome biogenesis in eukaryotes |
| 25 | 99 | 25 | 5.9e-02 | 04915 | Estrogen signaling pathway |
| 25 | 67 | 37 | 2.3e-04 | 04115 | p53 signaling pathway |
| 25 | 89 | 28 | 1.8e-02 | 04912 | GnRH signaling pathway |
| 25 | 82 | 30 | 5.9e-03 | 04210 | Apoptosis |
| 25 | 72 | 35 | 8.0e-04 | 04920 | Adipocytokine signaling pathway |
| 23 | 74 | 31 | 6.3e-03 | 04520 | Adherens junction |
| 23 | 87 | 26 | 4.3e-02 | 04012 | ErbB signaling pathway |
| 23 | 83 | 28 | 2.6e-02 | 03018 | RNA degradation |
| 23 | 82 | 28 | 2.2e-02 | 003320 | PPAR signaling pathway |
| 22 | 74 | 30 | 1.3e-02 | 04917 | Prolactin signaling pathway |
| 22 | 82 | 27 | 4.0e-02 | 04070 | Phosphatidylinositol signaling system |
| 22 | 66 | 33 | 2.9e-03 | 05230 | Central carbon metabolism in cancer |
| 21 | 82 | 26 | 6.9e-02 | 04350 | TGF-beta signaling pathwa |
| 20 | 79 | 25 | 8.2e-02 | 01230 | Biosynthesis of amino acids |
| 20 | 66 | 30 | 1.4e-02 | 00010 | Glycolysis / Gluconeogenesis |
| 20 | 61 | 33 | 5.4e-03 | 04150 | mTOR signaling pathway |
3.5. Functional analysis
A more stringent criteria was used to select genes for functional annotation: a fold change > 2 with a p-value < 0.01. There were 1861 probes satisfied this criteria. To control the false discovery rate (FDR), the Benjamini-Hocheberg procedure [5] was applied to select further: for the FDR at level q, find the largest k such that
| (6) |
then select i = 1...k, where pk is the p-value of m genes with lowest p-values. For a FDR of 0.01, 1860 out 1861 probes were selected (Fig. 1d). Among those were 656 up-regulated and 391 down-regulated uniquely identified genes (see files mypup.txt.2x0.01p and mypdown.txt.2x0.01p). The generated gene list was functionally annotated using DAVID 6.7 (http://david.abcc.ncifcrf.gov/, [6]) with default setting (online file: myfa_table_all.txt.2x0.01p). The functional groups of genes were extracted from the annotated file using the corresponding terms (e.g. “signal transduction,”) with our own script (online file: myfa.csh).
4. Results
4.1. Enriched expression in metabolic, cancer and PI3K signaling pathways
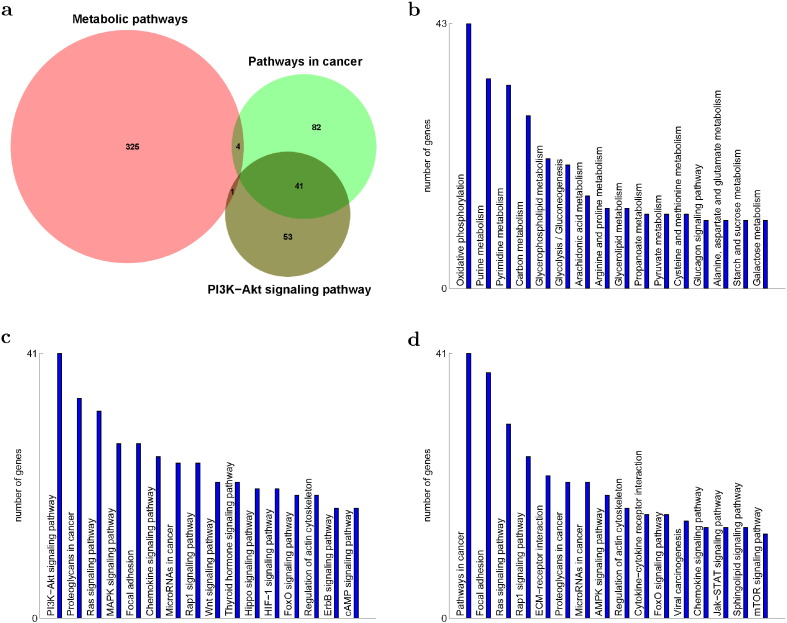
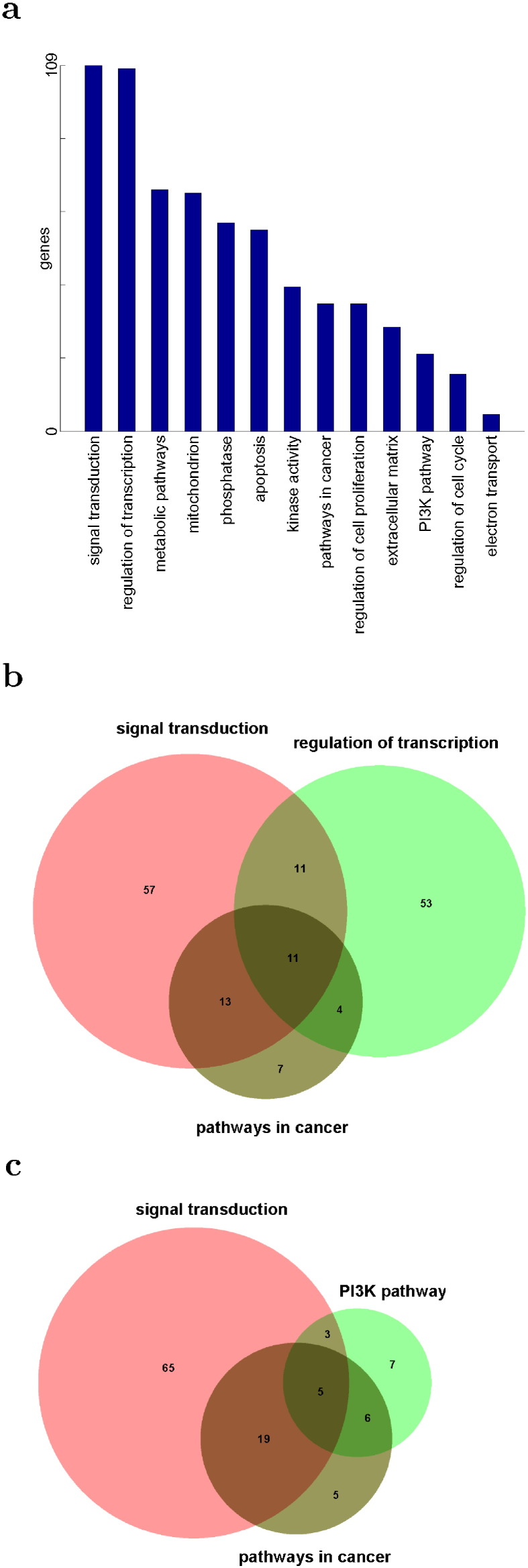
Metabolic Pathways, Pathways in Cancer and PI3K-Akt Signaling Pathway were the top three pathways enriched with the genes of significant expression change due to the CcO complex defect (Table 1). Among the three, Pathways in Cancer and PI3K-Akt Signaling Pathway had a lots of overlaps while they had little overlaps with Metabolic Pathways (Fig. 2a). The 330 genes enriching Metabolic Pathways were distributed in different pathways of metabolism (Fig. 2b). Oxidative Phosphorylation was the leading one. Another enriched one was Glycolysis/Gluconeogenesis. Glycolysis is known to be responsible for energy generation if Oxidative Phosphorylation is interrupted. The 127 genes enriching Pathways in Cancer were sub-divided into many pathways (Fig. 2c). Interestingly, genes in PI3K-Akt Signaling Pathway accounted for one third overall and contributed about half to others except Wnt and Hippo signaling pathways.
Fig. 2.
Three major pathways of significantly regulated genes of CcO4KD cells—Metabolic Pathways, Pathways in Cancer and PI3K-Akt Signaling Pathway: a, overlaps between the three; b, Metabolic Pathways' overlaps with others; c, Pathways in Cancer's overlaps with others; d, PI3K-Akt Signaling Pathway's overlaps with others.
The 95 genes enriching PI3K-Akt Signaling Pathway overlapped most significantly with pathways in cancer and also overlapped with additional signaling pathways (Fig. 2d). Especially interesting were AMPK and mTOR pathways, known to be activated during metabolic stress. Most of these pathways were among the top 64 enriched pathways and accounted for most of the signaling pathways of the top 64. One significant exception was Calcium Signaling Pathway (Table 1). It is significant because this pathway is involved in a unique retrograde signaling cascade responding to [Ca2+]c elevation during disruptions of the mitochondrial electron transport chain [7]. We show in the following sections with specific expression changes that the defect in CcO complex caused metabolic shift to glycolysis, induced calcium dependent retrograde signaling, activated PI3K pathway and resulted in carcinogenesis.
4.2. Disruption of CcO complex leading to metabolic shift to glycolysis
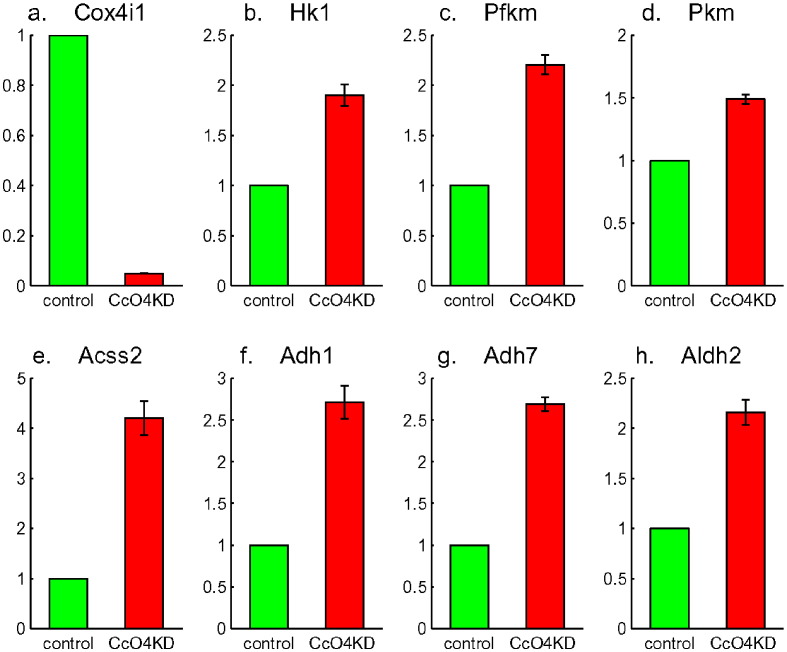
The expression of Cox4i1 in the cells with Cox4i1 silenced by shRNA, referred as CcO4KD cells throughout this paper, was less than 5% of the control with scrambled shRNA (Fig. 3a). The mRNA expression levels of the irreversible glycolytic enzymes, Hexokinase (Hk1, Fig. 3b), Phosphofructokinae (Pfkm, Fig. 3c) and Pyruvate kinase (Pkm, Fig. 3d) had approximately two-folds increase. Fig. 3 also shows the mRNA expression levels of four other enzymes in the glycolytic pathway with the most fold-changes (Acss2, Adh1, Adh7 and Aldh2 in Fig. 3e to h). They were all up-regulated in CcO4KD cells and are downstream of glycolysis—important for processing pyruvate produced during glycolysis. In agreement with these data, CcO activity was reduced by about 90%, the Hk and Pfk activities increased about two folds, and glucose uptake was more than two-fold higher in CcO4KD cells [2]. These indicated that silencing Cox4i1 lead to the disruption of CcO activity and caused metabolic shift to glycolysis.
Fig. 3.
Knock-down of the expression level of cytochrome c oxidase (Cox4i1 in a), and expression up-regulation of all three irreversible glycolytic enzymes Hexokinase (Hk1 in b), Phosphofructokinase (Pfkm in c) and Pyruvate Kinase (Pkm in d), as well as four other enzymes downstream of glycolysis: Acss2, Adh1, Adh7 and Aldh2 (e, f, g and h).
4.3. Activation of Calcium dependent retrograde signaling pathway
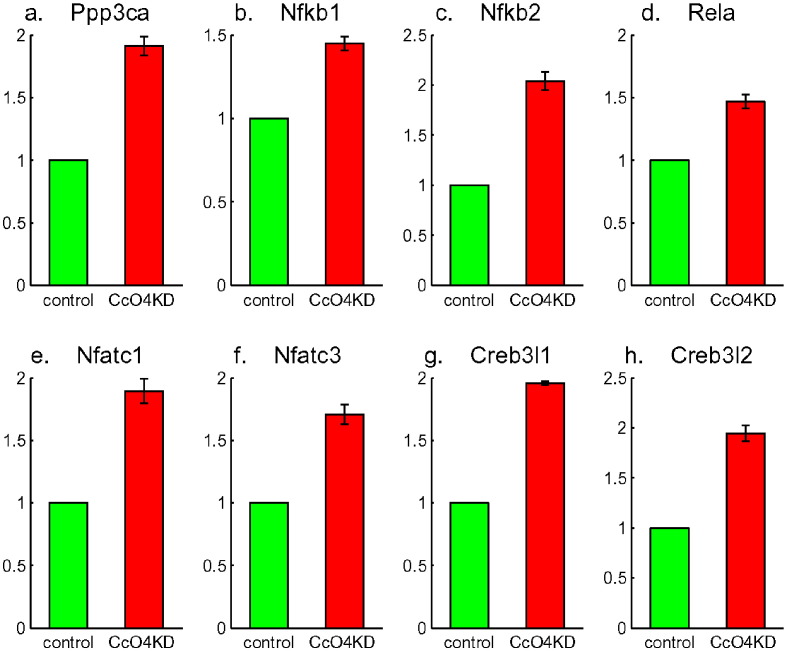
The mitochondrial stress and the loss of mitochondrial membrane potential were indicated by the enrichment of Calcium Signaling Pathway (Table 1) and in particular by a two-fold increase in the expression level of Ca2+ sensitive Calcineurin (Ppp3ca, Fig. 4a). This signal in response to sustained elevation of [Ca2+]c had been shown to activate a set of stress responsive transcription factors: NFkB, NFAT and CREB [7]. This was consistent with the increased expression levels of Nfkb1, Nfkb2, Rela, Nfatc1, Nfatc3, Creb3l1 and Creb3l2 (Fig. 4b to h). In addition to those, this retrograde stress signaling induced increased expression of many genes involved in tumor progression. In particular, Tgfb1 and Mmp16 had 2 to 3 folds expression increase (Fig. 5a and b). In agreement with these data, Calcineurin activity increased three folds in CcO4KD cells; while for the cells further treated with BAPTA, a Ca2+ chelator, or FK506, a specific Calcineurin inhibitor, the up-regulation of Tgfb1 and Mmp16 was significantly attenuated and the glucose uptake was significantly inhibited comparing to untreated cells [2]. These indicated that the Calcineurin mediated retrograde signaling pathway played an important role in metabolic shift to glycolysis and carcinogenesis.
Fig. 4.
Up-regulation of Calcineurin (Ppp3ca in a) and Calcineurin pathway: NFkB (Nfkb1 in b, Nfkb2 in c and Rela in d), NFAT (Nfatc1 in e and Nfatc3 in f), and CREB (Creb3l1 in g and Creb3l2 in h).
Fig. 5.
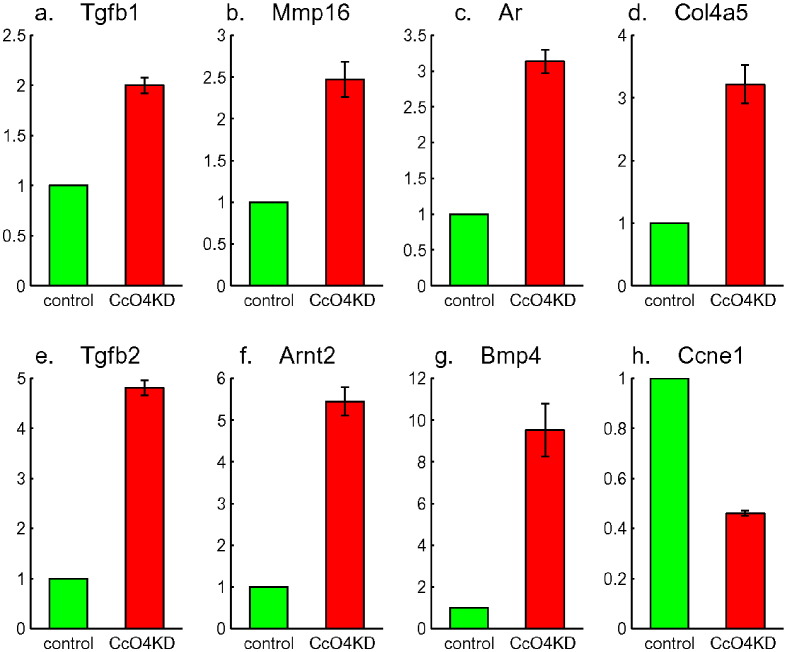
Up-regulation of seven genes related to cancer: Tgfb1 in a, Mmp16 in b, Ar in c, Col4a5 in d, Tgfb2 in e, Arnt2 in f and Bmp4 in g. Down-regulation of one gene related to cancer: Ccne1 in h.
4.4. Activation of PI3-kinase pathway and carcinogenesis
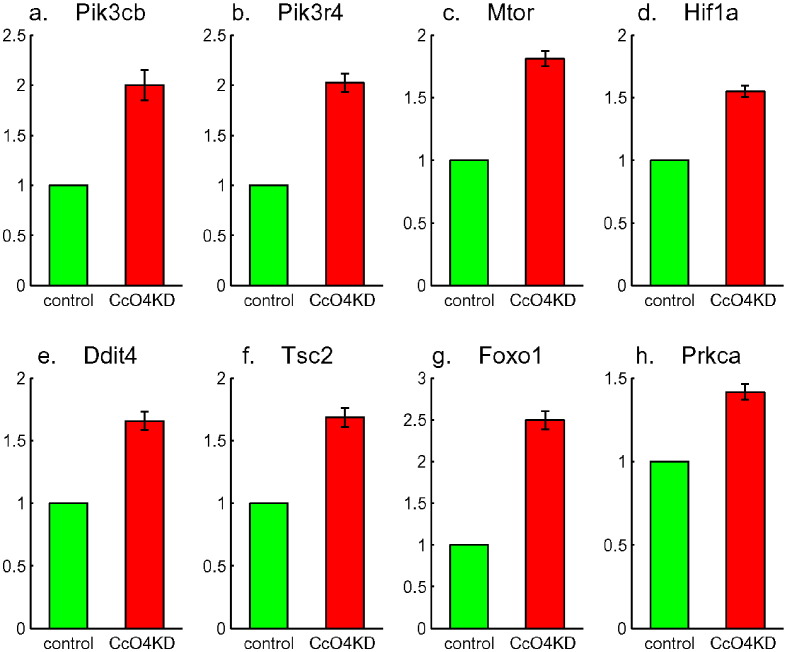
The activation of PI3-kinase pathway was indicated by a two-fold increase in expression level of PI3-kinase—of both catalytic and regulatory subunits Pik3cb and Pik3r4 (Fig. 6a and b). It has been shown that this pathway is a major determinant of the glycolytic phenotype through Akt1 and Mtor signaling and subsequent downstream Hif1a transcription factor activation [8]. In addition to increased expression of Mtor and Hif1a (Fig. 6c and d) in CcO4KD cells, there were also increased expression levels for genes further down stream [9] of this pathway including Redd1(Ddit4), Tsc2, Foxo1 and PKCα(Prkca) (Fig. 6e to h) as well as other genes related to pathways in cancer (Fig. 5). Aerobic glycolysis supports various biosynthetic pathways and, consequently, the metabolic requirements for proliferation. Consistent with this, the CcO4KD cells developed anchorage independent growth, an important hallmark of malignant cells, and further treatment with Wortmannin, a PI3-kinase inhibitor, attenuated the glucose uptake and inhibited the colony formation of those cells [2]. These indicated that the PI3-kinase pathway played a crucial role in metabolic shift to glycolysis and in tumor progression.
Fig. 6.
Up-regulation of PI3-kinase (Pik3cb in a and Pik3r4 in b) and PI3-kinase pathway: Mtor in c, Hif1a in d, Ddit4 in e, Tsc2 in f, Foxo1 in g, and Prkca in h.
4.5. Oncogenes enriched transcription profiles
Bmp4, Arnt2, Tgfb2, Col4a5 and Ar (shown in Fig. 5c to g) were the five most up-regulated genes in Pathways in Cancer. Ccne1 (shown in Fig. 5h) was the most down-regulated gene in Pathways in Cancer. Col4a5 and Ccne1 are in PI3K-Akt Pathway and their corresponding up and down regulation are consistent with their role in cancers. Furthermore, Ar, Bmp4 and Tgfb2 are involved in signal transduction and Ar, Arnt2 and Bmp4 are involved in regulation of transcription. To gain additional understanding of functional roles of the most significantly regulated genes of CcO4KD cells, we further narrowed down the list of genes and did functional annotation of the list. There were 1047 genes with 2-fold or more expression change and p-value less than 0.01. The distribution of those genes in the three dominate pathways and 10 different functional groups is shown in Fig. 7a. Among those were 38 significantly regulated genes in Pathways in Cancer (enrichment p-value less than 10− 6). Table 2 shows the fold changes and the p-values of the 38 genes. 35 genes in Pathways in Cancer were up-regulated (enrichment p-value less than 10− 5). 24, 15 and 11 of these up-regulated genes in pathways in cancer are in functional groups of signal transduction, regulation of transcription and PI3K pathway, respectively. There were extensive overlaps (11) between those in signal transduction and regulation of transcription (Fig. 7b). There were also overlaps (5) between those in signal transduction and PI3K pathway (Fig. 7c). There was only one (Rxra) overlap between those in regulation of transcription and PI3K pathway. Only two genes (Dapk2 and Ncoa4) in pathways in cancer were not in any of the other three functional groups. Interestingly, none of genes in pathways in cancer was in metabolic pathway. It appears that CcO defects induced Calcineurin/PI3K retrograde signaling which in turns induced up-regulation of genes involved in signal transduction and regulation of transcription, up-regulation of genes related to pathways in cancer, and carcinogenesis.
Fig. 7.
Functional groups of significantly regulated genes of CcO4KD cells: a, the number of genes with significant fold-changes in different functional groups; b, the Venn diagrams of up-regulated genes in signal transduction (pink), regulation of transcription (green) and pathways in cancer (brown); c, the Venn diagrams of up-regulated genes in signal transduction (pink), PI3K pathway (green) and pathways in cancer (brown).
Table 2.
38 significantly regulated genes in pathways in cancer. 35 were up-regulated in CcO4KD and only 3 were down-regulated (Ccne1, Ccne2 and Tceb1). The genes were selected with a fold change ≥ 2 and a p-value < 0.01. The fold change is expressed in log2 unit.
| Symbol | Fold | p-Value | Entrez | Gene name |
|---|---|---|---|---|
| Ar | 1.65 | 2e-04 | 11,835 | Androgen receptor |
| Arnt2 | 2.45 | 1e-03 | 11,864 | Aryl hydrocarbon receptor nuclear translocator 2 |
| Bmp4 | 3.25 | 3e-04 | 12,159 | Bone morphogenetic protein 4 |
| Ccne1 | − 1.12 | 1e-03 | 12,447 | Cyclin E1 |
| Ccne2 | − 1.09 | 3e-04 | 12,448 | Cyclin E2 |
| Cebpa | 1.54 | 2e-03 | 12,606 | CCAAT/enhancer binding protein (C/EBP), α |
| Col4a2 | 1.45 | 2e-04 | 12,827 | Collagen, type IV, α 2 |
| Col4a5 | 1.69 | 1e-03 | 12,830 | Collagen, type IV, α 5 |
| Col4a6 | 1.02 | 1e-04 | 94,216 | Collagen, type IV, α 6 |
| Dapk2 | 1.45 | 1e-03 | 13,143 | Death-associated protein kinase 2 |
| Ep300 | 1.03 | 2e-05 | 328,572 | E1A binding protein p300 |
| Figf | 1.31 | 1e-03 | 14,205 | c-fos induced growth factor |
| Foxo1 | 1.32 | 1e-03 | 56,458 | Forkhead box O1 |
| Fzd4 | 1.08 | 4e-03 | 14,366 | Frizzled homolog 4 (Drosophila) |
| Fzd6 | 1.56 | 4e-03 | 14,368 | Frizzled homolog 6 (Drosophila) |
| Gli2 | 1.07 | 5e-03 | 14,633 | GLI-Kruppel family member GLI2 |
| Jak1 | 1.02 | 2e-05 | 16,451 | Janus kinase 1 |
| Kitl | 1.46 | 2e-03 | 17,311 | Kit ligand |
| Lama2 | 1.49 | 1e-04 | 16,773 | Laminin, α 2 |
| Lamb3 | 1.35 | 6e-04 | 16,780 | Laminin, β 3 |
| Lamc1 | 1.18 | 3e-05 | 226,519 | Laminin, γ 1 |
| Met | 1.13 | 2e-03 | 17,295 | Met proto-oncogene |
| Ncoa4 | 1.05 | 2e-04 | 27,057 | Nuclear receptor coactivator 4 |
| Nfkb2 | 1.03 | 9e-04 | 18,034 | Nuclear factor of κ light polypeptide gene enhancer in B cells 2 |
| Pdgfb | 1.74 | 8e-04 | 18,591 | Platelet derived growth factor, B polypeptide |
| Ppard | 1.15 | 9e-04 | 19,015 | Peroxisome proliferator activator receptor delta |
| Rxra | 1.03 | 2e-04 | 20,181 | Retinoid X receptor α |
| Stat1 | 1.23 | 1e-03 | 20,846 | Signal transducer and activator of transcription 1 |
| Stat5a | 1.02 | 1e-04 | 20,850 | Signal transducer and activator of transcription 5A |
| Tceb1 | − 1.09 | 1e-03 | 67,923 | Transcription elongation factor B (SIII), polypeptide 1 |
| Tcf7 | 1.16 | 6e-03 | 21,414 | Transcription factor 7, T cell specific |
| Tcf7l2 | 1.05 | 4e-05 | 21,416 | Transcription factor 7 like 2, T cell specific, HMG box |
| Tgfb1 | 1.00 | 6e-04 | 21,803 | Transforming growth factor, β 1 |
| Tgfb2 | 2.27 | 1e-05 | 21,808 | Transforming growth factor, β 2 |
| Tgfbr2 | 1.36 | 4e-05 | 21,813 | Transforming growth factor, β receptor II |
| Wnt10a | 1.58 | 9e-04 | 22,409 | Wingless related MMTV integration site 10a |
| Wnt4 | 1.16 | 7e-03 | 22,417 | Wingless-related MMTV integration site 4 |
| Wnt7b | 1.40 | 3e-04 | 22,422 | Wingless-related MMTV integration site 7B |
5. Discussion
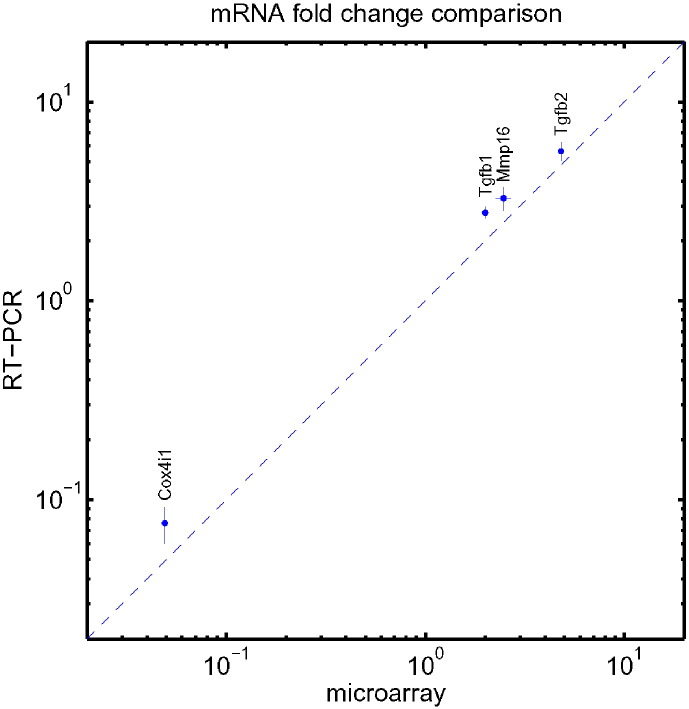
The whole genome microarray analysis of the knockdown and control cells gave the insight into the magnitude of systematic changes of gene expressions and also enabled the global analysis of pathways and functional groups of many genes efficiently. It is interesting to know the accuracy of individual fold change of gene expression estimated from the experimental array. For some of the genes, the relative mRNA levels were also measured by RT-PCR [2]. Fig. 8 shows the comparison between the results of microarray and RT-PCR. The agreement is quite good. The that fold change estimated by the array is slightly on the conservative side. This gave confidence to use the array not only for the global analysis but also for measuring relative change of individual gene's mRNA level.
Fig. 8.
The comparison between the expression fold changes of four genes from microarray and those measured by RT-PCR in a log-log plot. The straight line lies where they are equal.
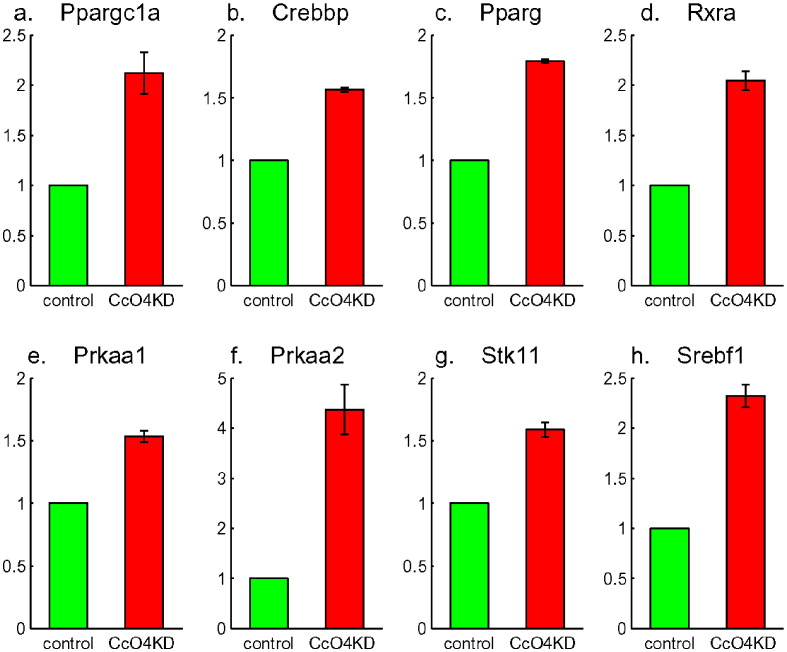
The pathway analysis also indicated that PPAR (PGC-1α) signaling pathway was activated. PGC-1α is another regulator of mitochondrial biogenesis and function. It has been shown that PGC-1α has a positive feedback to one of its upstream regulator: Calcineurin [10]. This protein can interact with, and regulate the activities of, cAMP response element-binding protein (CREB). Fig. 9a to d shows the up-regulation of PGC-1α (Ppargc1a) and its interaction partners Crebbp, Pparg, and Rxra. Another important pathway activated was the AMP-activated protein kinase (AMPK) signaling pathway. AMPK plays an important role in insulin signaling, glucose uptake, and energy homeostasis and is involved in PGC-1α-mediated mitochondrial biogenesis. The microarray analysis indicated up-regulation of AMPK (Prkaa1 and Prkaa2 in Fig. 9e and f) and its up-stream regulator LKB1 (Stk11, Fig. 9g) as well as two downstream genes (Srebf1, Fig. 9h; and Tsc2, Fig. 6f). This is puzzling since AMPK is supposed to down-regulated Srebf1 and Tsc2 and to be a metabolic checkpoint to control cell proliferation when activated under energy stress. But it was not doing any of those in CcO4KD cells. The role of AMPK pathway in Cox4i1 knockdown needs to be studied further.
Fig. 9.
Up-regulation of PGC-1α (Ppargc1a in a) and its interaction partners: Crebbp in b, Pparg in c, and Rxra in d. Up-regulation of AMPK (Prkaa1 in e and Prkaa2 in f), its upstream regulator LKB1 (Stk11 in g) and a downstream gene Srebf1 in h.
Our physiological experiments have shown that CcO dysfunction is a possible biomarker for cancer progression in digestive diseases such as esophageal tumors [2]. It will be very interesting to conduct a whole genome expression analysis on such tumors and to compare the results with those presented in this paper. Such comparison will help to further understand the pathways of this type of tumor progression and to find possible pathway-based methods of diagnosis and treatment.
Acknowledgments
This work was supported by NIH grant CA-22762, NIH/NIDDK Center grant P30DK050306, and a grant from Research Foundation, University of Pennsylvania.
References
- 1.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan S., Guha M., Dong D.W., Whelan K.A., Ruthel G., Uchikado Y., Natsugoe S., Nakagawa H., Avadhani N.G. Disruption of cytochrome c oxidase function induces Warburg effect and metabolic reprogramming. Oncogene. 2015 doi: 10.1038/onc.2015.227. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruxton G.D. The unequal variance t-test is an underused alternative to Student's t-test and the Mann-Whitney U test. Behav. Ecol. 2006;17:688–690. [Google Scholar]
- 4.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(1):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57(1):289–300. [Google Scholar]
- 6.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guha M., Srinivasan S., Biswas G., Avadhani N.G. Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J. Biol. Chem. 2007;282(19):14536–14546. doi: 10.1074/jbc.M611693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simons A.L., Orcutt K.P., Madsen J.M., Scarbrough P.M., Spitz D.R. Humana Press; 2012. Oxidative stress in cancer biology and therapy; pp. 21–46. [Google Scholar]
- 9.Brugarolas J., Lei K., Hurley R.L., Manning B.D., Reiling J.H., Hafen E., Witters L.A., Ellisen L.W., Kaelin J.W.G. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summermatter S., Thurnheer R., Santos G., Mosca B., Baum O., Treves S., Hoppeler H., Zorzato F., Handschin C. Remodeling of calcium handling in skeletal muscle through PGC-1α: impact on force, fatigability, and fiber type. Am. J. Physiol. Cell Physiol. 2012;302(1):C88–C99. doi: 10.1152/ajpcell.00190.2011. [DOI] [PubMed] [Google Scholar]