Abstract
To gain new insights about the genetic networks controlling hair cell (HC) development, we previously developed a direct genetic programming strategy to generate an inexhaustible supply of HC-like cells (induced HCs, iHCs) in vitro, starting from mouse embryonic stem cells (ESC). We found that combined activity of three transcription factors, Gfi1, Pou4f3, and Atoh1, can program ESC-derived progenitors towards HC fate with efficiencies of 55%–80%. These iHCs express several HC markers and exhibit polarized structures that are highly reminiscent of the mechanosensitive hair bundles, with many microvilli-like stereocilia. Here, we describe the experimental design, methodology, and data validation for the microarray analysis used to characterize the transcriptome profile of iHCs at different stages of their differentiation. This approach based on FACS sorting and microarray analysis revealed a highly similar iHC transcriptome to that of endogenous HCs in vivo. The data obtained in this study is available in the Gene Expression Omnibus (GEO) database (accession number GSE60352).
Keywords: Hair cells, Mouse embryonic stem cells, In vitro differentiation, Cell type programming, Transcriptome profiling
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus/mouse embryonic stem cells (iGPA-Myo7a:mVenus line) |
| Sex | N/A |
| Sequencer or array type | Affymetrix mouse gene 2.1 ST Array |
| Data format | Raw or analyzed |
| Experimental factors | iHCs or iHCs treated with retinoic acid at day 8 and day 12 vs untreated iGPA-Myo7a:mVenus-derived Embryoid Bodies at day 8 and day 12 |
| Experimental features | iGPA-Myo7a:mVenus-derived Embryoid Bodies treated with doxycycline (to activate GPA expression) or treated with doxycycline + retinoic acid during 4 or 8 days were sorted by FACS to purify iHCs (Myo7a:Venus positive cells). The iHCs were compared to untreated iGPA-Myo7a:mVenus-derived Embryoid Bodies at the corresponding time point to determine the transcriptional signature of iHCs. |
| Consent | N/A |
| Sample source location | Lisbon, Portugal |
1. Direct link to deposited data
The deposit data can be found at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60352.
2. Experimental design, materials and methods
2.1. Cell culture and embryoid bodies (EBs) formation
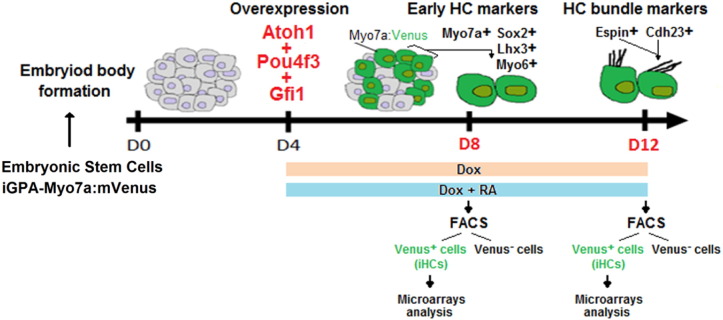
The embryonic stem cell (ESC) line iGPA-Myo7a:mVenus used in this study was generated from the Ainv15 ESC line [1] by two genetic engineering steps: I) a DNA fragment containing the three genes Gfi1-Pou4f3-Atoh1 (GPA) was inserted into the doxycycline (dox) inducible locus of the Ainv15 cells as previously described in [2] to produce the iGPA line. II) The mVenus fluorescent reporter under control of the promoter and regulatory regions of the incipient HC marker, Myo7a [3] was randomly inserted into the iGPA genome as described in [4], to generate the iGPA-Myo7a:mVenus line. These ESCs were routinely grown on gelatin-coated (0.1%) Nunc dishes at 37 °C in a 5% CO2 incubator with Dulbecco's Modified Eagles Medium 1 × (DMEM, GIBCO) supplemented with 10% of heat-inactivated fetal bovine serum (FBS) (GIBCO, ES-qualified), 2 mM glutamine (GIBCO), 1% penicillin/streptomycin (GIBCO), 1 mM sodium pyruvate (GIBCO), 1% MEM non-essential amino acids (GIBCO), 2 ng/ml Leukemia inhibitory factor (LIF) and 7 μm 2-mercaptoethanol (Sigma). All solutions were mixed and filtered through a 0.2 μm filter unit into a new sterile flask. Cells were passaged every other day, at constant plating density of 3 × 104 cells/cm2. To generate embryoid bodies (EBs), ESCs were dissociated into single cells (using 0025% trypsin at 37 °C for 3 min) and plated at low density (2 × 104 cells/cm2) with the same supplemented DMEM medium but without LIF, in 10 cm bacterial grade petri dishes to prevent attachment. EB formation was checked on day 1 and medium was replaced every two days (days 2, 4, 6, 8 and 10). Supplementation with 2 μg/ml doxycycline (Sigma, diluted in sterile PBS and filtered through a 0.2 μm filter unit), 1 μm retinoic acid (RA) (diluted in 0.01% DMSO, Sigma) was initiated at day 4 and maintained until the required time point for analysis (day 8 or day 12) (Fig. 1).
Fig. 1.
Schematic representation of the HC programming strategy used to generate and purify iHCs in vitro at an early (day 8) and later stage (day 12) of their development. FACS, fluorescence activated cell sorting.
2.2. iHC purification by FACS and isolation of total RNA
To determine the transcriptome profile of iHCs at different differentiation stages, we FACS-sorted Dox treated EBs at days 8 and 12 challenged with Dox or Dox + RA (Fig. 1). These EBs were collected into a conical tube and allowed to sediment (~ 5 min) at the bottom of the tube, the supernatant was removed and fresh PBS was added. EBs were washed twice in PBS, and after incubation with 1 ml of 0.25% trypsin for 5 min at 37 °C, DMEM media was added to stop the reaction. Cells were triturated 10–20 times using a P1000 Gilson pipette tip to obtain a single cell suspension. After centrifuge, cells were resuspended in FACS buffer (PBS with 4% FBS) and filtered using a cell strainer 70 μm (BD Bioscience). Cell sorting experiments were done on a FACS Aria cell sorter (Becton Dickinson). FACS-sorted cells were directly processed for RNA extraction using the High Pure RNA Isolation Kit (Roche), according to the manufacturer's instructions. For synthesis of cDNA, 1 μg of total RNA was used as a template for the reverse transcription performed with the Superscript II Reverse Transcriptase system (Invitrogen) in a final volume of 20 μl. Transcription was conducted with random primers. The absence of contaminating genomic DNA was confirmed for each RNA extraction by PCR amplification of GAPDH-specific product from reverse transcriptase negative samples (without Superscript II enzyme).
2.3. Sample validation
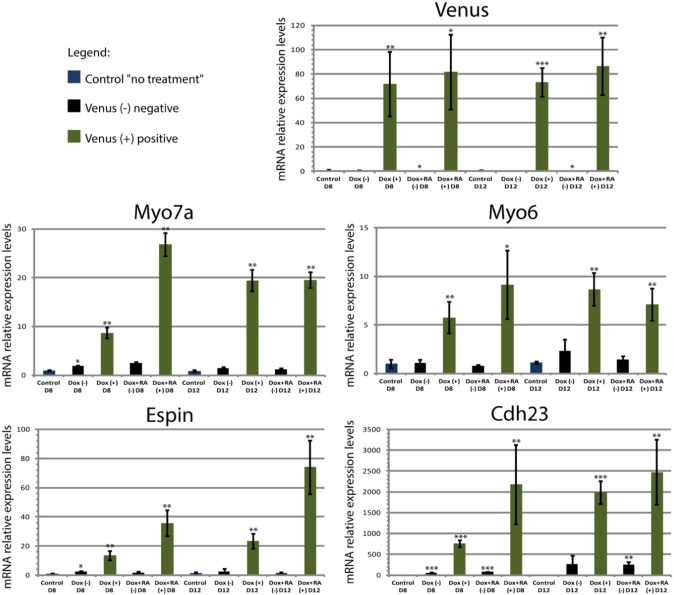
Sorted Venus-positive cell populations from EBs were collected with high purity (~ 98%) and the Venus-negative cell fraction was discarded for the microarray analysis. However, these nonfluorescent cells were used for quantitative-real time PCR (qPCR) analysis, to validate samples and to determine the enrichment of the iHC population in the Venus-positive fraction. The qPCR reaction was performed on 96-well plates (MicroAmp; Applied Biosystems) or 384-well plates (MicroAmp; Applied Biosystems) covered with optical adhesive covers (Applied Biosystems). The instruments used were Applied Biosystems 7500 Real-Time PCR or Applied Biosystems ViiA 7 Real-Time PCR. The Real-Time PCR was carried out using iTaq Universal Sybr Green Supermix (Bio-Rad), 2 μl of the retrotranscription cDNA template diluted 1:100 and 12,5 pmol of each primer. Reaction conditions were as follows: one step of 50 °C for 2 min, one 95 °C for 10 min, and 40 cycles of 95 °C for 15 s denaturation and 60 °C for 1 min annealing and extension. The cDNA was used as template for each pair of primers in a duplicate PCR reaction. GAPDH was used was a calibrator. Relative expression levels in the various Dox-treated samples were referred to the levels of expression in untreated control (without Dox), which were arbitrarily set to 1. Results are shown as averages ± standard error of mean (SEM) of three independent experiments. As expected, strong Venus expression was only observed among the sorted Venus-positive cells (Fig. 2). Myo7a expression was significantly higher in the Venus-positive sorted cells and HC markers such as Myo6 [5], Espin [6], and Cdh23 [7] were also enriched in this fraction (Fig. 2). Overall, the results confirmed the high degree of purity of the iHC-sorted cells.
Fig. 2.
Relative mRNA levels measured by qPCR for Venus and HC markers (Myo7a, Myo6, Espin and Cdh23) in the mVenus fluorescent reporter venus-positive (green) and venus-negative (black) sorted populations obtained from EBs treated with Dox or Dox + RA at day 8 and at day 12. Relative expression of each transcript is presented as fold change normalized to the mean of untreated EBs (arbitrary set to 1). Unpaired t-test was used for statistical analysis. *P < 0.05 **P < 0.01 ***P < 0.001 (n = 3).
3. Microarray sample preparation
RNA concentration and purity was determined by spectrophotometry, and integrity was confirmed using an Agilent 2100 Bioanalyzer with a RNA 6000 Nano Assay (Agilent Technologies, Palo Alto, CA). RNA was processed for use on Affymetrix (Santa Clara, CA, USA) Mouse Genome 2.1 ST Arrays Strip, by using the Ambion WT Expression Kit (Life Technologies, CA, USA) and Affymetrix GeneChip WT Terminal Labeling Kit, according to the manufacturer's protocols. Briefly, 100 ng of total RNA containing spiked in Poly-A RNA controls (GeneChip Expression GeneChip Eukaryotic Poly-A RNA Control Kit, Affymetrix) was used in a reverse transcription reaction (Ambion WT Expression Kit) to generate first-strand cDNA. After second-strand synthesis, double-stranded cDNA was used in an in vitro transcription (IVT) reaction to generate cRNA (Ambion WT Expression Kit). 15 μg of this cRNA was used for a second cycle of first-strand cDNA synthesis (Ambion WT Expression Kit). 5.5 μg of single stranded cDNA was fragmented and end-labeled (GeneChip WT Terminal Labeling Kit, Affymetrix). Size distribution of the fragmented and end-labeled cDNA, respectively, was assessed using an Agilent 2100 Bioanalyzer with a RNA 6000 Nano Assay. 3.5 μg of end-labeled, fragmented cDNA was used in a 150 μl hybridization cocktail containing added hybridization controls (GeneAtlas Hybridization, Wash, and Stain Kit for WT Array Strips, Affymetrix), of which 120 μl were hybridized on array strips for 20 h at 48 °C. Standard post-hybridization wash and double-stain protocols (GeneAtlas Hybridization, Wash, and Stain Kit for WT Array Strips, Affymetrix) were used on a GeneAtlas system (Affymetrix), followed by scanning of the array strips.
Three biologically independent replicates at day 8 and day 12 were analyzed for the Dox EBs-treated conditions (Dox and Dox + RA), and two independent RNA preparations for unsorted EBs “no treatment”. Thus, a dataset composed of 16 independent transcriptomes was generated.
4. Data analysis
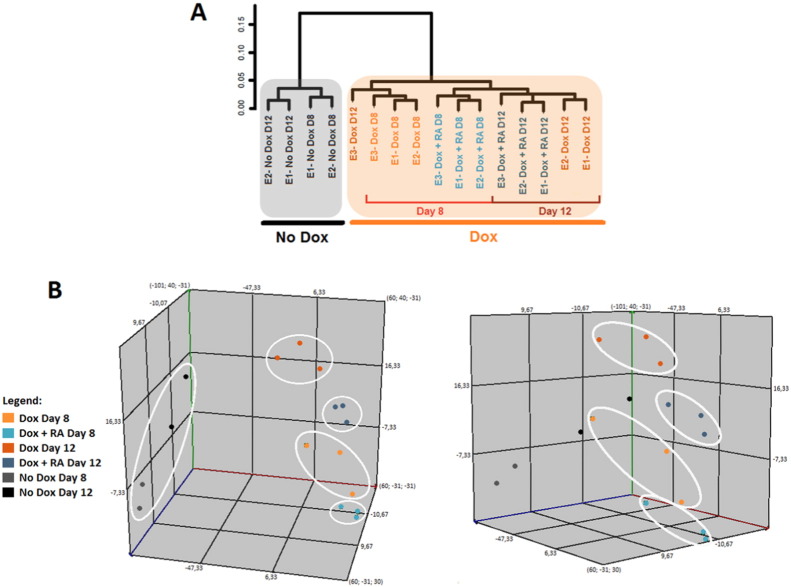
The 16 scanned arrays were analyzed first with Expression Console software (Affymetrix) using RMA to obtain expression values, and for quality control. Control probe sets were removed and log2 expression values of the remaining 33.710 transcripts were imported into Chipster 2.4 [8]. Unsupervised hierarchical clustering (cluster method: average linkage; distances: Pearson correlation) and principal component analysis of the 16 transcriptome datasets were performed using Chipster 2.4 software. This analysis clearly shows two main branches that correspond to the untreated conditions and to Dox-treated conditions (Fig. 3). The clear segregation between “No Dox“and “Dox” was expected, and validates this global dataset. We next examined the microarray datasets to identify genes whose expression vary significantly among the 4 iHC stages, when compared to the non-induced cells (without Dox) at the same time point. An empirical Bayes two-group test with Benjamini-Hochberg multiple testing correction (P-value < 0.01, expression fold change > 2) [9] was used to identify lists of differentially expressed genes in each stage. These lists are organized accordingly to the variations in expression (up- or down-regulated in relation to the untreated cells) (Table 1). To correlate these differentially expressed gene lists with biological function, we searched for enrichment of gene ontology (GO) functional groups in the up-regulated and down-regulated gene lists using DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/). We found that up-regulated genes are mainly involved in transmission of nerve impulses and sensory perception of mechanical stimulus in the 4 iHCs populations. In contrast, down-regulated genes in all lists are connected to cell cycle and cell division.
Fig. 3.
A) Dendrogram showing unsupervised hierarchical clustering of the various expression profiles (16 samples) obtain from iGBM-Myo7a:mVenus reporter-derived EBs (E1, E2, and E3 correspond to three biological replicates). B) Principal component analysis shows the distribution of the 16 expression profiles generated in this microarray. Each point corresponds to an individual sample and the three biological replicates for different Dox treatments are shown with the same color (“Dox”: orange, “Dox + RA”: blue and “No treatment”: black; day 8 and day 12 are labeled with light or dark colors respectively). White circles highlight the three biological replicates and show that these samples occupy similar state space indicating higher similarity in their gene expression profile.
Table 1.
Differentially expressed genes in each of the 4 different iHC stages, when compared to control, non-induced iGPA cells (P-value < 0.01, expression fold change > 2).
| Dox day 8 | Dox + RA day 8 | Dox day 12 | Dox + RA day 12 |
|---|---|---|---|
| Differentially expressed genes: | |||
| 2697 | 3652 | 3442 | 3961 |
| Up-regulated: 1207 | Up-regulated: 1550 | Up-regulated: 1492 | Up-regulated: 1705 |
| Down-regulated: 1490 | Down-regulated: 2102 | Down-regulated: 1950 | Down-regulated: 2266 |
Acknowledgements
We are indebted to Jörg Becker and IGC Gene Expression Unit for their technical support with microarray sample preparation and analysis. This work was supported by Fundação para a Ciência e Tecnologia, Portugal [PTDC/SAU-NEU/71310/2006, SFRH/BD/38461/2007 to A.C.]. A.C. was also a recipient of an EMBO Short-Term Fellowship.
Contributor Information
Aida Costa, Email: aida.costa@ed.ac.uk.
Domingos Henrique, Email: henrique@medicina.ulisboa.pt.
References
- 1.Kyba M., Perlingeiro R.C., Daley G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 2.Ting D.T., Kyba M., Daley G.Q. Inducible transgene expression in mouse stem cells. Methods Mol. Med. 2005;105(2005):23–46. doi: 10.1385/1-59259-826-9:023. [DOI] [PubMed] [Google Scholar]
- 3.Boeda B., Weil D., Petit C. A specific promoter of the sensory cells of the inner ear defined by transgenesis. Hum. Mol. Genet. 2001;10:1581–1589. doi: 10.1093/hmg/10.15.1581. [DOI] [PubMed] [Google Scholar]
- 4.Costa A., Sanchez-Guardado L., Juniat S., Gale J.E., Daudet N., Henrique D. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development. 2015;142:1948–1959. doi: 10.1242/dev.119149. [DOI] [PubMed] [Google Scholar]
- 5.Xiang M., Gao W.Q., Hasson T., Shin J.J. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development. 1998;125:3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- 6.Zheng L., Sekerková G., Vranich K., Tilney L.G., Mugnaini E., Bartles J.R. The deaf jerker mouse has a mutation in the gene encoding the espin actin-bundling proteins of hair cell stereocilia and lacks espins. Cell. 2000;102:377–385. doi: 10.1016/s0092-8674(00)00042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siemens J., Lillo C., Dumont R.A., Reynolds A., Williams D.S., Gillespie P.G., Müller U. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428:950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- 8.Kallio M.A., Tuimala J.T., Hupponen T., Klemela P., Gentile M., Scheinin I., Koski M., Kaki J., Korpelainen E.I. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:1544–6115. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]