Abstract
Fasciolosis is listed as one of the most important neglected tropical diseases according with the World Health Organization and is also considered as a reemerging disease in the human beings. Despite there are several studies describing the immune response induced by Fasciola hepatica in the mammalian host, investigations aimed at identifying the expression profile of genes involved in inducing hepatic injury are currently scarce. Data presented here belong to a time-course investigation of the gene expression profile in the liver of BALB/c mice infected with F. hepatica metacercariae at 7 and 21 days after experimental infection. The data published here have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE69588, previously published by Rojas-Caraballo et al. (2015) in PLoS One [1].
Keywords: Fasciolosis, Microarrays, Gene expression, Immune response
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus/liver |
| Sex | Female |
| Sequencer or array type | Mouse Gene 1.0 ST Array; Affymetrix |
| Data format | Raw |
| Experimental factors | Untreated vs. infected |
| Experimental features | Three experimental groups were used. Group 1 remains untreated and served as control; group 2 was orally infected with Fasciola hepatica metacercariae on day 0 and humanely euthanized and necropsied at 7 days post-infection; group 3 was infected as above and humanely euthanized and necropsied at 21 days post-infection. In necropsy, liver of each mouse was removed and the RNA isolated. We investigated the gene expression profile in both early and late stages of infection compared to untreated group. |
| Consent | N/A |
| Sample source location | N/A |
1. Direct link to deposited data
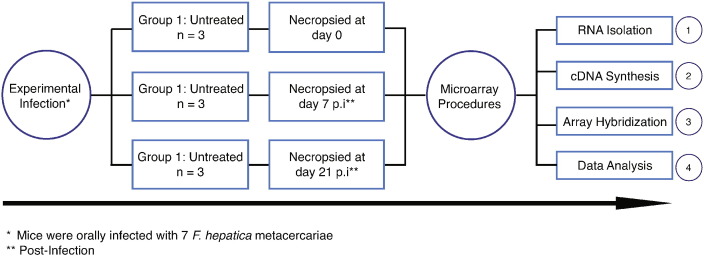
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69588 See Fig. 1.
Fig. 1.
Experimental rationale to investigate the gene expression profile during F. hepatica infection.
2. Experimental design, materials and methods
2.1. Experimental infection
We investigated the gene expression profile during early and late stages of Fasciola hepatica infection in BALB/c mice [1]. For this purpose, we have selected three experimental groups, each containing 3 mice. Mice from group 1 served as controls and did not receive any treatment; mice from groups 2 and 3 were orally infected with F. hepatica metacercariae and humanely euthanized and necropsied at 7 and 21 days after infection, respectively. We investigated the success of the infection by means of the detection of specific antibodies raised against the excretory/secretory antigen from F. hepatica at the time of necropsy.
2.2. Histological assessment
Histopathological studies were performed from representative samples from the right and left liver lobes coming from each individual mouse. Samples were fixed in 10% neutral-buffered formalin and embedded in paraffin wax. Sections (4 μm) were stained with hematoxylin and eosin to determine the liver damage areas.
2.3. Microarray procedures: RNA purification, cDNA synthesis and array hybridization
The Gen Elute Mammalian Total RNA Miniprep Kit (Sigma, San Louis, USA) was used to isolate the RNA, following the manufacturer's recommendations. Both the quantity and quality of each RNA sample was assessed with the Agilent 2100 Bioanalyzer (Agilent Tech, Palo Alto, CA, USA). Furthermore, only those samples with a RNA Integrity Number (RIN) higher than 7 were selected. The Ambion WT Expression Kit for Affymetrix GeneChip (Invitrogen) was used to amplify and label the RNA. After, it was hybridized to the GeneChip Mouse Gene 1.0 ST Array (Lot. No. 4138271; Affymetrix). Washing and scanning were performed using the GeneChip Instrument System of Affymetrix (GeneChip Hybridization Oven 640, GeneChip Fluidics Station 450 and GeneChip Scanner 3000 7 G). The 169-format GeneChip Mouse Gene 1.0 ST Array contained approximately 25 probes that were designed across the full length of 28,853 well-annotated genes for mouse. The design of the array was based on the February 2006 mouse genome sequence (UCSC mm8, NCBI build 36) with comprehensive coverage of RefSeq, putative complete CDS GenBank transcripts, all Ensembl transcript classes and synthetically mapped full-length mRNAs and RefSeq NMs from humans and rats. The GeneChip Mouse Gene 1.0 ST Array has 100% coverage of NM sequences present in the April 3, 2007 RefSeq database.
2.4. Microarray data analysis
The Babelomics V4.2 platform was used to analyze raw data (.CEL files), which were generated after hybridization scanning, and image acquisition [2]. After, the Affymetrix raw data files (.CEL files) were background-adjusted, normalized and log-transformed of the perfect match (PM) values using a robust multi-array average (RMA) methodology [3] for data normalization.
The intensity signal was standardized across arrays via the quantile normalization algorithm [4].
To assess differential gene expression, Limma moderated t-statistics [5], reporting a t-test statistic for each gene together with its corresponding p-value (generally called raw p-values) was used. These raw p-values were further corrected for multiple testing to minimize the number of false positives in the study. In this analysis, we used the conventional multiple testing p-value correction procedures proposed by Benjamini & Hochberg [6] to derive adjusted p-values. For functional profiling, each of the comparisons used here (t0 vs t7; t0 vs t21 and t7 vs t21) wasanalyzed using logistic regression models to identify the functional blocks that were enriched in any of the conditions [7], [8].
Finally, the functional blocks from two databases, GO Biological Process and KEGG Pathways, were used in the study. The Ingenuity Pathways Analysis (IPA) software v6.0 was used to organize the genes that were regulated during the infection into networks of interacting molecules. The gene identifiers of the genes with a statistically significant change in expression (p-value < 0.05) and with a calculated positive or negative fold change of at least two-fold were uploaded in the software. In addition, IPA was used for a functional analysis to identify the biological functions that were most significant to the uploaded datasets (Table 1).
Table 1.
The most representative up-regulated genes involved in liver injury.
| Gene | p-Value | Fold-change | Function |
|---|---|---|---|
| HMOX1 | 4.24E-0.6 | 2.82 | Liver necrosis, cell death |
| FOS | 5.57E-04 | 2.55 | Liver necrosis, cell death, liver proliferation |
| MYC | 1.61E-04 | 2.18 | Liver necrosis, cell death, liver proliferation |
| CD14 | 1.19E-04 | 3.09 | Liver necrosis, cell death, liver proliferation, increases liver damage, increases liver steatosis, hepatic cholestasis |
| Mt1 | 9.11E-04 | 2.55 | Liver necrosis, cell death, liver proliferation |
| NCF1 | 7.51E-05 | 2.09 | Liver necrosis, cell death, liver proliferation, increases liver damage |
| SELP | 1.21E-03 | 2.27 | Liver necrosis, cell death |
| SERPINE1 | 5.40E-05 | 4.59 | Liver necrosis, cell death, increases liver damage, increases liver hepatitis |
| SOCS3 | 9.27E-08 | 2.20 | Liver necrosis, cell death, liver proliferation |
| A2M | 1.77E-06 | 2.14 | Liver necrosis, cell death, increases liver hepatitis, hepatic fibrosis |
2.5. Microarray validation using PCR
The 1st Strand cDNA Synthesis Kit for RT-PCR (AMV) (Cat. no. 1148318801, Roche) was used to reverse transcribe each RNA sample, according to manufacturer's recommendations. For this purpose, six genes (up- and down-regulated) were randomly chosen and specific primers were designed using the Primer-Blast software (http://www.ncbi.nlm.nih.gov/tools/primer-blast). A touchdown PCR (TD-PCR) was performed with an initial denaturation at 94 °C for 1 min, which was followed by a touchdown program for 16 cycles with successive annealing temperature decrements of 1.0 °C every 2 cycles. For these 2 cycles, the reaction was denatured at 94 °C for 20 s, which was followed by annealing at 61 °C–54 °C for 30 s and polymerization at 72 °C for 30 s. The subsequent 15 cycles of amplification were similar, except that the annealing temperature was 53 °C. The final extension was performed at 72 °C for 10 min. The TD-PCR amplification was performed in a Mastercycler gradient (Eppendorf) and confirmed by electrophoresis. Genomic DNA contamination was ruled out for each RNA sample by amplification of each RNA sample without previous reverse-transcription step.
Acknowledgements
This work was supported by Fundación Ramón Areces (Madrid, Spain), Reference XV-2010-2013.
References
- 1.Rojas-Caraballo J., López-Abán J., Fernández-Soto P., Vicente B., Collía F., Muro A. Gene Expression Profile in the Liver of BALB/c Mice Infected with Fasciola hepatica. PLoS One. 2015;10(8):e0134910. doi: 10.1371/journal.pone.0134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medina I., Carbonell J., Pulido L., Madeira S.C., Goetz S., Conesa A. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic acids research. Jul 2010;38(Web Server issue):W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. Apr 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 4.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. Jan 22 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 5.Smyth G.K. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 6.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. Nov 1 2001;125(1–2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 7.Sartor M.A., Leikauf G.D., Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. Jan 15 2009;25(2):211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montaner D., Dopazo J. Multidimensional gene set analysis of genomic data. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010348. [DOI] [PMC free article] [PubMed] [Google Scholar]