Abstract
Transgenic crops have become a staple in modern agriculture, and are typically characterized using a variety of molecular techniques involving proteomics and metabolomics. Characterization of the transgene insertion site is of great interest, as disruptions, deletions, and genomic location can affect product selection and fitness, and identification of these regions and their integrity is required for regulatory agencies. Here, we present CONTRAILS (Characterization of Transgene Insertion Locations with Sequencing), a straightforward, rapid and reproducible method for the identification of transgene insertion sites in highly complex and repetitive genomes using low coverage paired-end Illumina sequencing and traditional PCR. This pipeline requires little to no troubleshooting and is not restricted to any genome type, allowing use for many molecular applications. Using whole genome sequencing of in-house transgenic Glycine max, a legume with a highly repetitive and complex genome, we used CONTRAILS to successfully identify the location of a single T-DNA insertion to single base resolution.
Abbreviations: FISH, Fluorescent In-situ Hybridization; hTG, human thyroglobulin; IGB, Integrated Genome Browser; NGS, Next-Generation Sequencing; T-DNA, Transfer DNA
Keywords: Transfer DNA, Insertion, Transformation, Junction sequences, Next generation sequencing, Agrobacterium
Highlights
-
•
We developed a pipeline for transgene identification using paired-end sequencing.
-
•
This method requires little troubleshooting and is applicable to any genome.
-
•
Identification of insertion sites is required for deregulation of modified food crops.
-
•
This assists in identifying potential genomic disruptions in transgenic events.
1. Introduction
Over the past two decades transgenic crops and foods have become integrated into worldwide agriculture, greatly increasing yields and easing cultivation labors through value added traits. Agrobacterium-mediated transformation and particle bombardment are common methods for creating crops to achieve this advancement. Current understanding indicates that transfer DNA (T-DNA) integration into the host's genome is a random process that has been reviewed extensively [1], [2], [3], [4]. Characterization of integration sites is of great interest, particularly if the host is to be deregulated for human consumption or for commercial applications to assess potential pleiotropic effects resulting from transformation and evaluate the potential for inadvertent mutagenesis [5], [6], [7].
T-DNA inserts have been reported in both transcriptionally active and repressed regions of chromatin [1], [2], [4], [8], [9]. Additionally, in some instances T-DNA sequences have been detected within host endogenous genes, including promoter and regulatory regions [4], [10]. Transgenic plants containing multiple copies of T-DNA sequences have also been reported, and these complex events can lead to silencing of the gene of interest [11] emphasizing the favorable selection of simple, single T-DNA insertion events. Single insertion events in transgenic plants can be generated through multi-generation propagation and are traditionally screened for complexity using Southern blots. While Southern blotting has been proven to be a reliable method for identifying copy numbers, no information regarding T-DNA insertion orientation, random DNA insertions or deletions at the insertion site, or the genomic location of the insert is revealed using this method. Furthermore, Southern blots can require extensive troubleshooting, may require radioactive materials, and can produce ambiguous results if the restriction enzymes exhibit star activity or digested genomic DNA products containing the transgene are similar in size. Thus, many alternative methods to estimate T-DNA copy number have been utilized but aren't without certain shortcomings.
Quantitative PCR analyses of transgene expression levels can be correlated with transgene copy numbers [12], [13], [14], although results from these methods are not always reliable due to other factors that could alter transgene expression independent of zygosity, such as gene silencing and truncation. Visualization methods such as Fluorescent In-Situ Hybridization (FISH) have been implemented for years to identify insertion regions on specific chromosomes [15], [16], [17], [18], however this is a relatively expensive visual technique and confers no information about the surrounding sequence of the insertion region, or if tandem insertions have occurred. FISH must be coupled with targeted PCR amplification of sequences spanning the observed integration region, followed by sequencing to identify more precise integration points.
PCR techniques designed for transposon characterization, such as splinkerette PCR and inverse PCR [19], [20], [21], can reveal detailed integration information and have proven accurate for transgene insertion characterization due to reliance on sequence specific initiation. Consequently, the presence of multiple or complex insertions, truncated transgene sequences, and highly repetitive genomes of host organisms can: a) prevent adequate detection, b) generate non-specific products, or c) fail to amplify products if primer targets are missing. Specialty restriction enzymes may also be required depending on the T-DNA fragment sequence (e.g.: methylation sensitivity, star activity), and a larger amount of genomic DNA is needed in order to visually verify digestion and ligation at each step. Genome walking has been employed effectively with universal primers [22], however as with the other PCR-based techniques, highly complex insertion events and repetitive genomic regions can potentially confound the results. In addition, larger T-DNA insertion sequences (e.g.: > 10 kb) are difficult to fully amplify in their entirety due to the limits of traditional polymerase activity; specialized polymerase varieties for longer amplification are available, but are more expensive than traditional polymerase, are subject to PCR-based assay complications, and can only extend amplification reliably to ~ 20–30 kb.
In order to address these limitations, many groups have utilized next-generation sequencing (NGS) to identify and validate transgene insertion events [23], [24], [25], [26], [27]. Within the past 10 years, sequencing costs have been significantly reduced, while throughput and efficiency have greatly increased. NGS has already proven to be a reliable and accurate method for rapid identification of transposon insertion locations [28]. In addition, further analyses may be conducted on the resulting stored datasets in future genomic studies, such as genome-wide single nucleotide polymorphism (SNP) profiling, updated gene models and fusions, and complete sequencing of the transgene fragment for verification of the insert's integrity. Recently, several reports have successfully used NGS to identify transgene insertion locations in various organisms [24], [25], [29], even at relatively low coverage (2–5 ×). The short turn-around time, coupled with the absence of a need for pre-experimental troubleshooting makes this a very attractive and cost-effective option for reliably identifying random transgene insertions. Furthermore, reference genomes for many species have been fully sequenced and are available for use, removing the need for complete genome de novo assembly of the resulting sequencing reads. This allows effective use of short read sequences in large and complex genomes, as efficient and accurate algorithms for such large de novo assemblies do not currently exist.
Here, we present and demonstrate CONTRAILS (Characterization of Transgene Insertion Locations with Sequencing): a pipeline using existing bioinformatics tools and paired-end Illumina next-generation genomic sequencing to identify and characterize transgene insertion locations in the highly complex and repetitive genome of the legume Glycine max (Fig. 1). Paired-end reads spanning the T-DNA insertion junction allow for one read to map to the reference genome, and the other to map to the transgene sequence. Using short insert (≤ 500 b.p.) paired-end reads allows the user to narrow the insertion site to a genomic region of 500 b.p. or less, provided assembly is assisted with an established reference genome. In some cases, it is possible for a single read to span both genomic and T-DNA sequences at the transgene insertion junction, giving immediate confirmation of insert location and neighboring sequences at single base resolution. However if this is not achieved, the matched paired-end reads will disclose the location well within conventional PCR amplification range for rapid characterization of the T-DNA junction sites. Using this technique, we have identified and characterized a single T-DNA insert site in a transgenic line expressing recombinant hTG protein [30] to single-base resolution. These results are consistent with previous Southern blot and western blot screens, confirming the findings of the NGS analysis. Using this pipeline in conjunction with event-specific PCR assays, we were able to fully characterize flanking genomic sequences surrounding the T-DNA location.
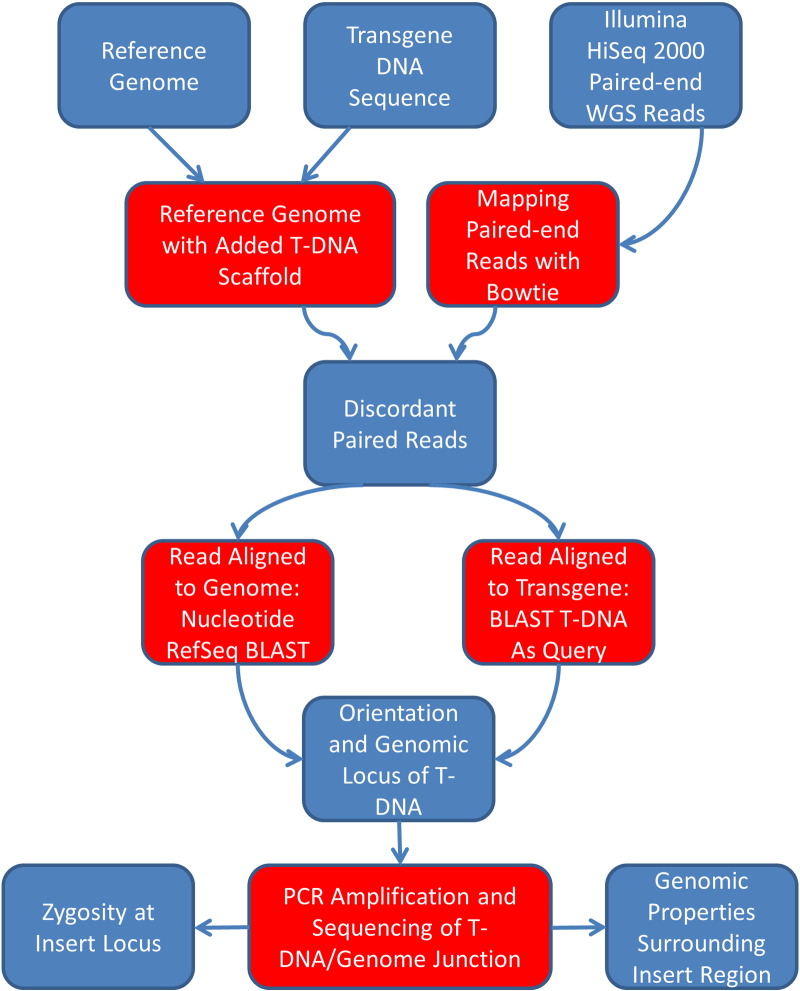
Fig. 1.
Experimental pipeline. Flowchart detailing each major step in the pipeline, from DNA extraction and sequencing to alignment to the reference genome and T-DNA sequence.
2. Methods
2.1. Genomic DNA extraction and preparation
Whole-seed genomic DNA was extracted from chips of cotyledon tissue using a Maxwell 16 instrument and DNA extraction kit (Promega, Madison WI). Extracts were cleaned by phenol–chloroform and precipitated with 100% ethanol. DNA concentrations and purity were assessed with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham MA) and agarose gels to ensure optimal quality and concentration (260/280 absorbance ratio 1.8–2.0, greater than 1 μg total DNA).
2.2. Illumina HiSeq 2000 library preparation, sequencing, and quality control
Library generation was conducted at the David H. Murdock Research Institute genomics department according to the Illumina (San Diego, CA) HiSeq protocol, generating reported insert sizes of 350 b.p. after quality control analysis. Paired-end sequencing was conducted on the Illumina HiSeq 2000 system. The soy sample ST77-KP2 characterized in this study was one of two pooled soy samples on a single lane sequenced to ~ 5 × theoretical genome-wide coverage with 100 base-pair reads. Low-quality reads were filtered out using in-house Illumina software and validated with FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc), showing the remaining read basecall quality scores all greater than 30.
2.3. Reference genome construction and read alignment
The soybean reference genome sequence version 2.75 was obtained from Phytozome [31] and amended with an extra chromosome scaffold containing the T-DNA sequence located between the left and right border repeat regions (Fig. 2) [30]. Paired sequence reads from the previously described seed genomic DNA sequencing were aligned to the constructed reference using Bowtie (ver. 2.2.1) [32] with parameters –un-conc to specify discordant read output. Default Bowtie search methods were used with zero allowed mismatches to limit ambiguous alignments due to the abundance of highly repetitive and homologous endogenous sequences, and in global mode to not trim read ends to enhance alignment scores.
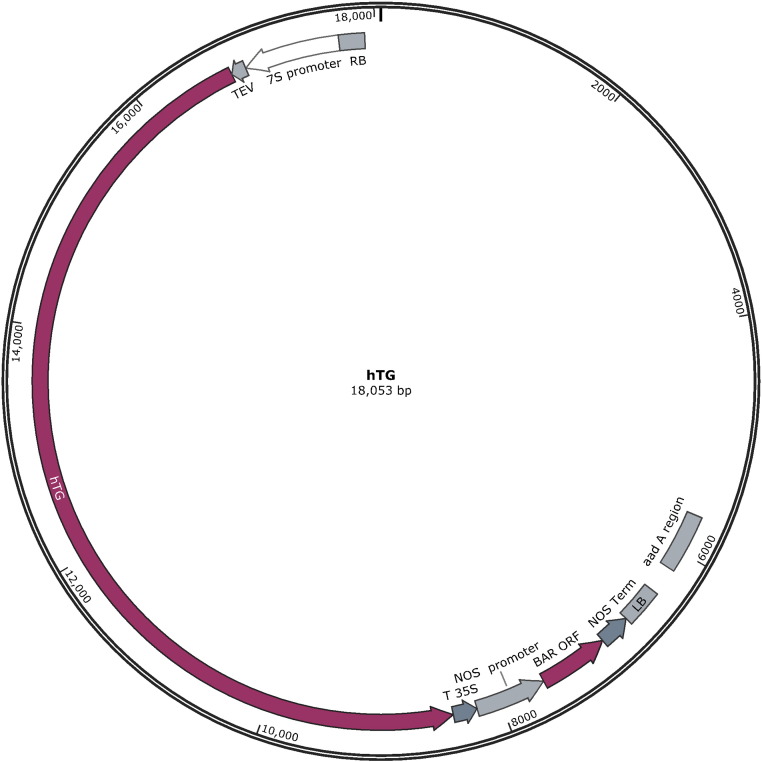
Fig. 2.
Plasmid map of hTG construct. The hTG plasmid map shows all regions included in the transformation plasmid utilized in the Agrobacterium transformation of the original ST77 event. The T-DNA construct contains the soybean β-conglycinin promoter (7S), tobacco etch virus translational enhancer element (TEV), human thyroglobulin gene (hTG), cauliflower mosaic virus terminator element (T35S) followed by the selectable marker cassette comprised of the nopaline synthase promoter (NOS promoter), phosphinothricin acetyltransferase gene (BAR ORF), and nopaline synthase terminator element (NOS Term). The aad A region of the vector confers antibiotic resistance to spectinomycin and streptomycin for selection of Agrobacterium.
2.4. Identification of the transgene insertion site
Fragments in which one read aligned to known genomic reference sequence and the other read aligned to T-DNA sequence were flagged and separated from reads that aligned strictly to the known soybean reference sequence. Each enriched discordant read sequence was aligned against both the G. max reference genome using the “refseq_genomic” function in BLAST [33] and the T-DNA sequence, and matching mates were selected for further characterization. Reads matching the endogenous 7S glycinin promoter were detected in the filtered output and were excluded as illegitimate insertion sites. The genomic read furthest upstream and downstream from the T-DNA read pairs was selected for PCR amplification of the insert junctions to ensure that the anticipated fragment was included within the selected genomic region.
2.5. Validation of T-DNA insertion via PCR
Primers were designed to generate an amplicon that spans the genomic region and into both the right and left border sequences: genomic right border forward (5′-AGGATGACCCGACATGTCTCTTAG-3′), T-DNA right border reverse (5′-CAAATGAAGGGCATGGATCCTGC-3′), T-DNA left border forward (5′-CGGTTTGCGTATTGGCTAGAGC-3′), and genomic left border reverse (5′-GCCCGTCCTGAGCCTAAAATTG-3′). PCR amplification of the right and left border junction sequences consisted of an initial 5 minute denaturation step (95 °C), followed by 35 cycles of 95 °C for 30 s, 54 °C for 30 s, and 72 °C for 1 min and a final extension at 72 °C for 5 min. Wild type soy DNA was used as a negative control in reactions containing both border primer pairs, as well as with the right border forward and left border reverse primers as a positive control to amplify the native genomic locus. Amplified products were separated and visualized on 1% agarose gels stained with ethidium bromide.
2.6. Sequencing of border and junction sequences
PCR reactions were cleaned in preparation for sequencing with phenol chloroform/3 M sodium acetate containing glycogen as a carrier and precipitated with 100% ethanol at − 80 °C for 1 h. Extracts were spun at 21,000 × g for 15 min, washed twice with 70% ethanol and air dried for 10 min. Cleaned precipitated DNA pellets were re-suspended in molecular grade water and quantified with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham MA). Concentrations of PCR product were adjusted based on product size according to the recommendations provided by the University of California, Davis sequencing center (2 ng/μL/100 bp). Based on estimated sizes from migration in 1% agarose gels, the right border product was supplied at 8 ng/μL (~ 400 bases) and the left border product was supplied at 11 ng/μL (~ 550 bases), giving both forward and reverse strand sequences for each junction. Primers were provided at a concentration of 3 μM for sequencing.
2.7. Sequence alignment and characterization
Sequences obtained from Davis showed a right border junction product of 373 bases and a left border junction product of 530 bases. Both sequences were BLASTed against the soybean reference genome, the T-DNA construct sequence for ST77, and against each other. Aligned regions were then extrapolated and examined for overlap by viewing the genomic sequence using the Integrated Genome Browser (IGB) [34] and the T-DNA sequence with SnapGene software (from GSL Biotech; available at snapgene.com).
3. Results
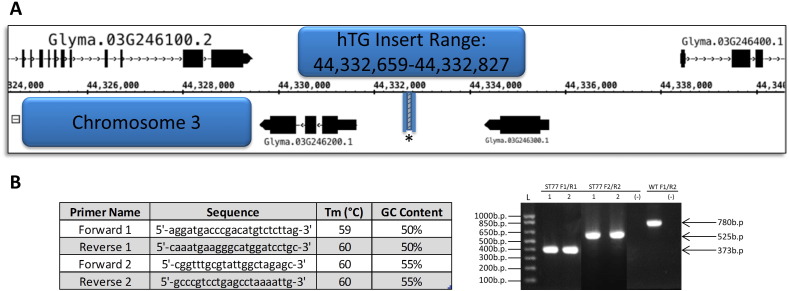
Illumina sequencing for the ST77-KP2 hTG sample generated 27,983,663 reads after quality filtering. Bowtie mapped 96.01% of the paired reads to the soybean reference sequence generating a theoretical whole-genome coverage of ~ 5 ×, establishing 8 total discordant read pairs mapping across the right and left border ends of the T-DNA sequence. Reads mapping to the genomic reference corresponded to sequences at a single locus on chromosome 3, with upstream reads beginning at bases 44,332,446 and 44,332,559 paired with reads 187 and 226 base pairs into the right border, respectively. Likewise, two reads within the left border region of the T-DNA at bases 11,269 and 11,433 paired with reads in downstream genomic sequence at bases 44,332,927 and 44,332,928 respectively. All discordant reads and respective information are shown in Table 1. This indicates a narrow region where the insertion occurred (base 44,332,659–44,332,927 shown in Fig. 3A), and illustrates that the right border of the T-DNA is oriented towards upstream genomic sequences in the 5′ to 3′ direction. Once this narrowed region was identified, primer design for genomic upstream and downstream sequences was facilitated utilizing the most recent G. max reference genome build in conjunction with visualization in IGB to achieve products within range for normal PCR amplification.
Table 1.
Discordant read pairs and sequences. All discordant read pairs for ST77 and the position of the start of the read are shown, as well as their mated sequence and pair relationship.
| Read origin | Start base | Mate pair relationship | Read sequence |
|---|---|---|---|
| Chr03 | 44,332,446 | Mate1; other read matches reverse reference; this read is one of a pair | ATTAGGATGACCCGACATGTCTCTTAGAATGAGTAACATAAAAC1TAGAATTATGGAAATTAGAATATTTCAAGAGCCTTTCACTTCAACTGATTATAAG |
| scaffold ST77 | 187 | Mate2; this read matches reverse reference; this read is one of a pair | AGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCATGCCTGCAGGATCCATGCCCTTCATTTGCCGCrTATTAATTAATTTGGTAACAGTCCGT |
| scaffold ST77 | 11,433 | Mate1; other read matches reverse reference; this read is one of a pair | CGGCGTTAATTCAGTACATTAAAAACGTCCGCAATGTGTTATTAAGTTGTCTAAGCGTCAATTTGTTTACACCACAATATATCCTGTTCAACATTCAACA |
| Chr03 | 44,332,928 | Mate2; this read matches reverse reference; this read is one of a pair | TAATAATAAAACAAGTAGTCCTTGGCTAGTTGGCTTACTTTTCATGTTTTAAGGAAACAAGTTGAGGAAGGGAAAAAATGTTGATACTGCTGCrCGTACG |
| Chr03 | 44,332,927 | Mate1; this read matches reverse reference; this read is one of a pair | ATAATAATAAAACAAGTAGTCCTTGGCTAGTTGGCTTACTTTTCATGTTTTAAGGAAACAAGTTGAGGAAGGGAAAAAATGTTGATACTACTACTCGTAC |
| scaffold ST77 | 11,269 | Mate2; other read matches reverse reference; this read is one of a pair | AAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCmCCAGTCGGGAAACCTGTCGTGC |
| scaffold ST77 | 226 | Mate1; this read matches reverse reference; this read is one of a pair | CATGCCTGCAGGATCCATGCCCTTCATTTGCCGCTTATTAATTAATTTGCTAACAGTCCGTACTAATCAGTTACTTATCCrTCCTGCATCATAATTAATC |
| Chr03 | 44,332,559 | Mate2; other read matches reverse reference; this read is one of a pair | ATTTAGTTAATACAACGTGGATGAAGAAAGGAAAGACATTAGAGAAAGAGTAAGCAAATAACGCACTCGATTTGTTATCTAATTAGTATGCTGTTGTACC |
Fig. 3.
Insert location range and PCR verification. (A) The established maximum range of the location of the T-DNA insert based on discordant paired-end read mates. The discordant paired read reported farthest upstream began at base 44,332,659. The discordant paired read reported farthest downstream began at base 44,332,827. (B) Primer sequences and attributes used in the amplification of right and left border T-DNA junction sequences. The resulting products and their sizes are shown for the transgenic sample analyzed in duplicate, including a wild-type control using primers F1 and R2 to amplify the genomic insert locus in the absence of the hTG T-DNA.
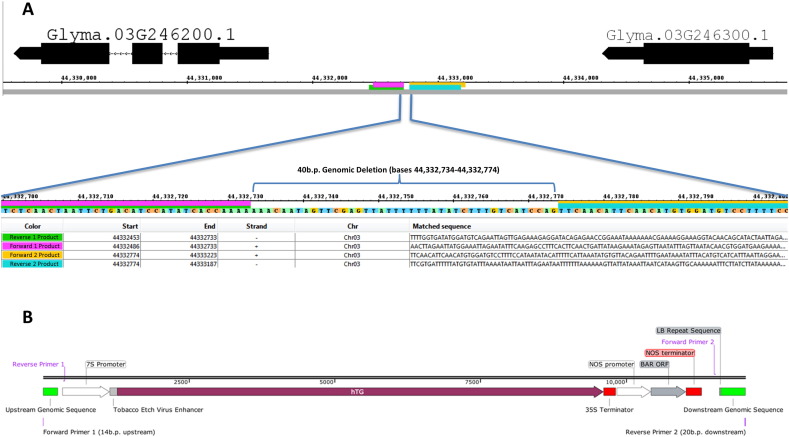
Junction sites were amplified for both the left and right border sequences generating products of ~ 400 bases and ~ 550 bases respectively. Sequencing results identified products of 373 and 530 bases for the right and left border PCR amplicons, respectively. The primers used for amplification, their attributes and the sequences generated are shown in Fig. 3B. Alignments of these sequences to both the soybean genome reference and the T-DNA sequence identified the insertion site to single-base resolution at base 44,332,733. Furthermore, alignments revealed a 40 base pair deletion at the insertion locus on chromosome 3 as shown in Fig. 4A. This deleted sequence was not part of an existing regulatory region, exon, or gene. In addition, 159 bases were deleted from the 5′ end of the right border region from the T-DNA, but left the 7S promoter intact. From the junction sequencing data, we constructed a consensus sequence of the insert relative to the genome which is illustrated in Fig. 4B.
Fig. 4.
Aligned sequenced PCR products and insert layout. (A) Section of the insert location between two soybean genes. Colored bars represent sequences from the PCR amplicons of the junction sites that aligned to the soybean reference genome on chromosome 3. Purple is the product from primer F1, green from primer R1, yellow from primer F2, and blue from primer R2. 40 bases of genomic DNA have been deleted as a result of the insertion, shown as the uncolored region between the primer products. Start bases for each primer product are shown, as well as their alignment to either the sense or antisense DNA strand. (B) Illustration of the constructed consensus sequence of the T-DNA insert locus, showing the location of the primers used for junction characterization, flanking genomic DNA sequences, and inserted T-DNA elements.
4. Discussion
Previously our group has demonstrated the efficacy of transgenic G. max as a cost-effective expression and storage system for recombinant proteins that are expensive to manufacture and/or difficult to generate in traditional systems [30], [35], [36], [37]. Until now, we have determined zygosity of transgenic events based on Mendelian inheritance, and western and Southern blotting. However these techniques reveal no characteristics of the T-DNA genomic insertion site, potential disruptions of endogenous genes, or truncation of the transgene and/or border sequences. Due to the highly repetitive nature of the soybean genome, our previous characterization attempts of the T-DNA using PCR-based techniques have failed to produce verifiable products.
Next-generation sequencing technologies offer a multitude of advantages when compared to traditional molecular characterization techniques, including rapid results, precise datasets that can be repurposed, exceptional consistency, and little experimental troubleshooting. Furthermore, sequencing costs are consistently decreasing every year making NGS methods more accessible to a larger range of investigators. In this study, low coverage paired-end genomic sequencing using the Illumina HiSeq 2000 platform was able to locate and identify a single copy transgene insertion in a highly complex and repetitive genome. The ability to use lower coverage is assisted with the existence of a reference genome to facilitate alignments. The absence of such a reference in a different organism would likely require higher coverage for confidence in the resulting assembly, however further optimization of de novo assembly algorithms will be the more likely technical bottleneck. The ability to pool multiple samples together on a single lane drastically reduces sequencing costs; however caution must be used to not dilute potential reads too extensively to avoid the possibility of a large coverage gap over the insert location, especially in organisms with particularly sizeable or complex genomes.
Insertion site identification exemplifies many properties of the transgene structure and can identify problematic or non-desirable transgenic events early in a production pipeline. Locations within interspersed repeat regions, or regions of heavy methylation and dense chromatin may exhibit lower than expected expression of the transgene. Likewise, transformation events containing multiple copies of the T-DNA may show promise in molecular characterizations (e.g.: increased expression of the transgene), but are not ideal for the generation of homozygous events. Verification of the insertion site with PCR is rapid and straightforward to design using the genomic sequencing information, and can be used to screen other siblings from a particular event to assist in assessing zygosity for each specific locus. Quality control following library generation will report the total fragment size for each library, which can be used in conjunction with the genomic locations of the discordant reads to predict the size of the PCR products from the junction sites. Deviations from the reported insert size are not uncommon; extreme variances in the size of the amplified product may reveal a genomic deletion or insertion in the insert region that would otherwise remain undetected. The actual T-DNA sequence transferred to the host is contained between the right border and left border repeat regions, which act as cleavage signals for internal virulence factors in Agrobacterium. While designing PCR primers, it is prudent to choose sites well within the border boundaries to create a margin of safety against nucleolytic truncation and potential deletion of primer annealing sites. In addition, the raw aligned reads from the sequencing output may be consulted to verify the integration of these sequences and bolster confidence in the presence of primer annealing locations.
In some instances, illegitimate insert locations may be reported in the discordant read output if the T-DNA sequence contains promoter regions or other elements that are native in the target host (e.g.: glycinin promoters in soybean). In these cases, it would be beneficial to know the genomic location of these elements in the host genome prior to designing PCR assays for junction sequencing, as this will aid in the selection of read pairs representing true insertion locations and prevent attempts to amplify an absent sequence.
In the case of multiple T-DNA copy events, screens for tandem T-DNA inserts are easily implemented using the same forward primers designed for amplification of left border junction sequences in conjunction with the reverse primers used for right border amplification. Likewise, reversed and inverted tandem insert junctions with the left border integrated in the 5′ direction should form self-amplified products utilizing only the left border forward primer in the PCR reaction. Reversed tandem inserts may require an additional primer annealing to the lagging strand of the left border for amplification. Information pertaining to the orientation of the T-DNA at the identified locus is easily evaluated by comparing which region of the T-DNA is paired with the upstream and/or downstream genomic reads.
Assembly of reads spanning the T-DNA sequence can also be aligned to the reference construct sequence to assess total insert integrity without the use of step-wise PCR techniques. Fragmented or truncated inserts are easily identified in this way, preventing propagation of incomplete or partially transformed events. In addition, it is a relatively common occurrence for Agrobacterium to incompletely nick the T-DNA leading to read-through at the left border, possibly integrating vector features into the host [38]. The inclusion of vector backbone sequences in the T-DNA scaffold supplemented in the reference genome will allow for their detection as an integrated step.
Native endogenous gene disruption is moderately prevalent following Agrobacterium transformation via base inserts/deletions at the integration site, or direct insertion of the transgene into native exons. Gene disruption can induce pleiotropic effects on the host, many of which may cause adverse effects that might not be phenotypically identifiable. Identification of these modifications as a result of integration breakpoints is a crucial advantage of CONTRAILS in candidate products for commercialization.
An indirect advantage to NGS-based approaches is the generation of permanent datasets containing extensive genomic sequence information. Soft data results are easily and rapidly referable, non-consumable, and are preserved indefinitely unlike biological samples. Collaborative efforts and the interpretation of results greatly benefit from shared digital datasets on cloud-based storage, and current organism-specific databases (e.g.: Soybase, the Soy Knowledge Base, Wormbase) and public repositories welcome the addition of new data. Further expansion of these freely accessible databases as genomics studies advance is crucial, and will serve as invaluable references for current and future genetic and molecular investigations.
5. Conclusions
Here we have demonstrated a cost-effective, rapid method for identification and characterization of transgene insertion locations in the complex, repetitive genome of transgenic G. max. Utilizing next-generation genomic sequencing and conventional PCR verification techniques, this method may be employed for many applications and genomes of varying complexity, with little to no time required for laboratory troubleshooting, using benchtop computational power in a straightforward pipeline. Considerable time savings from a universally applicable process, in conjunction with the generation of extensive genomic datasets for future analyses, make this a valuable resource for genomics analysis of all organisms containing DNA insertions.
6. Availability of supporting data
Genomic sequencing files associated with the ST77 transgenic event described herein are available at the NCBI Short Read Archive under the Biosample accession number SRR2180176.
Author's contributions
KCL designed and conducted PCR analysis, analyzed aligned border sequences, and generated figures. AMW processed sequencing files, conducted genomic sequence alignments and generated paired discordant read pools. KCL, KLB, and KJP wrote and edited the manuscript with input from all authors.
Competing interests
The authors of this manuscript declare no conflict of interest.
Acknowledgments
The authors acknowledge the contributions of Mike Wang at the David H. Murdock Research Institute for conducting the genomic sequencing and library preparation, Dr. Elizabeth Scholl (NC State BCSC) for advice on sequence alignment parameters, Raymond Fernalld (Soymeds) for running the initial ST77 PCR reaction, and Drs. Adam Reitzel (UNCC) and Linda Robles (Soymeds) for providing editing advice to improve the manuscript.
Contributor Information
Kevin C. Lambirth, Email: kclambirth@uncc.edu.
Adam M. Whaley, Email: awhaley9@uncc.edu.
Jessica A. Schlueter, Email: jschluet@uncc.edu.
Kenneth L. Bost, Email: klbost@uncc.edu.
Kenneth J. Piller, Email: kjpiller@uncc.edu.
References
- 1.Lacroix B., Citovsky V. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int. J. Dev. Biol. 2013;57(6–8):467–481. doi: 10.1387/ijdb.130199bl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gheysen G., Montagu M.V., Zambryski P. Integration of Agrobacterium tumefaciens transfer DNA (T-DNA) involves rearrangements of target plant DNA sequences. Proc. Natl. Acad. Sci. U. S. A. 1987;84(17):6169–6173. doi: 10.1073/pnas.84.17.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kononov M.E., Bassuner B., Gelvin S.B. Integration of T-DNA binary vector ‘backbone’ sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J. 1997;11(5):945–957. doi: 10.1046/j.1365-313x.1997.11050945.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim S.I., Veena G.G.S.B. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007;51(5):779–791. doi: 10.1111/j.1365-313X.2007.03183.x. [DOI] [PubMed] [Google Scholar]
- 5.Houshyani B., van der Krol A.R., Bino R.J., Bouwmeester H.J. Assessment of pleiotropic transcriptome perturbations in Arabidopsis engineered for indirect insect defence. BMC Plant Biol. 2014;14:170. doi: 10.1186/1471-2229-14-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darmency H. Pleiotropic effects of herbicide-resistance genes on crop yield: a review. Pest Manag. Sci. 2013;69(8):897–904. doi: 10.1002/ps.3522. [DOI] [PubMed] [Google Scholar]
- 7.Filipecki M., Malepszy S. Unintended consequences of plant transformation: a molecular insight. J. Appl. Genet. 2006;47(4):277–286. doi: 10.1007/BF03194637. [DOI] [PubMed] [Google Scholar]
- 8.Magori S., Citovsky V. Epigenetic control of Agrobacterium T-DNA integration. Biochim. Biophys. Acta. 2011;1809(8):388–394. doi: 10.1016/j.bbagrm.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitzschke A., Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;29(6):1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krysan P.J., Young J.C., Tax F., Sussman M.R. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. U. S. A. 1996;93(15):8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy M.S., Dinkins R.D., Collins G.B. Gene silencing in transgenic soybean plants transformed via particle bombardment. Plant Cell Rep. 2003;21(7):676–683. doi: 10.1007/s00299-002-0567-4. [DOI] [PubMed] [Google Scholar]
- 12.Mason G., Provero P., Vaira A.M., Accotto G.P. Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol. 2002;2:20. doi: 10.1186/1472-6750-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingham D.J., Beer S., Money S., Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques. 2001;31(1):132–134. doi: 10.2144/01311rr04. 136-140. [DOI] [PubMed] [Google Scholar]
- 14.Honda M., Muramoto Y., Kuzuguchi T., Sawano S., Machida M., Koyama H. Determination of gene copy number and genotype of transgenic Arabidopsis thaliana by competitive PCR. J. Exp. Bot. 2002;53(373):1515–1520. [PubMed] [Google Scholar]
- 15.Lattenmayer C., Loeschel M., Steinfellner W., Trummer E., Mueller D., Schriebl K., Vorauer-Uhl K., Katinger H., Kunert R. Identification of transgene integration loci of different highly expressing recombinant CHO cell lines by FISH. Cytotechnology. 2006;51(3):171–182. doi: 10.1007/s10616-006-9029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulnane L.S., Lehman E.J., Hock B.J., Tsuchiya K.D., Lamb B.T. Rapid and efficient detection of transgene homozygosity by FISH of mouse fibroblasts. Mamm. Genome. 2002;13(4):223–226. doi: 10.1007/s00335-001-2128-5. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi T., Kuroiwa A., Yamada S., Isotani A., Yamashita A., Tairaka A., Hayashi T., Takagi T., Ikawa M., Matsuda Y. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics. 2002;80(6):564–574. doi: 10.1006/geno.2002.7008. [DOI] [PubMed] [Google Scholar]
- 18.Moscone E.A., Matzke M.A., Matzke A.J. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma. 1996;105(4):231–236. [PubMed] [Google Scholar]
- 19.Potter C.J., Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlopoulos A. Identification of DNA sequences that flank a known region by inverse PCR. Methods Mol. Biol. 2011;772:267–275. doi: 10.1007/978-1-61779-228-1_16. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B., Huang J.Q., Wei Z.M. A quick method to estimate the T-DNA copy number in transgenic rice using inverse PCR (IPCR) Shi Yan Sheng Wu Xue Bao. 1999;32(2):207–211. [PubMed] [Google Scholar]
- 22.Leoni C., Gallerani R., Ceci L.R. A genome walking strategy for the identification of eukaryotic nucleotide sequences adjacent to known regions. Biotechniques. 2008;44(2):229. doi: 10.2144/000112680. 232-225. [DOI] [PubMed] [Google Scholar]
- 23.Chambers K., Lowe R., Howlett B., Zander M., Batley J., Van de Wouw A., Elliott C. Next-generation genome sequencing can be used to rapidly characterise sequences flanking T-DNA insertions in random insertional mutants of Leptosphaeria maculans. Fungal Biol. Biotechnol. 2014;1(1):10. doi: 10.1186/s40694-014-0010-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava A., Philip V.M., Greenstein I., Rowe L.B., Barter M., Lutz C., Reinholdt L.G. Discovery of transgene insertion sites by high throughput sequencing of mate pair libraries. BMC Genomics. 2014;15:367. doi: 10.1186/1471-2164-15-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Y., Abrams N., Zhu W., Salinas E., Yu Z., Palmer D.C., Jailwala P., Franco Z., Roychoudhuri R., Stahlberg E. Identification of the genomic insertion site of Pmel-1 TCR alpha and beta transgenes by next-generation sequencing. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0096650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepage E., Zampini E., Boyle B., Brisson N. Time- and cost-efficient identification of T-DNA insertion sites through targeted genomic sequencing. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovalic D., Garnaat C., Guo L., Yan Y.P., Groat J., Silvanovich A., Ralston L., Huang M.Y., Tian Q., Christian A. The use of next generation sequencing and junction sequence analysis bioinformatics to achieve molecular characterization of crops improved through modern biotechnology. Plant Genome. 2012;5(3):149–163. [Google Scholar]
- 28.Jiang C., Chen C., Huang Z., Liu R., Verdier J. ITIS, a bioinformatics tool for accurate identification of transposon insertion sites using next-generation sequencing data. BMC Bioinforma. 2015;16(1):72. doi: 10.1186/s12859-015-0507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R., Yin Y., Zhang Y., Li K., Zhu H., Gong Q., Wang J., Hu X., Li N. Molecular characterization of transgene integration by next-generation sequencing in transgenic cattle. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0050348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell R., Hudson L.C., Lambirth K.C., Luth D., Wang K., Bost K.L., Piller K.J. Recombinant expression of homodimeric 660 kDa human thyroglobulin in soybean seeds: an alternative source of human thyroglobulin. Plant Cell Rep. 2011;30(7):1327–1338. doi: 10.1007/s00299-011-1044-8. [DOI] [PubMed] [Google Scholar]
- 31.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7(1–2):203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 34.Nicol J.W., Helt G.A., Blanchard S.G., Jr., Raja A., Loraine A.E. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25(20):2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson L.C., Garg R., Bost K.L., Piller K.J. Soybean seeds: a practical host for the production of functional subunit vaccines. 2014;2014:340804. doi: 10.1155/2014/340804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bost K.L., Lambirth K.C., Hudson L.C., Piller K.J. Soybean-derived thyroglobulin as an analyte specific reagent for in vitro diagnostic tests and devices. Adv. Med. Biol. 2014;80 [Google Scholar]
- 37.Hudson L.C., Seabolt B.S., Odle J., Bost K.L., Stahl C.H., Piller K.J. Sublethal staphylococcal enterotoxin B challenge model in pigs to evaluate protection following immunization with a soybean-derived vaccine. Clin. Vaccine Immunol. 2013;20(1):24–32. doi: 10.1128/CVI.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Buck S. T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol. Breed. 2000;6(5):459–468. [Google Scholar]