Abstract
The vertebrate heart develops from two distinct lineages of cardiomyocytes that arise from the first and second heart fields (FHF and SHF, respectively). The FHF forms the primitive heart tube, while adding cells from the SHF allows elongation at both poles of the tube. Initially seen as an exclusive characteristic of higher vertebrates, recent work has demonstrated the presence of a distinct FHF and SHF in lower vertebrates, including zebrafish. We found that key transcription factors that regulate septation and chamber formation in higher vertebrates, including Tbx5 and Pitx2, influence relative FHF and SHF contributions to the zebrafish heart tube. To identify molecular modulators of heart field migration, we used microarray-based expression profiling following inhibition of tbx5a and pitx2ab in embryonic zebrafish (Mosimann & Panakova, et al, 2015; GSE70750). Here, we describe in more detail the procedure used to process, prioritize, and analyze the expression data for functional enrichment.
Keywords: Genomics, Zebrafish, Heart development, tbx5, pitx2
| Specifications | |
|---|---|
| Organism/cell line/tissue | Zebrafish |
| Sex | Mix |
| Sequencer or array type | Affymetrix array (Zebrafish Gene 1.0 ST Array) |
| Data format | Raw (CEL Files) |
| Experimental factors | Comparison of whole-embryo transcript expression from embryos injected with morpholinos targeting pitx2ab or tbx5a, versus control embryos |
| Experimental features | Extracted RNA from at least 40 embryos per experimental condition (in triplicate) was used to generate cDNA libraries that were hybridized to an Affymetrix array. Data were subsequently background subtracted, normalized, and analyzed for enrichment of functional gene sets using Gene Set Enrichment Analysis (GSEA). |
| Consent | NA |
| Sample source location | All samples were generated using embryos from fish colonies maintained at the Children's Hospital zebrafish facility, Boston, MA, USA |
1. Direct link to deposited data
2. Experimental design
Value of the data.
-
–
The raw data files linked here provide unique expression profiles from two conditions highly relevant to vertebrate developmental biology.
-
–
The scripts included in the supplement not only provide the means to replicate the referenced study, but also are immediately applicable to any dataset available in the Gene Expression Omnibus [1] (GEO).
-
–
The resulting ranked list is a resource of potential markers of the cardiomyocyte lineages.
3. Data, materials and methods
3.1. Generation of microarray expression data
To generate the expression data accompanying our initial study [2], adult wild-type (AB) zebrafish were kept in breeding cages overnight with dividers separating males and females, then allowed to breed the following morning. Resulting embryos were collected over a short time window (approximately 30 minutes), and injected into the yolk with a morpholino targeting either tbx5a (5-CCTGTACGATGTCTACCGTGAGGC-3; as previously designed [3]) or pitx2ab (5′-TGGGAGTCCATTTAGTAGGTTATAT-3′) before the 4-cell developmental stage using a standard microinjection platform. At least 40 embryos were injected per morpholino treatment, with the experiment being performed in triplicate (at least 120 embryos total per group). At 56 hours post fertilization embryos were visually inspected for efficiency of morpholino-based knockdown by assessing their resulting developmental phenotypes; tbx5a morphants lack pectoral fins and develop cardiac edema, while pitx2ab morphants display left-right asymmetry defects (i. e. heart looping randomized). Occasional wildtype-looking and thus possibly inefficiently injected embryos in clutches of morpholino-injected cohorts were manually sorted out. Embryos were homogenized in Trizol LS (Life Technologies) using a tissue ruptor (Qiagen), and mRNA extracted using a standard phenol/chloroform extraction protocol as per manufacturer's instructions with an extra chloroform clean-up step thereafter. Resulting total RNA was checked for quality using a Bioanalyzer nano RNA chip, and 100 ng total RNA went into a standard Affymetrix pipeline for probe synthesis, labeling, hybridization and scanning of the hybridized array at the Boston Children's Hospital Molecular Genetics Core.
3.2. Normalization and processing of microarray data
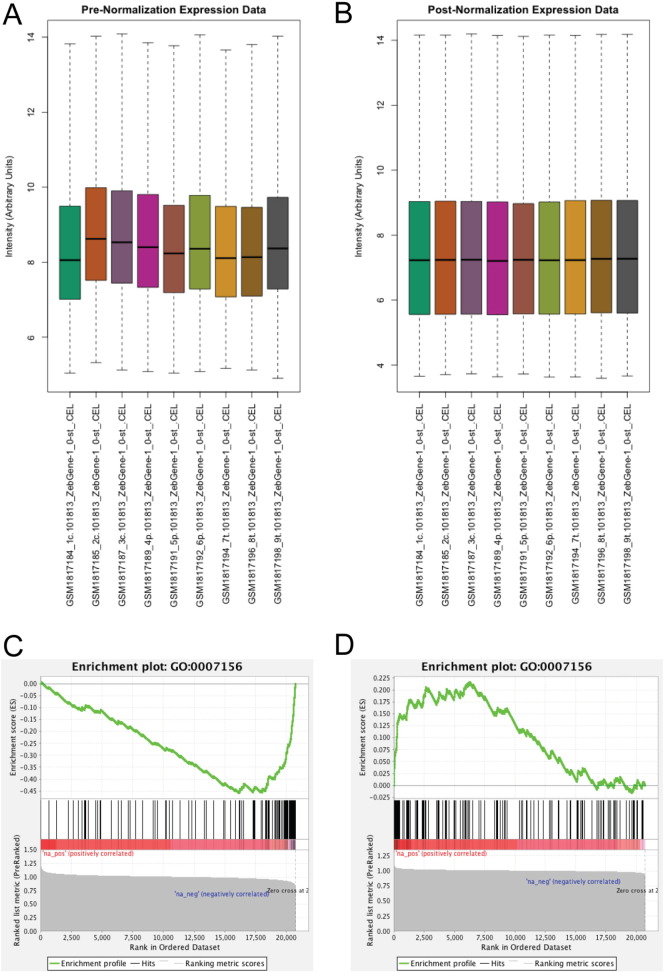
Raw CEL files were processed using the Oligo package [4] as part of the Bioconductor suite (www.bioconductor.org) in the R statistical framework (www.r-project.org), using a customized script (Supplementary Script 1). Specifically, background subtraction and normalization were performed using the Robust Multiarray Average (RMA) method implemented in the Oligo package. Boxplots of intensity values were compared for all chips before and after normalization to visualize the corresponding effects on mean and quartile values (Fig. 1A–B). Following normalization, probeset IDs were matched to corresponding transcript IDs. Specifically, the zebrafish 1.0 ST array NetAffx annotation file was downloaded in CSV format from the Affymetrix website (www.affymetrix.com), and transcript/gene IDs corresponding to given probe IDs were extracted (Supplementary Table 1). In the interest of representing the data using a single gene annotation framework, all transcript and gene IDs were mapped to corresponding Ensemble gene IDs. For Ensemble transcript IDs, corresponding Ensemble gene IDs were obtained using the BioMart community portal [5]. The Synergizer web application [6] was used to convert gene IDs from other annotation frameworks to Ensemble gene IDs. The resulting table (Supplementary Table 2) was then merged with the table of probeset expression data. In instances where a gene ID matched multiple probesets, probeset values were averaged (see Supplementary Script 1).
Fig. 1.
Normalization and GSEA enrichment analysis results. Boxplots (A & B) show the expression intensity values for each CEL file both before (A) and after (B) RMA normalization. Plots outputted by GSEA (C & D) show enrichment for genes annotated as being involved in homophilic cell adhesion following pitx2 (C) and tbx5 (D) knockdown.
3.3. Examination for highly expressed subgroups using GSEA pre-ranked analysis
Transcript data were averaged for each gene over the three experimental replicates. Genes were then ranked by fold-change in experimental conditions: pitx2ab morpholino injected divided by control, and tbx5a morpholino injected divided by control. These pre-ranked datasets were used as the input for gene set enrichment analysis [7] (GSEA). GSEA pre-ranked analysis also requires a list of gene sets that will be examined for enrichment at the top or bottom of the ranked expression datasets. Gene Ontology [8] (GO) annotations were used for this purpose. GO SLIM biological process, molecular function, and cellular compartment annotations for zebrafish were downloaded from the BioMart community portal [5] and converted to GSEA-compatible GMT format using a Perl script (Supplementary Script 2). GSEA pre-ranked analysis (1000 permutations, minimum term size of 5, maximum term size of 500) was implemented using the stand-alone GSEA tool [2]. GSEA analysis found genes annotated with the term ‘homophilic cell adhesion’ (GO:0007156) as being significantly (q < 0.001) down regulated in expression following inhibition of pitx2, and up regulated following inhibition of tbx5 (Fig. 1C–D). This reinforced the notion that pitx2 and tbx5 control alternate aspects of cardiac cell adhesion and migration, and provided a novel group of genes for experimental follow-up that, notably, would not have been identified through traditional univariate statistical analysis. Ongoing experimental work is aimed at experimentally determining how downstream targets of pitx2 and tbx5 mitigate homophlic (like-to-like) cell adhesion, and the consequent physiological effects on cellular function.
The following are the supplementary data related to this article.
Supplementary Table 1 — probe IDs and corresponding transcript/gene IDs, as mined from raw Affymetrix files.
Supplementary Table 2 — probe IDs and corresponding Ensemble gene IDs.
Supplementary Table 3 — GO term annotations as downloaded from the BioMart community portal.
(processSamples.R) — R script using the Oligo package and Bioconductor framework to download CEL files from GEO, normalize, and process for analysis. The Oligo and GEOquery packages must be installed prior to use, instructions are within the script. To use, from within the directory containing the script, type at the R prompt: source(“processSamples.R”). Table of probeset/gene IDs (Supplemental File 1) must also be included in the same directory. Output file names/locations, as well as GEO IDs to be used to retrieve data can be modified within the script to work with a new dataset. This script was developed using R version 3.2.0.
(processSamples.R) — R script using the Oligo package and Bioconductor framework to download CEL files from GEO, normalize, and process for analysis. The Oligo and GEOquery packages must be installed prior to use, instructions are within the script. To use, from within the directory containing the script, type at the R prompt: source(“processSamples.R”). Table of probeset/gene IDs (Supplemental File 1) must also be included in the same directory. Output file names/locations, as well as GEO IDs to be used to retrieve data can be modified within the script to work with a new dataset. This script was developed using R version 3.2.0.
(convertListToGMT.pl) — Perl script used to convert two-column Gene-Annotation files into GSEA-compatible GMT files. To use, specify the input and output filenames, and the minimum number of gene-term associations to be considered a gene set in GSEA analysis (optional, default is set to 5). The input file should be a 3-column format including gene names (column 1), descriptions (column 2), and term IDs (column 3). The input file should be tab delimited, if descriptions are not available the second column can be left blank. The input file used for the analysis presented here can be obtained by converting Supplementary Table 3 to tab delimited text format. To use, from within the installed directory type: “perl convertListToGMP.pl inputFile.txt outputFile.gmt 5”.
(probeAndGeneIDs.txt) — Text file of probe ID and Ensemble Gene ID mapping. Required for Supplemental Script 1, please transfer to the working directory.
Acknowledgments
C.M. received support through an EMBO long-term fellowship, an HFSP long-term fellowship, an SNSF advanced researcher fellowship, an SNF professorship, and a Marie Curie CIG; D.P. received support through an HFSP long-term fellowship, a Helmholtz Young Investigator Program and a Marie Curie CIG; C.A.M. received support from the March of Dimes, the Harvard Stem Cell Institute and the Leducq Foundation; L.I.Z. is supported by HHMI and NIH 5U01HL10001-06, 5P30 DK49216-20, 5PO1HL32262-32, and 5R01HL04880-22.
References
- 1.Barrett T., Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosimann C., Panakova D., Werdich A.A., Musso G., Burger A., Lawson K.L., Carr L.A., Nevis K.R., Sabeh M.K., Zhou Y., Davidson A.J., DiBiase A., Burns C.E., Burns C.G., MacRae C.A. Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 2015;6:8146. doi: 10.1038/ncomms9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn D.G., Kourakis M.J., Rohde L.A., Silver L.M., Ho R.K. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G., Bardou P., Beck T., Blake A., Bonierbale M., Brookes A.J. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berriz G.F., Roth F.P. The Synergizer service for translating gene, protein and other biological identifiers. Bioinformatics. 2008;24:2272–2273. doi: 10.1093/bioinformatics/btn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 — probe IDs and corresponding transcript/gene IDs, as mined from raw Affymetrix files.
Supplementary Table 2 — probe IDs and corresponding Ensemble gene IDs.
Supplementary Table 3 — GO term annotations as downloaded from the BioMart community portal.
(processSamples.R) — R script using the Oligo package and Bioconductor framework to download CEL files from GEO, normalize, and process for analysis. The Oligo and GEOquery packages must be installed prior to use, instructions are within the script. To use, from within the directory containing the script, type at the R prompt: source(“processSamples.R”). Table of probeset/gene IDs (Supplemental File 1) must also be included in the same directory. Output file names/locations, as well as GEO IDs to be used to retrieve data can be modified within the script to work with a new dataset. This script was developed using R version 3.2.0.
(processSamples.R) — R script using the Oligo package and Bioconductor framework to download CEL files from GEO, normalize, and process for analysis. The Oligo and GEOquery packages must be installed prior to use, instructions are within the script. To use, from within the directory containing the script, type at the R prompt: source(“processSamples.R”). Table of probeset/gene IDs (Supplemental File 1) must also be included in the same directory. Output file names/locations, as well as GEO IDs to be used to retrieve data can be modified within the script to work with a new dataset. This script was developed using R version 3.2.0.
(convertListToGMT.pl) — Perl script used to convert two-column Gene-Annotation files into GSEA-compatible GMT files. To use, specify the input and output filenames, and the minimum number of gene-term associations to be considered a gene set in GSEA analysis (optional, default is set to 5). The input file should be a 3-column format including gene names (column 1), descriptions (column 2), and term IDs (column 3). The input file should be tab delimited, if descriptions are not available the second column can be left blank. The input file used for the analysis presented here can be obtained by converting Supplementary Table 3 to tab delimited text format. To use, from within the installed directory type: “perl convertListToGMP.pl inputFile.txt outputFile.gmt 5”.
(probeAndGeneIDs.txt) — Text file of probe ID and Ensemble Gene ID mapping. Required for Supplemental Script 1, please transfer to the working directory.