Abstract
16S rRNA deep sequencing analysis, targeting V3 region was performed using Illumina bar coded sequencing. Sediment samples from two hot springs (Atri and Taptapani) were collected. Atri and Taptapani metagenomes were classified into 50 and 51 bacterial phyla. Proteobacteria (45.17%) dominated the Taptapani sample metagenome followed by Bacteriodetes (23.43%) and Cyanobacteria (10.48%) while in the Atri sample, Chloroflexi (52.39%), Nitrospirae (10.93%) and Proteobacteria (9.98%) dominated. A large number of sequences remained taxonomically unresolved in both hot springs, indicating the presence of potentially novel microbes in these two unique habitats thus unraveling the importance of the current study. Metagenome sequence information is now available at NCBI, SRA database accession no. SRP057428.
Keywords: Hot springs, Atri, Taptapani, 16S rRNA, Illumina sequencing, Bacterial community
| Specification | |
|---|---|
| Organism/cell line/tissue | Soil metagenome of Taptapani hot spring |
| Sex | Not applicable |
| Sequencer/array type | Illumina GAIIx |
| Data format | Analyzed |
| Experimental factors | Environmental sample |
| Experimental features | V3 hypervariable region of 16S rDNA was sequenced using paired end Illumina Mi-Seq technology and the sequence was analyzed using QIIME data analysis package. |
| Consent | Not applicable |
| Sample source location | Sediment sample, Taptapani hot spring, Atri hot spring Odisha, India |
1. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/sra/ SRP057428.
Hot springs have been of utmost importance because of the unique thermophilic properties of the organisms thriving in these niches, the description of an increasing figure of new thermophilic species and thermophilic biocatalysts they produce [1]. Thermal stability of enzymes like protease, lipase, amylase, cellulase, phosphatase, asperginase, esters, carboxylase etc. from thermophiles is gaining importance because of a number of commercial applications. Studies in the last two decades have revealed that less than 1% of these organisms of a community are possibly cultured in vitro with known cultivation techniques, but 99% still remain unexplored [2]. According to the geological survey report, in Odisha (East India), eight thermal hot springs have been located in different districts viz.; Atri and Badaberena of Khurda; Taptapani of Ganjam; Tarobalo of Nayagarh; Deulijhari and Magarmuhan of Anugul; Bankhol of Cuttack; and Boden of Nuapada [3]. Recently, Sen and Maiti [4] reported the bacterial diversity of Atri, Taptapani and Tarobalo hot springs by conventional culture dependent approach. However, the majority of the bacterial isolates were affiliated with the single genus Bacillus that further emphasized the need of more in-depth analysis to understand the total bacterial diversity of these hot springs. To our knowledge, this study describes the first effort to explore the bacterial diversity of Atri and Taptapani hot springs using the Illumina platform.
Sediment samples were collected from two different sulfur hot springs of Odisha: Atri (Latitude: 20°13′32.784″N, Longitude: 85°29′58.485″E), and Taptapani (Latitude: 19°30′16.8″N, Longitude: 84°24′4.6″E). Samples were collected in triplicate. Metagenomic DNA of sediment samples (5 g) were extracted as described by Kumar and Khanna (2014). The V3 region of the 16S rRNA was amplified using primers 341F, 5′-CCTACGGGAGGCAGCAG-3′ and 518R, 5′-ATTACCGCGGCTGCTGG-3′ with 50 ng of metagenomic DNA. The amplified PCR product was purified by gel elution using minelute columns (Qiagen, India) and subjected to 150 nucleotide paired-end multiplex sequencing using Illumina GAIIx sequencer at Genotypic Technology Pvt. Ltd. (Bangalore, India). Bioinformatic analysis was carried out by QIIME data analysis package [5]. Krona graphs were plotted using Krona [6].
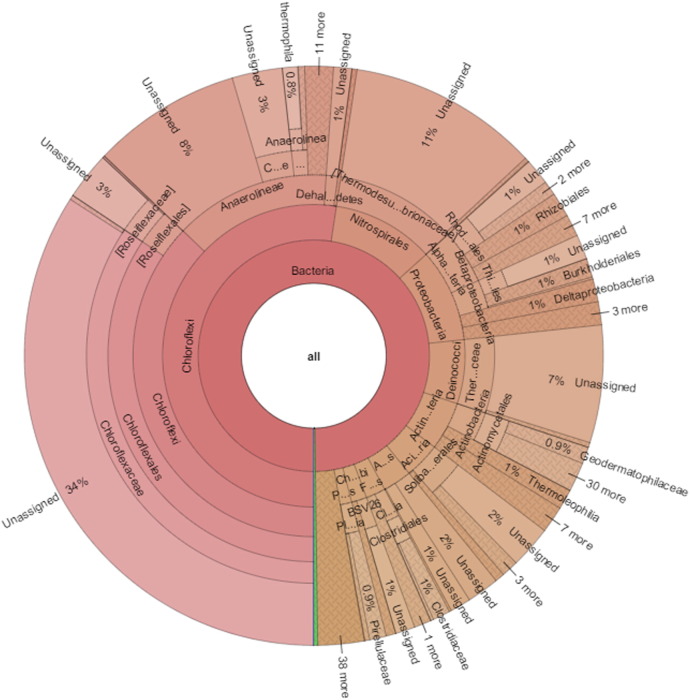
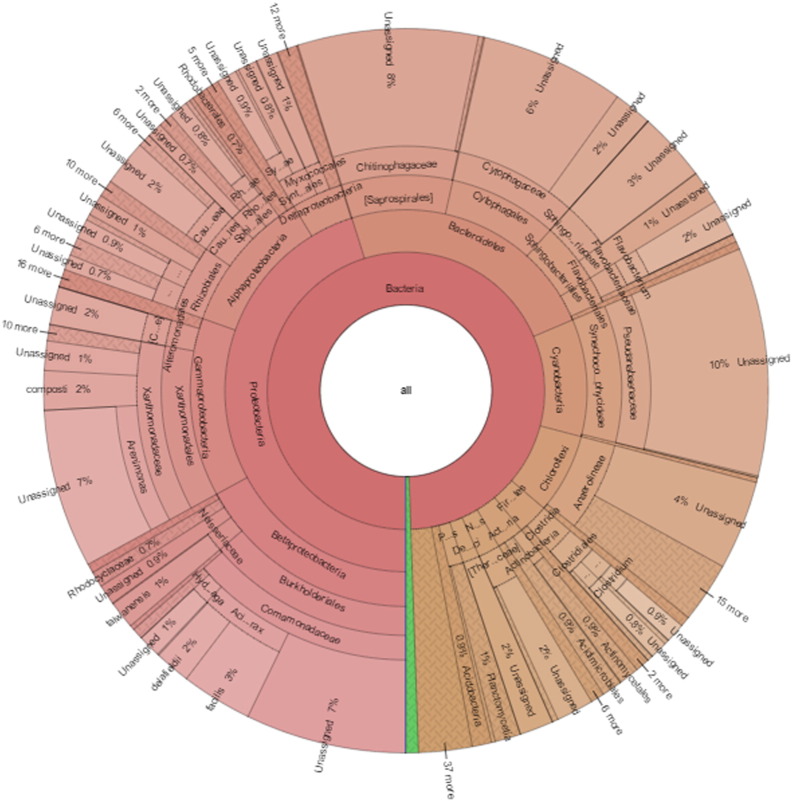
The output data comprised 186,292 and 333,276 high quality reads for Atri and Taptapani samples respectively were used for analysis. These stringent measures removed nearly 65% (Atri) and 51% (Taptapani) of the initial reads. At 80% confidence threshold in the RDP classifier, the 16S rRNA gene sequencing reads of Atri and Taptapani samples were classified into 50 and 51 bacterial phyla. In the Atri sample, most sequences were affiliated predominantly with Chloroflexi (52.39%), Nitrospirae (10.93%), Proteobacteria (9.98%), [Thermi] (6.84%), Actinobacteria (5.19%), Acidobacteria (3.84%), Armatimonadetes (3.09%) and Firmicutes (2.09%) that together accounted for ~ 95% of all sequence reads (Fig. 1). On the other hand, Proteobacteria dominated the Taptapani sample (45.17%) followed by Bacteriodetes (23.43%) and Cyanobacteria (10.48%) (Fig. 2). The other dominant phyla in Taptapani sample were Chloroflexi (7.16%), Firmicutes (2.76%) and Actinobacteria (2.44%) (Fig. 2). Out of 814 genera assigned to Taptapani hot spring, only 51% were identified at the genus level. The same trend was followed in Atri sample.
Fig. 1.
Community composition in Atri hot spring metagenome.
Fig. 2.
Community composition in Taptapani hot spring metagenome
A large proportion of phylotypes obtained in this study was not assigned to known taxonomic groups and so these data contribute significantly to the known diversity of hot spring environmental phylotypes. Consequently, Our results indicate that both hot springs (Atri and Taptapani) harbor complex and distinct bacterial community which were not reported before.
Acknowledgments
The Department of Biotechnology, Government of India is greatly acknowledged for funding (BT/PR7944/BCE/8/1036/2013) the present study. We are also grateful to our colleagues Mr. Champakraj Kar for the support during sampling and Mr. Mahendra Gaur for his computational assistance.
Contributor Information
Enketeswara Subudhi, Email: enketeswarasubudhi@soauniversity.ac.in.
Mohit Kumar, Email: kumarmohit@yahoo.com.
References
- 1.Tomova I., Stoilova-Disheva M., Lyutskanova D., Pascual J., Petrov P., Kambourova M. Phylogenetic analysis of the bacterial community in a geothermal spring, rupi basin, Bulgaria. World J. Microbiol. Biotechnol. 2010;26:2019–2028. [Google Scholar]
- 2.Kumar M., Khanna S. Shift in microbial population in response to crystalline cellulose degradation during enrichment with a semi-desert soil. Int. Biodeterior. Biodegrad. 2014;88:134–141. [Google Scholar]
- 3.Mahala S., Singh S.C., Das P., Acharya M. Genesis of thermal springs of Odisha, India. Int. J. Earth Sci. Eng. 2012;05:1572–1577. [Google Scholar]
- 4.Sen R., Maiti N.K. Genomic and functional diversity of bacteria isolated from hot springs in Odisha, India. Geomicrobiol J. 2014;31:541–550. [Google Scholar]
- 5.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]