Abstract
Acidithiobacillus ferrooxidans YQH-1 is a moderate acidophilic bacterium isolated from a river in a volcano of Northeast China. Here, we describe the draft genome of strain YQH-1, which was assembled into 123 contigs containing 3,111,222 bp with a G + C content of 58.63%. A large number of genes related to carbon dioxide fixation, dinitrogen fixation, pH tolerance, heavy metal detoxification, and oxidative stress defense were detected. The genome sequence can be accessed at DDBJ/EMBL/GenBank under the accession no. LJBT00000000.
Keywords: Acidithiobacillus ferrooxidans, Genome, Iron-oxidizing bacteria, Bioleaching, Volcano
| Specifications | |
|---|---|
| Organism/cell line/tissue | Acidithiobacillus ferrooxidans |
| Strain (s) | YQH-1 |
| Sequencer or array type | Sequencer; Illumina MiSeq |
| Data format | Processed |
| Experimental factors | Microbial strains |
| Experimental features | Draft genome sequence of Acidithiobacillus ferrooxidans YQH-1 assembly and annotation |
| Consent | N/A |
| Sample source location | Volcanic river in the Wudalianchi volcano, China |
1. Direct link to deposited data
2. Experimental design, materials and methods
Acidithiobacillus ferrooxidans is a Gram-negative, motile, moderately thermophilic (20 to 40 °C), acidophilic (pH 1.0 to 4.5) and strictly chemolithoautotrophic bacterium [1]. It utilizes ferrous iron or reduced inorganic sulfur compounds as energy source and fixes carbon dioxide from the atmosphere as a carbon source [2], [3]. Strains of A. ferrooxidans are abundant in iron and sulfur bearing acidic environments and considered as important participants in the iron, sulfur and carbon cycles of the acid mine drainage ecosystem [4].
Research on A. ferrooxidans has been a promising area of study as this bacterium is known to be an important member of microbial consortia that is used to recover metals via a process known as bioleaching or biomining [3]. In this study, A. ferrooxidans YQH-1 was isolated from a river in Wudalianchi volcano, Northeast China (48o 40′ 14″ N, 126o 10′ 16″ E). The 16S rRNA sequence of YQH-1available in GenBank database (accession number: KT633236) showed 99.64% identity with that of A. ferrooxidans ATCC 19859T (accession number: AJ457808). To better understand the important information regarding the bioleaching capabilities, we performed a genomic analysis of A. ferrooxidans YQH-1.
Genomic DNAs were extracted using a Bacterial Genomic DNA Extraction Kit (Biomed, Beijing, China) according to the manufacturer's instructions. The genome of YQH-1 was sequenced using the Illumina MiSeq platform at the Biomarker Technologies Co., Ltd. (Beijing, China). A library with a fragment length of 500 bp was constructed, and a total of 854,755,347 bp paired-end reads of 300 bp were generated. The high-quality reads, which provided an approximately 285-fold depth of coverage were assembled with Velvet Version 1.2.10 [5]. Protein-coding sequences were predicted by Glimmer software version 3.0 [6], while Ribosomal RNA (rRNA) and transfer RNA (tRNA) genes were predicted using an RNAmmer 1.2 server [7] and tRNAscan-SE Search Server version 1.21 [8], respectively. Tandem repeats were predicted using Tandem Repeats Finder Version 4.04. Genes annotated using BLAST searches of nonredundant protein sequences from the NCBI, Swiss-Prot, NCBI Refseq, COG [9], and Rapid Annotation using Subsystem Technology (RAST) Version 2.0.
A total of 123 contigs were generated with N50 size of 69,681 bp. The assembled genome size of A. ferrooxidans YQH-1 was estimated to be 3,111,222 bp from 96 scaffolds with a mean G + C content of 58.63% (Table 1). In the whole genome, 2281 genes (73.84%) that encode known function proteins and 635 (20.55%) genes were considered to encode hypothetical proteins. Of the total, 173 (5.61%) genes have no database match.
Table 1.
Genome features of A. ferrooxidans YQH-1 draft genome.
| Attributes | Values |
|---|---|
| Genome size (bp) | 3,111,222 |
| Number of scaffolds | 96 |
| Scaffold N50 (bp) | 99,137 |
| G + C content (%) | 58.63 |
| Number of contigs | 123 |
| Contig N50 (bp) | 69,681 |
| CDS | 3089 |
| rRNAs | 10 |
| tRNAs | 42 |
| Pseudo genes | 64 |
| Tandem repeat sequence | 115 |
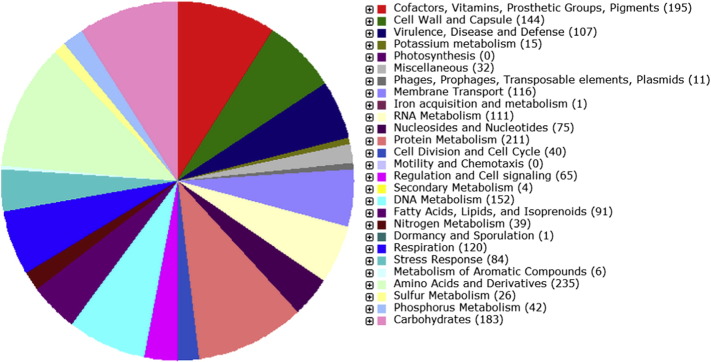
RAST functional annotation predicted 183 genes were linked to carbohydrate metabolism, out of which 58 genes were predicted to be involved in carbon dioxide fixation (Fig. 1). The presence of these genes may explain the ability of YQH-1 in the natural carbon cycle. A. ferrooxidans YQH-1 contains several genes involved in iron and sulfur metabolism, such as encapsulating protein for DyP-type peroxidase and ferritin-like protein oligomers, sulfate and thiosulfate import ATP-binding protein CysA, sulfate transport system permease protein CysW, Sulfate and thiosulfate binding protein CysP et al. (Fig. 1). They may be responsible for the oxidation of ferrous iron and reduced inorganic sulfur compounds.
Fig. 1.
Subsystem category distribution of A. ferrooxidans YQH-1 (based on RAST annotation server).
Additionally, 23 Nif genes involved in nitrogen metabolism were found, which indicated that A. ferrooxidans YQH-1 has nitrogen-fixation potential (Fig. 1). The Nif genes were also detected in the genomic sequences of A. ferrooxidans ATCC 23270 [4] and A. ferrooxidans ATCC 53993 (CP001132). These indicated that A. ferrooxidans has the genes involved in dinitrogen fixation and they are able to grow in diazotrophic conditions.
Fifteen genes related to potassium metabolism were detected, of which 14 were related to potassium homeostasis (Fig. 1), which may be responsible for the strain YQH-1 inhabiting an extreme acid environment [4]. There are 41 genes involved in heavy metal resistance and 35 genes involved in oxidative stress response. The presence of these genes may explain why A. ferrooxidans can live in hostile condition with high concentrations of heavy metal ions.
3. Nucleotide sequence accession number
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LJBT00000000. The version described in this paper is LJBT00000000.
Acknowledgments
This work was supported by the Natural Science Foundation of Heilongjiang Province of China (C201442).
References
- 1.Jensen A.B., Webb C. Ferrous sulphate oxidation using Thiobacillus ferrooxidans: a review. Process Biochem. 1995;30:225–236. [Google Scholar]
- 2.Esparza M., Cárdenas J.P., Bowien B., Jedlicki E., Holmes D.S. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans. BMC Microbiol. 2010;10:229. doi: 10.1186/1471-2180-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan L., Yin H., Zhang S., Leng F., Nan W., Li H. Biosorption of inorganic and organic arsenic from aqueous solution by Acidithiobacillus ferrooxidans BY-3. J. Hazard. Mater. 2010;178:209–217. doi: 10.1016/j.jhazmat.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 4.Valdés J., Pedroso I., Quatrini R., Dodson R.J., Tettelin H., Blake R., Eisen J.A., Holmes D.S. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics. 2008;9:597. doi: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.-H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N. The COG database: an updated version includes eukaryotes. BMC Bioinf. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]