Abstract
Citrobacter freundii strain ST2, isolated from the algae bloom sample, possesses an N-acylhomoserine lactone (AHL) production activity that secretes short-chain AHL molecules. In this study, we sequenced the complete genome of C. freundii strain ST2 to understand the molecular regulation of the AHL system and to search for the AHL gene in this bacterium. The results show that the genome size is 4.89 Mb with a G + C content of 51.96%. 4626 function proteins were predicted and 3647 proteins were assigned to COG functional categories. A predicted AHL-coding gene LuxR was found at contig 4 and the length was 1541 bp. The strain temporary deposited at Shenzhen Public Platform of Screening & Application of Marine Microbial Resources (Shenzhen, China), and the genome sequence can be accessed at GenBank under the accession no. LJSQ00000000.
Keywords: Citrobacter freundii, Genome sequence, N-acylhomoserine lactone, LuxR
| Specification | |
|---|---|
| Organism | Citrobacter freundii |
| Strain | ST2 |
| Sequencer | Illumina Hiseq 2000 |
| Data format | Processed |
| Experimental factor | Microbial strain |
| Experimental features | Whole genome sequence of Citrobacter freundii ST2, assembly and annotation |
| Consent | N/A |
| Sample source location | Marine spring algae bloom, Shenzhen coast, China. |
1. Direct link to deposited data
2. Experimental design, materials and methods
Quorum sensing (QS, e.g., AHL molecules), a common form of cell signaling, is gaining significant interest in marine ecology. Quorum sensing is a form of intercellular communication within microbial populations that occurs in a density-dependent manner. QS substance can result in downstream changes in gene regulation and modulation of bioluminescence, virulence, biofilm formation, and secondary metabolite production [1]. Chemical mediators of AHL and molecular crosstalk between bacteria and eukaryotes have been described for a wide range of symbiotic organisms [2]. In algal–bacterial microenvironments, previous works have investigated the crosstalk signaling between algae and bacteria, which can broaden understanding of the complex multi-species interactions [3], [4], [5]. In this work, we found the Citrobacter freundii strain ST2, which was isolated from the dinoflagellate (Scrippsiella trochoidea) and possessed AHL activities to produce N-acylhomoserine lactones. The genome sequence of C. freundii strain ST2 may provide insights on the regulation of quorum sensing mechanisms, and will help us understand the ecological behavior of this specie in algae–bacteria symbiosis.
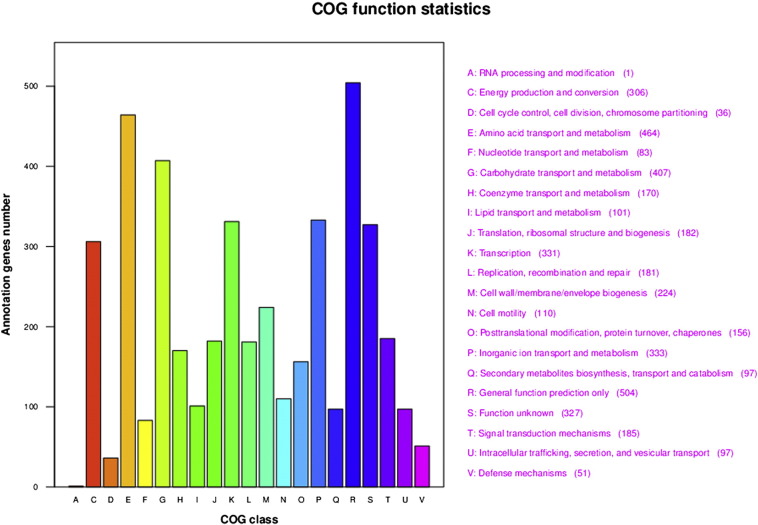
For the above-mentioned reasons, the draft genome sequence of C. freundii strain ST2 was determined. Genomic DNA of this bacterium was isolated using a DNA kit (Mo Bio, CA, USA) according the instructions of the manufacturer. The quality of DNA was examined using a Nanodrop spectrophotometer (Thermo Scientific, USA) and gel electrophoresis (Bio-Rad, USA). The whole-genome sequencing of C. freundii ST2 was performed using the Illumina Hiseq 2000 platform after the construction of the sequencing library using a Truseq DNA sample preparation kit, V2 (Illumina Inc. CA). After filtering the low-quality data, we assembled the short reads into a genome sequence using SOAP denovo v1.05, and the scaffolds were manually connected based on paired-end relationships. The gaps were filled using GapCloser with read mapping information. This resulted in 4,313,757 filtered reads and approximately 50-fold coverage. The filtered reads were de novo assembled with the CLC Genomic workstation v5.1, generating 21 contigs, containing 4,896,351 bp. The G + C content of this genome was 51.96%. Gene prediction was performed using prodigal (v260), and 4626 open reading frames were predicted [6]. Homologous comparisons by BLAST showed 3647 CDS involving the 21 functional COG groups and a part of the CDS involving the 34 metabolic pathway KEGG groups (Fig. 1). Annotation was performed using Blast2GO [7] against NCBI-NR sequence database [8]. One complete rRNA operon and one copy each of nine 5 s-rRNA gene, seven 16 s-rRNA gene, and six 23 s-rRNA gene were identified using RNAmmer [9]. A total of 79 tRNAs were identified using tRNAscan-SE [10]. In addition, 57 minisatellite DNAs and 3 microsatellite DNAs were determined based on the Repbase transposable elements library. The genome features of C. freundii strain ST2 were summarized in Table 1.
Fig. 1.
Function category distribution of Citrobacter freundii strain ST2 strain (based on COG function statistics).
Table 1.
Genome features of Citrobacter freundii strain ST2.
| Attributes | Values |
|---|---|
| Genome size | 4.89 Mb |
| GC content % | 51.96% |
| Number of Contigs | 21 |
| Total contig size | 4,885,161 |
| Scafflods | 6 |
| Total scaffold size | 4,896,351 |
| Protein encoding genes | 4626 |
| tRNAs | 79 |
| rRNAs | 21 |
| Predicted AHL gene LuxR site | Contig 4 |
| Encoding-AHL gene length | 1541 bp |
Based on Blast2GO analysis, the LuxR gene of C. freundii strain ST2 was found in contig 4, and the gene length was 1541 bp. It has a highly identity of the LuxR gene of another Enterobacteriaceae family member (C. freundii UCI 31, GenBank: EJF23932.1). Furthermore, a putative AI-2 (autoinducer-2) synthase gene with a metallo-beta-lactamase domain was also found in silico contig 4.
The complete genome of C. freundii strain ST2 provides novel insights into the quorum-sensing and signaling confusion genomic features of strain ST2. Further work needs to be conducted to study the roles of this bacterium in the phycosphere environment under AHL molecular regulation.
3. Nucleotide sequence accession number
The genome sequences were deposited in GenBank with the accession number LJSQ0000000. The version described in this paper is the first version.
Conflict of interest
The authors declare that there is no conflict of interests on work published in this paper.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (41476092), S&T Projects of Shenzhen Science and Technology Innovation Committee (JCYJ20140417115840286, CXZZ20150529165045063), Innovation Project of Science and Technology of Nanshan District Shenzhen (KC2013JSCX0003A), and the Project of Marine Fishery Science and Technology & Industry Development of Administration of Ocean and Fisheries of Guangdong Province (A201503D07).
References
- 1.Pacheco A.R., Sperandio V. Inter-kingdom signaling: chemical language between bacteria and host. Curr. Opin. Microbiol. 2012;12:192–198. doi: 10.1016/j.mib.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangwani N., Dash H.R., Chauhan A., Das S. Bacterial quorum sensing: functional features and potential applications in biotechnology. J. Mol. Microbiol. Biotechnol. 2012;22:215–227. doi: 10.1159/000341847. [DOI] [PubMed] [Google Scholar]
- 3.Amin S.A., Parker M.S., Armbrust E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012;76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin S.A., Hmelo L.R., van Tol H.M., Durham B.P., Carlson L.T., Heal K.R., Morales R.L., Berthiaume C.T., Parker M.S., Djunaedi B., Ingalls A.E., Parsek M.R., Moran M.A., Armbrust E.V. Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;7554:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson P.R., Pavia H., Toth G. Formation of harmful algal blooms cannot be explained by allelopathic interactions. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11177–11182. doi: 10.1073/pnas.0900964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]