The active-site glutamate 14 residues of the homodimeric multidrug transporter EmrE have distinct macroscopic pKa values, raising new questions regarding the coupling of proton import to drug efflux.
Abstract
The small multidrug resistance transporter EmrE is a homodimer that uses energy provided by the proton motive force to drive the efflux of drug substrates. The pKa values of its “active-site” residues—glutamate 14 (Glu14) from each subunit—must be poised around physiological pH values to efficiently couple proton import to drug export in vivo. To assess the protonation of EmrE, pH titrations were conducted with 1H-15N TROSY-HSQC nuclear magnetic resonance (NMR) spectra. Analysis of these spectra indicates that the Glu14 residues have asymmetric pKa values of 7.0 ± 0.1 and 8.2 ± 0.3 at 45°C and 6.8 ± 0.1 and 8.5 ± 0.2 at 25°C. These pKa values are substantially increased compared with typical pKa values for solvent-exposed glutamates but are within the range of published Glu14 pKa values inferred from the pH dependence of substrate binding and transport assays. The active-site mutant, E14D-EmrE, has pKa values below the physiological pH range, consistent with its impaired transport activity. The NMR spectra demonstrate that the protonation states of the active-site Glu14 residues determine both the global structure and the rate of conformational exchange between inward- and outward-facing EmrE. Thus, the pKa values of the asymmetric active-site Glu14 residues are key for proper coupling of proton import to multidrug efflux. However, the results raise new questions regarding the coupling mechanism because they show that EmrE exists in a mixture of protonation states near neutral pH and can interconvert between inward- and outward-facing forms in multiple different protonation states.
INTRODUCTION
The ionization state of amino acid side chains in proteins can dramatically affect important biological processes, such as enzymatic activity or protein folding and stability. In the case of secondary active transporters, ionizable groups are critical for harnessing the proton motive force needed to drive transport of another substrate against its electrochemical gradient. Microscopically, the energetically downhill movement of protons across the membrane occurs through protonation and deprotonation of ionizable residue(s) in the transporter. Although these ionizable residues must be accessible to the aqueous compartments on either side of the membrane as the transporter opens and closes, their environment deep within the transmembrane (TM) helices is unique. As a result, these residues may have pKa values dramatically shifted from the pKa values of similar groups exposed to water in peptides or soluble proteins (Castañeda et al., 2009; Grimsley et al., 2009; Farrell et al., 2010; Platzer et al., 2014). Accurate measurement of these pKa values is necessary to understand the mechanism of proton-coupled transport in secondary active transporters, such as EmrE.
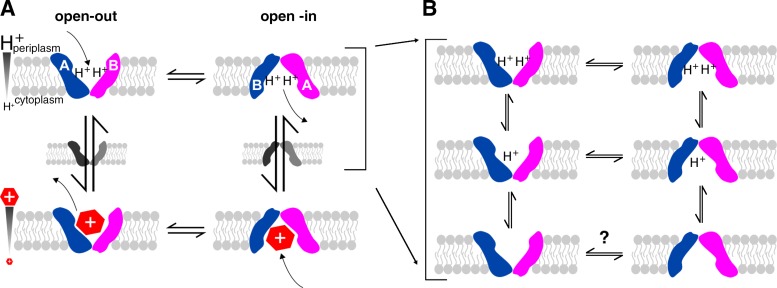
EmrE is a small multidrug resistance transporter in the inner membrane of Escherichia coli that exports polyaromatic cation substrates, conferring resistance to compounds of this chemical type. It is a secondary active transporter, importing protons to drive drug efflux. EmrE has long been thought to function by a single-site alternating access mechanism (Fig. 1 A; Yerushalmi and Schuldiner, 2000a). The highly conserved glutamate 14 (Glu14) is the only charged residue located in a TM helix. It is positioned in the middle of TM1 and is an essential residue necessary for binding both polyaromatic cation substrates and the counter-transported protons. As a result, Glu14 is central to coupling drug export to proton import to drive active efflux (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Soskine et al., 2004). It defines the “single-site” in the single-site alternating access model of EmrE transport activity. We have previously shown that drug-bound EmrE interconverts between inward- and outward-facing forms in a simple two-state exchange process with a rate that is determined by the identity of the bound substrate (Morrison et al., 2012; Morrison and Henzler-Wildman, 2014). Here, we focus on the proton side of the transport cycle to investigate how EmrE performs coupled antiport of drug and protons (Fig. 1).
Figure 1.
The mechanism of EmrE transport. EmrE is a homodimer composed of two subunits (blue and pink) oriented antiparallel to each other and in unique conformations (distinct shapes, labeled A and B). The subunits exchange conformations to switch between open-in and open-out forms. (A) In the standard single-site alternating access model, a single active site located between the two subunits alternates between binding protons or drug. Coupling between protons and drug is achieved by preventing exchange in the apo state and only allowing one substrate to bind at a time. (B) Drug-free EmrE has more states than represented in the basic model shown in A and most, if not all, of these states exchange.
Although the current structural data available for EmrE is of modest resolution, it is clear that EmrE forms an asymmetric, antiparallel homodimer (Chen et al., 2007; Morrison et al., 2012). This naturally raises the question of asymmetry in the pKa values of Glu14 from each subunit, particularly because Glu14 itself has been shown to be asymmetric both in the presence and absence of substrate (Lehner et al., 2008). However, these experiments were performed only at pH 8.0 and did not explore the pKa of Glu14.
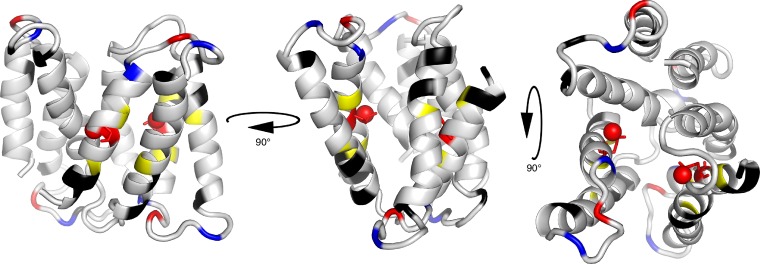
Estimates of the pKa of Glu14 have been reported based on substrate binding and transport assays as a function of pH. The published Glu14 pKa values range from 6.8 to 8.5 (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Yerushalmi et al., 2001; Soskine et al., 2004; Adam et al., 2007), with more recent studies centering on a value of 7.3. Each of these experiments monitored a single bulk parameter reflecting EmrE function, either transport activity or substrate binding. These assays included both equilibrium and kinetic studies of transport or binding of radioactive substrates, proton release upon substrate binding monitored through direct pH measurement or pH-sensitive dyes, or substrate binding monitored through intrinsic tryptophan fluorescence. In all cases, the data were analyzed using a single pKa value, either as a simplifying assumption based on the stoichiometry of EmrE antiport (Rotem and Schuldiner, 2004; Soskine et al., 2004) and the “functional equivalence” of Glu14 in modification studies (Weinglass et al., 2005), or because the data could not justify the assignment of more than one pKa value (Adam et al., 2007). This long-held model of a single pKa implies that the two Glu14 residues, one from each subunit, are simultaneously protonated or deprotonated in a highly cooperative single event. However, consideration of the close proximity of the two ionizable Glu14 residues in the hydrophobic substrate-binding pocket of EmrE (Fig. 2) suggests the possibility of negative cooperativity in the deprotonation of the two Glu14 residues. Furthermore, non-identical pKa values for the two Glu14 residues might be expected as a result of the structural asymmetry of Glu14 (Chen et al., 2007; Lehner et al., 2008).
Figure 2.
Structure of homodimeric TPP+-bound EmrE with pH-sensitive residues highlighted. Basic residues are colored blue, and acidic residues are colored red. TPP+ is not shown. Note that Glu14, shown as sticks with the protonatable oxygen represented as a ball, is the only charged residue in the TM regions. Yellow residues (Ala10, Gly17, and Ser43) were used for the pKa fits and are located near the Glu14 active site. Black residues are more remote from the active site but also fit to the same two pKa values determined using the yellow residues close to Glu14 (Protein Data Bank accession no. 3B5D).
The importance of Glu14 is demonstrated by the effects on EmrE activity of mutating this residue. Mutation of Glu14 to Cys abolishes EmrE activity (Yerushalmi and Schuldiner, 2000b). Mutation of Glu14 to Asp (E14D-EmrE) creates a transporter that does not confer drug resistance in E. coli but is able to transport drugs with a reduced rate and a shifted pH dependence of substrate binding and transport in liposomes (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b). These effects are explained by a reduced substrate affinity and a significant downward shift of the pKa of residue 14 to ≈5.0–6.7 in E14D-EmrE (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Soskine et al., 2004; Adam et al., 2007). This moves the pKa out of the physiological range and prevents coupling of drug export to proton import in cells.
Accurate measurement of Glu14 pKa value(s) for EmrE is also important for identifying conditions suitable for further study of both protonated EmrE and deprotonated (truly apo) EmrE. In particular, deprotonated EmrE represents an ideal target for structural studies, as conformational exchange between open-in and open-out EmrE should not occur under these conditions according to the single-site alternating access model for coupled antiport (Fig. 1 A). High resolution structures can provide mechanistic insights and generate hypotheses for refining mechanistic models of proton-coupled transport. Thus, higher resolution structures of EmrE have been a long-standing goal for the field. Along with its small size and limited water-exposed surface area, the highly dynamic nature of EmrE has likely impeded progress toward this goal. Thus, a trapped, static structure is highly sought after, and the deprotonated state is predicted to be such a state.
When analyzing pH-dependent effects on EmrE, it is necessary to keep track of multiple structural states and exchange processes. EmrE is an asymmetric antiparallel homodimer, with a single dimer structure when bound to the substrate tetraphenylphosphonium (TPP+). Each subunit in the asymmetric homodimer has a distinct conformation. The two subunits swap conformations as the homodimer interconverts between the open-in and open-out forms, and this process is referred to as conformational exchange (Fig. 1). Whether EmrE is open-in or open-out, the dimer structure is identical except for its orientation in the membrane. In accord with the single-site alternating access model for proton-coupled antiport, we hypothesized that different protonation states of EmrE would have different structures. We also expected that the rate of conformational exchange between inward- and outward-facing EmrE would depend on protonation state.
Nuclear magnetic resonance (NMR) is a well-established tool for directly measuring pKa values of ionizable groups in proteins (Nielsen, 2008; Castañeda et al., 2009; Farrell et al., 2010; McIntosh et al., 2011; Reijenga et al., 2013). The strength of NMR lies in the sensitivity of the chemical shift to the electrostatic properties of its environment and the ability to directly and simultaneously monitor many positions throughout the protein. These features have led NMR to become the method of choice for accurate measurement of pKa values for residues critical to enzyme function and protein folding. NMR has been used previously to determine the pKa value of important residues in several integral membrane proteins and peptides in membrane-mimetic environments, such as the M2 channel (Hu et al., 2006, 2012) and Neu TM domain (Smith et al., 1996). However, to our knowledge NMR has not been used previously to determine pKa values of the critical residue in a proton-coupled transporter. Here, we have used NMR pH titrations of EmrE solubilized in isotropic bicelles to address the following questions: How many distinct protonation states are observed for Glu14 in EmrE? Is the protonation state coupled to broader changes in EmrE structure? Do different protonation states/structures of EmrE have different rates of conformational exchange between open-in and open-out forms?
MATERIALS AND METHODS
Protein purification and sample preparation
The pET 15b-EmrE plasmid was provided by G. Chang (University of California, San Diego, Skaggs School of Pharmacy and Pharmaceutical Sciences, La Jolla, CA). EmrE was expressed, purified, and reconstituted into isotropic bicelles as described previously (Morrison and Henzler-Wildman, 2012). In brief, 6xHis-2H/15N-EmrE was purified from BL21 cultures grown in D2O M9 media supplemented with one generic multivitamin, 2 g glucose, 1 g 15NH4Cl, and 0.5 g 2H/15N ISOGRO (Sigma-Aldrich) per liter. 6xHis-2H/15N-EmrE was purified in n-decyl-β-d-maltopyranoside (Anatrace) with metal-affinity chromatography followed by size-exclusion chromatography. The 6xHis tag was cleaved with thrombin before reconstitution at a mole ratio of 70–100:1 long chain lipid/EmrE monomer into 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC)/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) (unless noted as 1,2-dimyristoyl-sn-glycero-3-phosphocholine [DMPC]; Avanti Polar Lipids, Inc.) isotropic bicelles (q = 0.33 long chain/short chain). EmrE has the same binding affinity for TPP+ and the same TPP+-bound conformational exchange rate when reconstituted in both DLPC and DMPC bicelles (Morrison et al., 2012).
NMR spectroscopy
Samples for NMR experiments contained 0.8–1 mM of 15N-labeled and extensively deuterated EmrE in isotropic bicelles. WT EmrE samples were buffered with 20 mM acetate and 50 mM MOPS, with the addition of 50 mM bicine at high pH. E14D-EmrE was buffered with 30 mM potassium phosphate plus 25 mM acetate. The EmrE spectrum is identical in each of these buffers alone or in combination. Additionally, NMR samples contained 20 mM NaCl, 5–10% D2O, 0.05% NaN3, and 2 mM 3-(trimethylsilyl)-1-propanesulfonic acid for chemical-shift referencing.
To accurately adjust the pH of NMR samples for data collected on WT EmrE at 45°C, pH electrode calibration buffers and the NMR sample were placed in a water bath equilibrated at 45°C. The pH electrode (BioTrode; Hamilton Company) was then calibrated using these prewarmed buffers, and the pH of the NMR sample was likewise adjusted at temperature. For E14D-EmrE, sample pH was adjusted at room temperature and later corrected to account for the temperature dependence of the buffer. To do this, NMR buffer was measured at both 25 and 45°C, with the pH electrode correspondingly calibrated at these temperatures, to make a calibration curve for correcting the pH for temperature effects. Sample pH was measured before and after each NMR spectrum to account for any pH drift. pH drift larger than the error in measurement was only observed for values above pH 8.0 and is included in the x-axis error bars. A single sample was used for each complete titration series, and buffer was exchanged once if needed to reduce salt midway through the titration. 1H-15N transverse relaxation optimized spectroscopy (TROSY)–heteronuclear single quantum coherence (HSQC) spectra were acquired starting from neutral pH, first raising, then lowering, and then raising the pH to finish near neutral. The consistency of the spectra acquired “out of order” confirms the sample stability throughout the titration. The temperature of the probe was calibrated using 100% methanol and ethylene glycol.
NMR spectra were collected on a 700-MHz spectrometer (Varian Inova) with a room temperature probe. 1H-15N TROSY-HSQCs were collected with 24 scans and 128 increments, with recycle delays of 2 s. The TROSY-selected ZZ-exchange experiment (Li and Palmer, 2009) was modified and run as described previously (Morrison et al., 2012). NMR spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed in CcpNmr analysis (Vranken et al., 2005). Assignments were transferred from TPP+-bound EmrE (Morrison et al., 2012) to apo EmrE based on the strong similarity between the high pH apo EmrE and TPP+-bound EmrE spectra. The chemical shifts were followed along the pH titration trajectory.
Data analysis
Chemical-shift data as a function of pH was analyzed in IgorPro (WaveMetrics), and data were plotted onto the protein structure using the PyMOL Molecular Graphics System (Schrödinger, LLC). Data for each titration was fit to either one or two global pKa values using the following equations. Amide proton and nitrogen chemical shifts were fit separately, rather than the overall chemical-shift change, as amide proton and nitrogen shifts have different sensitivity to through-bond and through-space effects. Not combining the amide proton and nitrogen chemical shifts allows greater insight into the pH-dependent effects on the protein (Tomlinson et al., 2010). For a single pKa, the modified Henderson–Hasselbalch equation describes the chemical shift of each residue (δ) as a function of pH (Shrager et al., 1972; McIntosh et al., 2011):
| (1) |
where δH and δD are the chemical shift of the protonated and deprotonated states of that residue. For two pKa values, a general macroscopic model was used (McIntosh et al., 2011):
| (2) |
where δH is the chemical shift of the doubly protonated species, δI is the chemical shift of the intermediate singly protonated species, and δD is the chemical shift of the fully deprotonated species. Eq. 2 describes the average overall behavior of the system as observed by NMR, with the chemical shift of a single residue dependent on two distinct ionization events. The same macroscopic behavior can arise from several different microscopic situations, which are mathematically identical and differ only in the physical interpretation of the parameters (McIntosh et al., 2011).
Online supplemental material
Fig. S1 shows an overlay of the full spectra of WT EmrE at all pH values in the 45°C titration. Fig. S2 shows an overlay of the full spectra of E14D-EmrE at all pH values in the titration. Fig. S3 shows an overlay of the full spectra of WT EmrE at all pH values in the 25°C titration. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201511404/DC1.
RESULTS
Using NMR to monitor protonation of Glu14
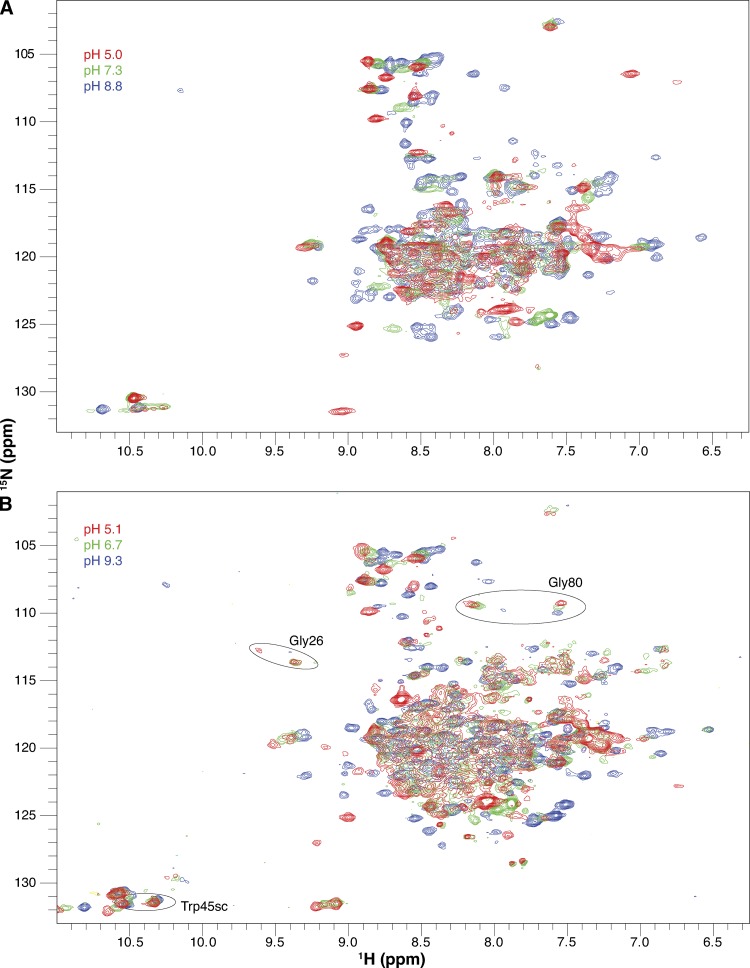
Ideally, the pKa of ionizable residues is measured by following the 13C and 1H chemical shifts of the side-chain carbons that are closest to the ionizable group in each titratable residue (Lindman et al., 2006; Platzer et al., 2014). Unfortunately, we were not able to do this because of technical challenges. Limited side-chain assignments and chemical-shift overlap in 1H-13C HSQC spectra prevent direct monitoring of the Glu14 side chain in uniformly labeled EmrE. Amino acid–specific labeling of glutamate when EmrE is expressed in its native E. coli is precluded as a result of the central role of glutamate in metabolism. Elegant NMR experiments designed to selectively monitor aspartate and glutamate side chains (Pellecchia et al., 1997; Castañeda et al., 2009) in uniformly labeled samples did not have sufficient sensitivity when applied to EmrE solubilized in isotropic bicelles. Therefore, we have used simple 1H-15N TROSY-HSQC spectra to monitor the pH-dependent chemical shift of backbone amides in EmrE (Figs. 3 and S1).
Figure 3.
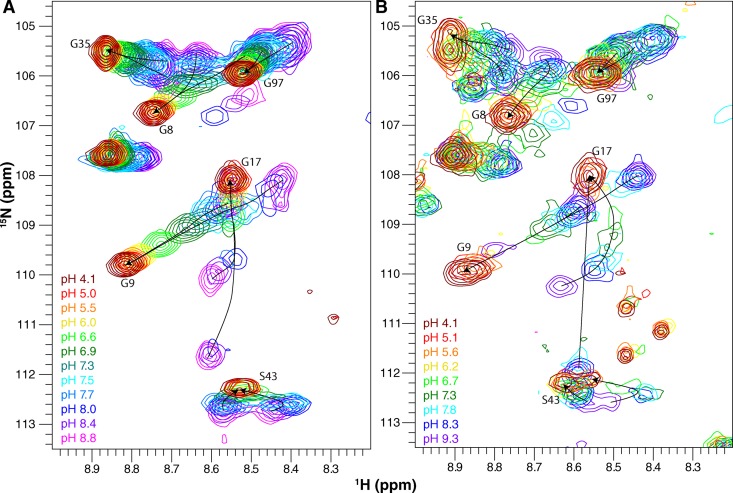
Drug-free EmrE is sensitive to pH. Full 1H-15N TROSY-HSQC spectra of drug-free WT EmrE in DLPC/DHPC isotropic bicelles at three pH values collected at 45°C (A) and 25°C (B). The pH of each spectrum is indicated by its color as designated in the figure. All spectra in the full pH titration datasets are shown in Figs. S1 (45°C) and S3 (25°C). Circles highlight several residues that remain in the slow exchange regime at 25°C at low pH.
Although not as precise as direct measurement of the ionizable side chain itself, monitoring pH titrations via the backbone amide resonances is a widely used strategy that provides more localized information on the pKa values of ionizable side chains than the single global readout provided by bulk biochemical experiments. The spectra in Fig. 3 reveal that many residues titrate near neutral pH in bicelle-solubilized EmrE, not just those located in close proximity to Glu14 (Fig. 2). Because the direct electrochemical effects of protonation will only be felt at close range, what is the origin of these extensive pH-dependent chemical-shift changes? One possibility is that protonation of Glu14 is coupled to global structural changes. This would not be particularly surprising given that the single-site alternating access model predicts that binding of either substrate (protons or drug) facilitates conformational exchange between open-in and open-out states, which is a global process in EmrE. An alternative possibility is that all of the Asp (Asp84), His (His110), and Glu (Glu14 and Glu25) residues in EmrE happen to have pKa values in the same near-neutral pH range, and we are observing local effects from all of these titration events simultaneously. We can experimentally distinguish between these two possibilities by considering the distribution of ionizable residues in EmrE and performing mutagenesis of Glu14.
First, all of the titratable residues in EmrE are located in loop regions except for Glu14 (Fig. 2), making it less likely that all titratable residues will have similarly shifted pKa values. Furthermore, if we are observing local effects of independent protonation events at each of these sites, then only nearby loop residues should be sensitive to pH. However, several residues in the TM helices that are not close to E14 or the titratable residues in the loop regions do titrate with pH, as shown in black in Fig. 2. Although these residues are clearly not close to Glu14 or any other titratable residues, it is difficult to determine precise distances because of the low resolution of the crystal structure. An example of just how few residues have pH-dependent chemical shifts caused by local effects of protonation/deprotonation is shown in the section on E14D below.
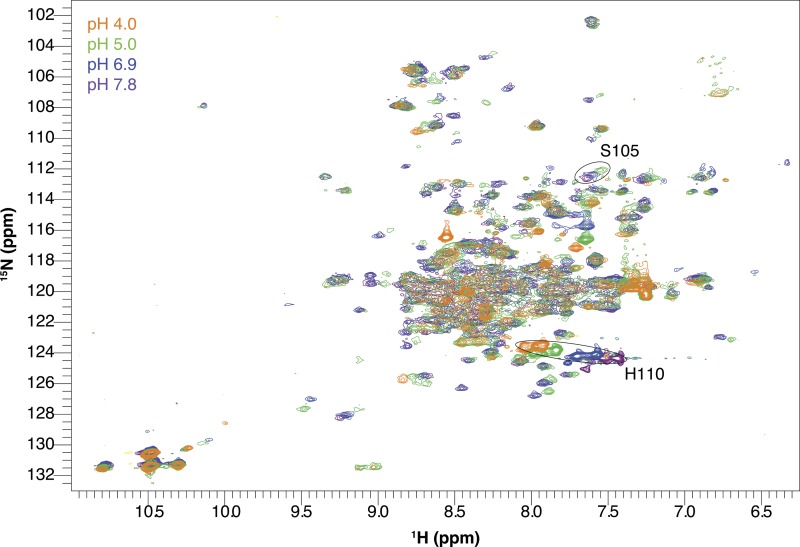
Second, we can perform a pH titration of a Glu14 mutant to confirm that we are specifically detecting protonation of Glu14 (Figs. 4 and S2). If the hypothesis that we are observing a global structural change upon Glu14 protonation is true, then (a) all residues should sense a single identical pKa, and (b) mutation of Glu14 to another residue should change the pH dependence of all residues in an identical manner. If the alternative is true and the NMR experiments detect only local effects of many separate protonation events at individual Glu, Asp, and His residues, then (a) different pKa values should be observed for each local region, and (b) mutation of Glu14 should only cause pH-dependent chemical-shift changes for residues in close proximity. To test the effect of mutation of Glu14, we chose E14D-EmrE because it is the only E14 mutation that retains any function (Yerushalmi and Schuldiner, 2000b). All other mutations of Glu14 render EmrE unable to transport or even bind substrate, so it is more difficult to assess that the protein is properly folded and otherwise well behaved. Based on previous biochemical estimates, the pKa of the active site is shifted from ≈7.3 in WT-EmrE to ≈5.4 in E14D-EmrE (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b), a significant shift that is easily detectable in our titrations. This is exactly what is observed in the spectra (Figs. 4 and S2). The same residues in EmrE still titrate, but the pH-dependent chemical-shift changes occur at more acidic pH, exactly as expected. These results show that the NMR titrations are monitoring the protonation state of Glu14 coupled to a global conformational change. Quantitative analysis of all of the titrations is discussed below.
Figure 4.
E14D-EmrE has a shifted pH sensitivity. 1H-15N TROSY-HSQC spectra of drug-free E14D-EmrE in DMPC/DHPC isotropic bicelles at 45°C and varying pH values. Note that chemical shifts of His110 and Ser105 (circled peaks) titrate with a higher pKa than the rest of the protein. The pH of each spectrum is indicated by its color as designated in the figure. The full set of pH titration spectra are shown in Fig. S2.
pH-Dependent conformational exchange rates in drug-free EmrE
Closer inspection of the titration patterns of individual residues, particularly Ala10 (Fig. 5) and the glycine region (Fig. 6), reveals that the conformational exchange rate between open-in and open-out states of EmrE is also pH dependent. At high pH, two peaks are observed for each residue of EmrE at 25 and 45°C (Fig. 3). The two peaks present in the spectra of drug-free EmrE at high pH have very similar chemical shifts to the drug-bound spectra of EmrE that we have analyzed previously (Morrison et al., 2012; Morrison and Henzler-Wildman, 2014) and can be assigned to subunit A and subunit B of the asymmetric homodimer. We have shown previously that the two subunits swap conformations as drug-bound EmrE converts from open-in to open-out (Morrison et al., 2012). Whether the two subunits swap conformations at high pH for drug-free EmrE is discussed below in the section, Is apo EmrE still able to interconvert?
Figure 5.
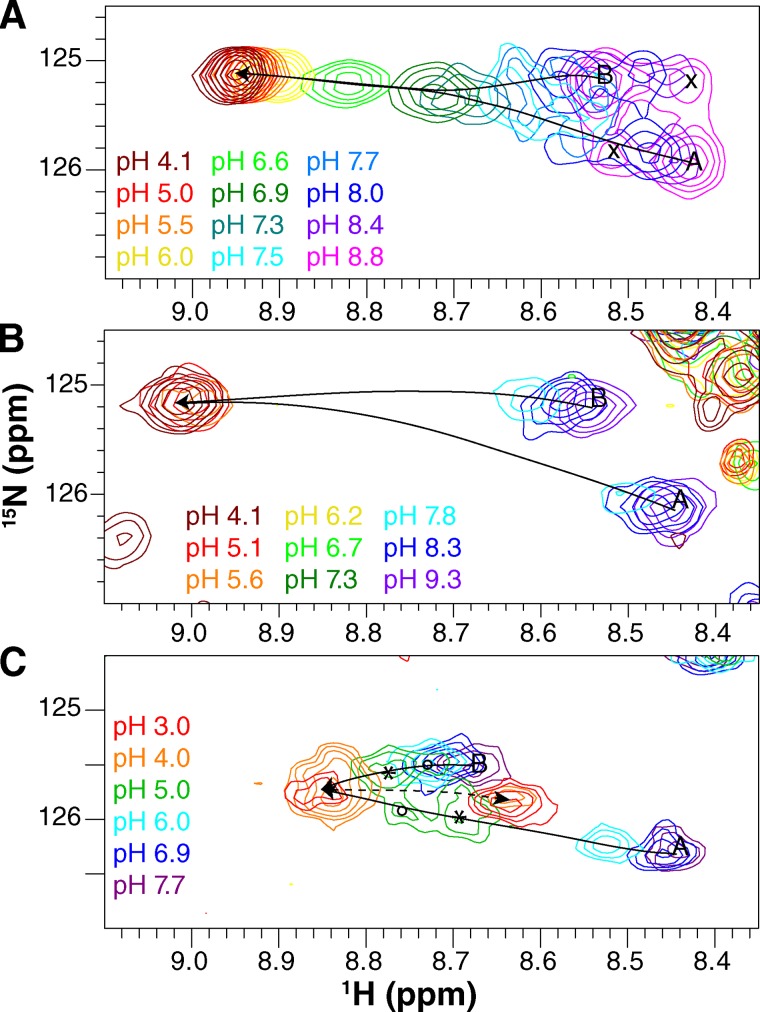
Ala10 senses the protonation state of EmrE. Enlarged region of the 1H-15N TROSY-HSQC spectra showing the Ala10 peaks for both WT EmrE (A and B) and E14D-EmrE (C). Spectra were collected at 45°C (A and C) and 25°C (B). The pH of each spectrum is indicated by its color as designated in the figure. Lines highlight the movement of peaks as the pH is lowered. When two distinct subunit peaks can be distinguished, the assignment of each subunit is labeled as A and B. (B) Exchange cross peaks are marked with an “x.” (C) The four peaks at pH 5.0 (green spectrum) are interpreted as two states of the E14D-EmrE dimer, indicated with ○ and *. These peaks do not reflect conformational exchange because they do not align in a square pattern. Both subunits have a distinct conformation in each state. The dashed line highlights the additional slow-exchange process at low pH.
Figure 6.
Gly17 senses the protonation state of EmrE. Enlarged glycine region of the 1H-15N TROSY-HSQC spectrum for WT EmrE. Spectra were collected at 45°C (A) and 25°C (B). The pH of each spectrum is indicated by its color as designated in the figure. Lines highlight the movement of the peaks as the pH is lowered.
As the pH is lowered at 45°C, the two peaks corresponding to subunit A and subunit B merge into a single peak for many residues, such as Ala10 (Figs. 3 A, 5 A, and 6 A). A single peak for each residue would be observed if either (a) the exchange between conformations increases and becomes fast on the NMR timescale (i.e., the exchange rate is significantly larger than the chemical-shift difference; see Fig. 7 A), or (b) asymmetry is lost and the two subunits have identical conformations. We can differentiate between these two options by looking at spectra collected at a lower temperature (25°C; Figs. 3 B, 5 B, and 6 B), which decreases the rate of conformational exchange. Some residues such as Gly26 or Gly80 still have two distinct peaks visible at low pH at 25°C (Fig. 3 B). The reason that some residues have a single peak at low pH (such as Ala10; Fig. 5 B) and some have two peaks at low pH in this 25°C spectrum is because of the definition of the NMR “timescale” (Fig. 7 A), which depends on the exchange rate relative to the chemical-shift difference. Although there is a single global conformational exchange process (subunits A and B swapping conformations to convert from the open-in to open-out state) with a single global rate, the chemical-shift difference between the subunit A and subunit B peaks is a residue-specific property. Some residues in EmrE exist in very similar environments in subunits A and B, whereas other residues have very different environments in the two subunits and thus large chemical-shift differences (see Morrison et al., 2012, for further discussion of the chemical-shift differences between the two subunits in the asymmetric EmrE homodimer). For the same global exchange rate, some residues with large chemical-shift differences will fall in the slow exchange regime, and two peaks will be observed even at low pH, as for Gly26 and Gly80 at 25°C (Fig. 3 B). Residues with smaller chemical-shift differences will fall in the fast exchange regime, and only a single peak will be observed at low pH, as for Ala10 (Fig. 5 B). Even for residues in the slow exchange regime, the two peaks visible at low pH are broader and weaker than the two peaks at high pH for the same residue, reflecting an increased exchange rate. Thus, the data appear to report the behavior predicted by the single-site alternating access model for coupled antiport: EmrE should interconvert rapidly when bound to substrate (protons) and should not exchange in the absence of substrate (high pH, deprotonated). However, closer inspection of the spectra reveals greater complexity.
Figure 7.
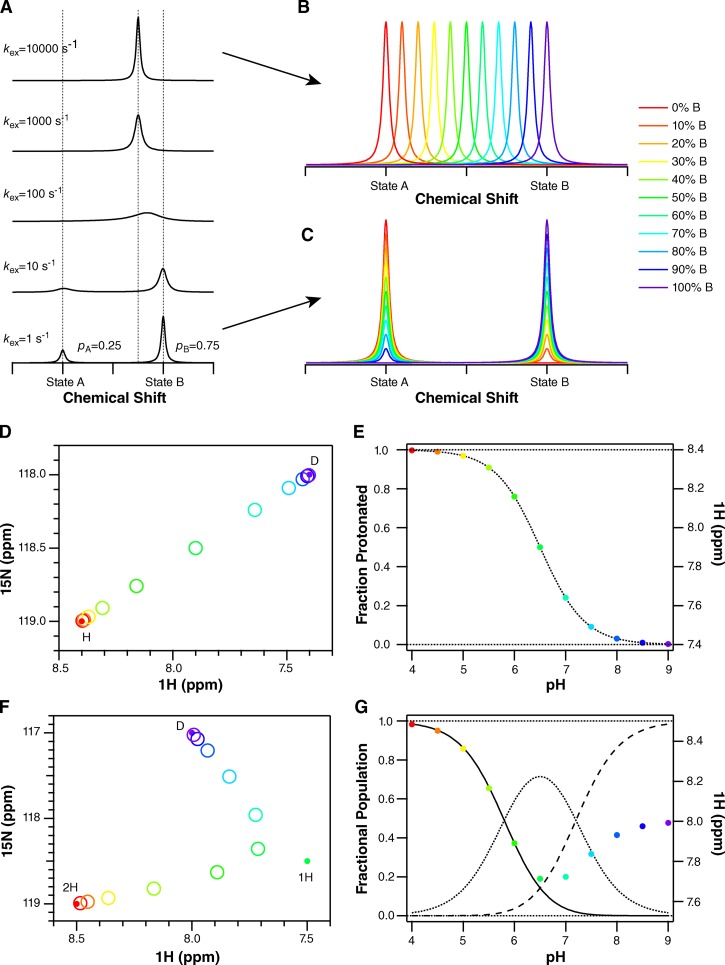
Simulation of different NMR titration patterns. (A) For a single residue, each unique conformation or state of a protein will have a distinct chemical shift. When these conformations or states are able to exchange, the pattern observed in the NMR spectrum depends on the rate of the exchange process (kex) relative to the frequency difference between the chemical shifts of the two species (Δω). In A, one-dimensional NMR spectra are simulated for Δω = 100 Hz. When kex is much slower than Δω (bottom, slow-exchange regime), two peaks are observed at the unique chemical shifts of these two conformations or states. The area under each peak reflects the relative population of each, 25%/75% in this example. As the exchange rate increases, the peaks broaden and merge, eventually resulting in a single narrow peak at the population-weighted average chemical shift of the exchanging species (top, fast-exchange regime). NMR spectra are usually reported with axes in units of parts per million (ppm) to remove their dependence on the spectrometer field strength. However, the chemical shift actually corresponds to a frequency. For the spectra presented here, acquired on a 700-MHz NMR spectrometer, 1 ppm in proton corresponds to 700 Hz. (B) In the fast-exchange regime, where kex is fast compared with Δω, titration results in a shift in peak position, from the free to bound state as ligand is added and the relative population of the free and bound states changes. (C) In the slow-exchange regime, the free state disappears and the bound state appears during the course of a titration as ligand is added and the population shifts from free to bound. Intermediate exchange will result in a combination of peak shifting and broadening. Because proton on/off is generally fast, we expect (and observe) spectra where peaks shift position with pH. (D) In the case of two-state exchange from a protonated state, marked H, to a deprotonated state, marked D, the peak position in a two-dimensional spectrum will move along a line connecting the peaks corresponding to the fully protonated and fully deprotonated states as pH is changed. (E) Plotting the position of the peak (in ppm) as a function of pH will result in a classical binding curve, reflecting the nonlinear dependence of the fraction protonated on pH. Thus, the chemical shift can be analyzed in the same way as any other protein property that is sensitive to protonation-state changes. The exact equations are given in Materials and methods. (F) If the protein is exchanging between three states (2H, two protons bound; 1H, one proton bound; D, deprotonated, no protons bound), each with unique chemical shifts, then the peak position will reflect the population-weighted average of all three chemical shifts at each pH value. An example is shown for two protonation steps assuming non-interacting sites with pKa values separated by 1.4 pH units. The averaging of three states results in a curved path of the peak across the spectrum. (G) The fraction doubly protonated (solid line), singly protonated (dotted), and deprotonated (dashed) are shown along with the peak position (in parts per million) as a function of pH. Both transitions are clearly observed in the peak position, although the chemical-shift difference between states 1H and D is smaller along the proton dimension. Eq. 2 in Materials and methods takes into account the relative chemical shifts of all three states.
Asymmetry in the pKa of the EmrE active-site residue
The pattern of peak movement across the NMR spectrum during a pH titration depends on the number of different protonation states occurring over the course of the titration. Analysis of the titration patterns of residues near Glu14 (Ala10, Gly17, and Ser43) reveals that the movement of the peaks across the NMR spectra during the pH titration is curved, particularly for Gly17 and Ser43 (Fig. 6). This is important because it demonstrates that EmrE must exist in more than two different protonation states. Proton on- and off-rates are generally fast, and thus the position of each peak in the NMR spectrum represents a population-weighted average of all protonation states present at that pH (Fig. 7 B). If there is only a single protonation event, there are only two possible states: protonated and deprotonated. In the case of EmrE, this would correspond to identical pKa values for both Glu14 residues in the asymmetric homodimer or fully cooperative binding of the two protons to the two Glu14 residues, resulting in EmrE existing only in the doubly protonated or fully deprotonated state at Glu14. In this two-state scenario, the position of a peak will move along the line between the endpoints (from 0 to 100% protonated as pH is decreased) upon pH titration (Fig. 7 D), and the position along this line will reflect the relative population of protonated and deprotonated states. Thus, although each peak will move across the NMR spectrum along a line, the relative position of the peak along that line will be a nonlinear function of pH that reflects the usual binding equations (Fig. 7 E).
Curvature in the path of the peaks across the NMR spectrum upon titration indicates that more than two states are contributing to the population-weighted average. In the case of three states, the peak position at any pH must fall within the triangle defined by 100% population in any one of these states, and the position within this triangle will reflect the population-weighted average of all three states (Fig. 7 F). These populations will again follow the usual nonlinear binding equilibrium equations (Fig. 7 G). The curvature observed in the titration patterns of Gly17, and Ser43 (Fig. 6), among others, demonstrates that more than two protonation states exist for the active-site Glu14 residues. Given the asymmetric environment of the antiparallel EmrE homodimer, it is not surprising that the two Glu14 residues have distinct pKa values.
Quantitative determination of the Glu14 pKa values in WT EmrE
To quantitatively determine the pKa value of Glu14 in EmrE, it is necessary to isolate the protonation state–dependent chemical-shift changes. Ideally, we would like to follow the residues from each subunit independently to determine the pKa of subunits A and B directly, and for peaks that remain in the slow exchange regime throughout the titration, we have taken this approach. However, the protonation-dependent changes in peak position of many residues are complicated by pH-dependent changes in conformational exchange rate, as discussed above. As the exchange rate increases at lower pH, the peaks corresponding to the two conformations broaden and move toward each other and finally merge. To avoid interpreting this exchange-induced change in peak position as protonation, we averaged both the proton chemical shifts and the nitrogen chemical shifts of the two peaks at high pH. This effectively simulates fast exchange between open-in and open-out at high pH to match the conditions at low pH and allows us to monitor the single average peak position for each residue as a function of pH. Although this discards potential subunit-specific information, residues in close proximity to Glu14 from one subunit are also likely to be in close proximity to Glu14 from the other subunit as a result of the small size of EmrE and thus may sense both protonation events anyway.
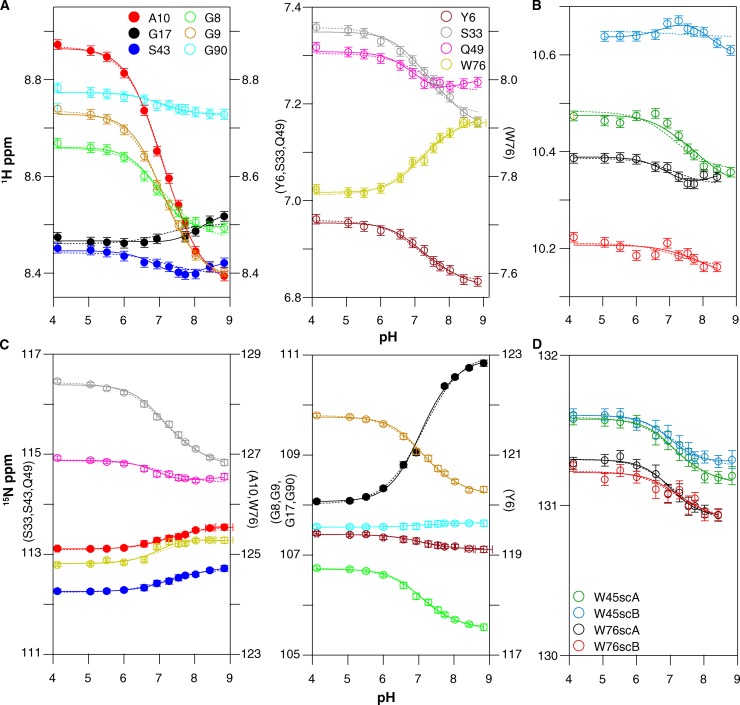
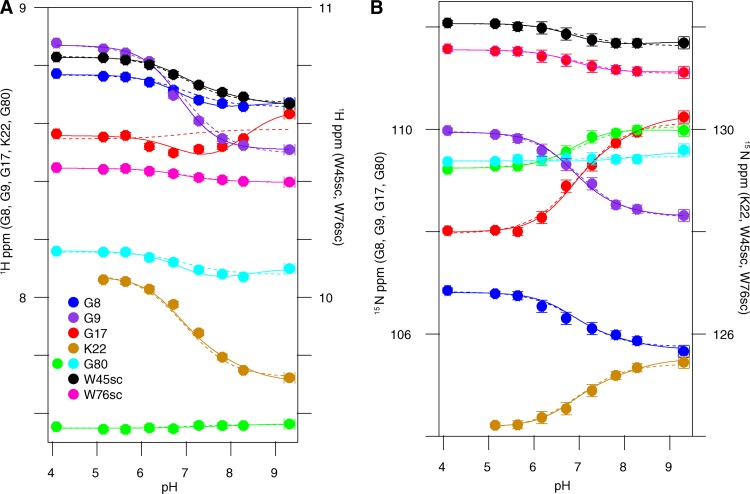
The accuracy of pKa values determined from pH-dependent chemical-shift changes of more remote nuclei, those not in the side chain of the titrating residue itself, is greatly improved if the monitored nuclei are close to the titrating residue and removed from other ionizable groups (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Soskine et al., 2004; Webb et al., 2011). Thus, to determine the pKa of Glu14 as accurately as possible, we focus on residues within the transport pore in the vicinity of Glu14 and globally fit the amide proton and nitrogen titration curves (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Yerushalmi et al., 2001; Soskine et al., 2004; Adam et al., 2007; Tomlinson et al., 2010; McIntosh et al., 2011). As shown in Fig. 2, Ala10, Gly17, and Ser43 are close to Glu14 and far removed from other ionizable residues located in the water-exposed loops of EmrE. These residues close to Glu14 will most directly sense the unique local changes in electrostatic environment occurring upon protonation/deprotonation of each Glu14 residue in the homodimer. These three residues also have well-resolved peaks in the spectra, enabling accurate analysis. Because of the curvature in the peak positions as a function of pH, fitting to a single pKa value (Eq. 1) does not yield a good fit (Fig. 8, dotted lines). However, the peak position as a function of pH for all three residues can be simultaneously fit quite well with two macroscopic pKa values (Eq. 2; Fig. 8, solid lines). This yields pKa values of 7.0 ± 0.1 and 8.2 ± 0.3 at 45°C. These pKa values are highly shifted from the standard pKa of 4.2 ± 0.1 for a water-exposed glutamate side chain (Castañeda et al., 2009; Grimsley et al., 2009; Farrell et al., 2010; Platzer et al., 2014), but well within the range of the functional pKa values reported previously for EmrE. This is consistent with the expectation that Ala10, Gly17, and Ser43 report on the protonation of Glu14.
Figure 8.
The pH-dependent chemical shifts of drug-free WT EmrE at 45°C fit to two pKa values. Proton (A and B) and nitrogen (C and D) chemical shifts were plotted independently as a function of pH for both backbone amides (A and C) and tryptophan side chains (sc) (B and D). Each y axis is labeled with the residues it represents. The residues are indicated by color as displayed in the figure. The data for residues marked with closed symbols were fit to both a single pKa model (dotted lines; pKa = 7.1 ± 0.1; n = 0.7) and a double pKa model (solid lines; pKa = 7.0 ± 0.1 and 8.2 ± 0.3). Open symbols indicate residues that were not used to determine the pKa values but can be fit to the same pKa values.
We also tested whether other residues could be fit by these same pKa values, as tight coupling between the E14 protonation state and the global conformational change of EmrE should result in all residues titrating with the same Glu14 pKa values. The pH-dependent chemical-shift changes for noncharged residues with resolved chemical shifts are also shown in Fig. 8 (open symbols). These residues are well fit with the two pKa values determined from Ala10, Gly17, and Ser43 (Fig. 8, solid lines), despite the fact that these residues are remote from Glu14. This confirms that global conformational changes are coupled to Glu14 protonation.
We repeated the pH titration of drug-free EmrE at 25°C to confirm our results, as the pH titration of Glu14 is complicated by tight coupling to global structural and dynamics changes. Changing the temperature helps separate the open-in to open-out conformational exchange processes, which are highly temperature dependent, from protonation/deprotonation of Glu14, which is much less temperature dependent (Chen et al., 2007; Nagai et al., 2008; Pace et al., 2009). At 25°C, slower overall tumbling and changes in the pH-dependent conformational exchange rates between open-in and open-out EmrE lead to increased line broadening, particularly near neutral pH (Figs. 3 B, 5 B, and 6 B). In addition, temperature-dependent chemical-shift changes cause a different subset of residues to be well resolved. As a result, Gly17 is the only residue near Glu14 for which pH-dependent chemical shifts can be measured across the pH titration. Because the 45°C data demonstrated that remote residues accurately sense the Glu14 protonation state, we globally fit the pH-dependent chemical-shift changes of all the well-resolved noncharged residues that were not severely line broadened. The only well-resolved residues that were excluded were His110 and Lys22, as these residues contain titratable side chains. Once again, the data are best fit by two pKa values of 6.8 ± 0.1 and 8.5 ± 0.2 (Fig. 9, solid lines). These are very similar to the pKa values at 45°C, confirming that Glu14 protonation is relatively insensitive to temperature, as expected for a glutamate side chain (Nagai et al., 2008; Smirnova et al., 2008).
Figure 9.
The pH-dependent chemical shifts of drug-free WT EmrE at 25°C fit to two pKa values, similar to those at 45°C. Proton (A) and nitrogen (B) chemical shifts were plotted independently as a function of pH for both backbone amides and tryptophan side chains (sc). Each y axis is labeled with the residues it represents. The residues are indicated by color as displayed in the figure. The data were globally fit to both a single pKa model (dotted lines; pKa = 6.9 ± 0.1; n = 0.8) and a double pKa model (solid lines; pKa = 6.8 ± 0.1 and 8.5 ± 0.2).
Interestingly, at both temperatures, many residues can also be fit reasonably well with a single global pKa (Figs. 8 and 9, dotted lines), although this requires a Hill coefficient significantly <1. A low Hill coefficient is consistent with multiple titrating groups, which may be electrostatically coupled (Lindman et al., 2006; Mager et al., 2011). These residues are mostly remote from Glu14 and detect Glu14 protonation through the coupled global structural change. The fact that the titration of these residues does not obviously detect the asymmetric protonation of Glu14 highlights the difficulty in detecting multiple pKa values that are only separated by ∼1 pH unit. Only residues in close proximity to Glu14, such as Gly17, Ser43, and the Trp45 side chain, sense distinct chemical-shift effects from protonation of Glu14 in each subunit. This also illustrates the improved accuracy in pKa determination provided by monitoring the chemical shifts of nuclei close enough to the site of titration to sense changes in electrostatic environment directly.
Protonation of E14D
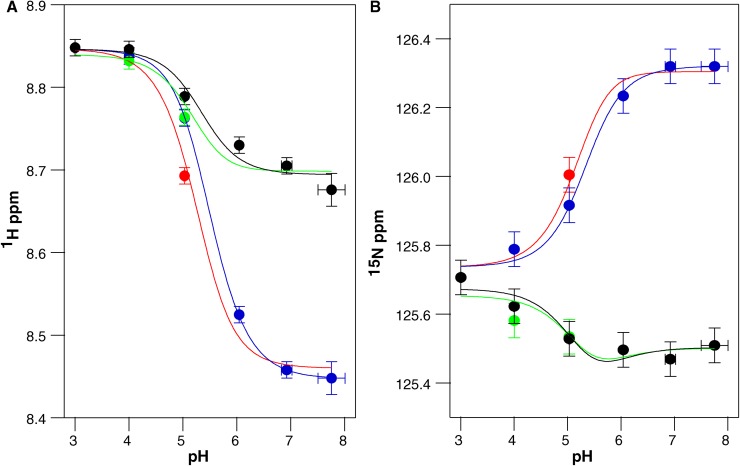
Close examination of the E14D pH titration (Figs. 4 and 5 C) reveals a significant difference in the relative timescales of the protonation and conformational exchange processes compared with WT EmrE. It is easiest to see the pattern in the Ala10 peaks. At high pH, two peaks corresponding to the two subunits in the homodimer are visible as in WT. This indicates that conformational exchange between open-in and open-out E14D-EmrE is slow at high pH (deprotonated EmrE). As the pH decreases, the peaks move along a trajectory (Fig. 5 C, solid arrows), as expected for fast proton on/off, analogous to WT EmrE. However, at pH 5, near the pKa reported from binding assays, four distinct peaks are visible (Fig. 5 C, green spectrum, pH 5) as two pairs of peaks (*) and (○), with each pair having relatively equal intensity. One peak from each pair lies along the titration path for subunit A, and one peak from each pair lies along the titration path for subunit B. This suggests that there may be two states of the EmrE dimer (* and ○) resulting in two conformations of subunit A and two conformations of subunit B. Based on the relative peak intensities, these two states of the dimer have unequal populations. Given this understanding of the unique pattern of peak movement, the chemical shifts of Ala10 as a function of pH can be fit to two pKa values of 5.2 ± 0.1 and 5.5 ± 0.1 for the major and minor peaks, respectively (Fig. 10).
Figure 10.
The pH-dependent chemical shifts of Ala10 from drug-free E14D-EmrE fit to two significantly shifted pKa values. Proton (A) and nitrogen (B) chemical shifts were plotted independently as a function of pH. Blue and green symbols represent subunit A, and black and red symbols represent subunit B. Near the pKa, each subunit splits into a major and minor peak resulting in the multiple colors to represent the two states of each subunit. The data were globally fit to a double pKa model (solid lines; pKa = 5.2 ± 0.1 and 5.5 ± 0.1). The same pKa values are obtained if the major and minor states are independently fit to single pKa values.
As the pH is lowered further, the peaks merge and then switch exchange regimes: instead of continuing to shift position with pH, the major peak at pH 4.0 disappears and a new peak grows in (Fig. 5 C, dotted arrow). This behavior represents slow exchange on the NMR timescale (Fig. 7 C), indicating that the exchange rate is much less than the chemical-shift difference between these two peaks. Because these spectra were acquired on a 700-MHz spectrometer and these peaks are separated by 0.2 ppm in the proton dimension, the exchange rate must be at least an order of magnitude slower than 140 Hz. The pH titration is reversible, indicating that the low pH state is not a result of denaturation and aggregation of the protein. It may correspond to the doubly protonated state of E14D-EmrE, but this would indicate that the proton on- and off-rates for the addition of the second proton are extremely slow. Additional experiments to further characterize this state were not performed, as it is only observed in the mutant at extremely low pH.
EmrE contains one histidine residue at the C terminus of the protein, His110. Because water-exposed histidine residues typically have pKa values near neutral pH, a potential concern was separating His110 protonation from Glu14 protonation. To avoid this, His110 was excluded from the pKa fits above (Figs. 8–10), along with all other charged residues. We can directly observe His110 throughout both the WT and E14D pH titrations. In WT EmrE (Fig. 3), His110 titrates in the same pH range as the other residues and can be fit with the global pKa values determined above. In E14D-EmrE, His110 has a titration profile that is clearly distinct from most other residues (Fig. 4). Although His110 titrates at low pH along with the rest of the protein, it also titrates in the near neutral pH range with a pKa of 7.3 ± 0.2. Examination of the E14D-EmrE titration shows that only other residues in the C-terminal tail near His110, such as Ser105, titrate in this pH range (Fig. 4). Thus, although His110 does titrate in the same pH range as Glu14, protonation of His110 occurs independently of the active site Glu/Asp, and only the C-terminal tail senses His110 protonation. This nicely illustrates how few residues sense the local effects of protonation/deprotonation.
Is apo EmrE still able to interconvert?
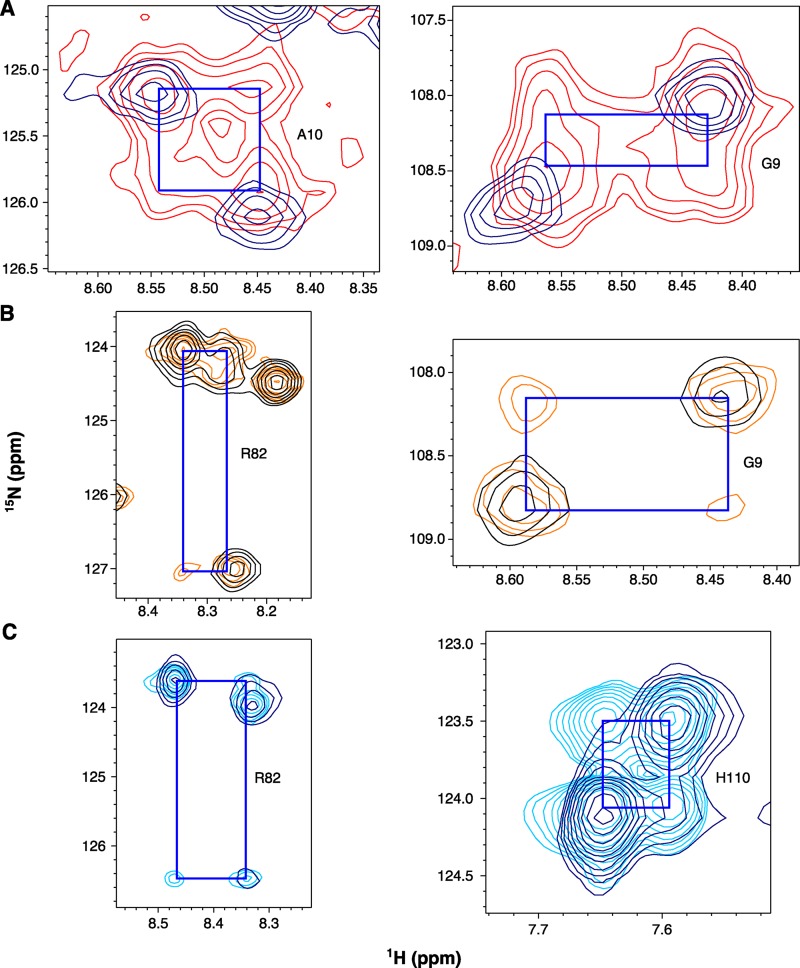
The single-site alternating access model predicts that truly apo EmrE should not interconvert between open-in and open-out. In theory, EmrE at high pH in the absence of drug should exist in a true apo state. However, based on the pKa values determined above, we estimate that at pH 8.8 (the highest pH studied), EmrE is ∼80% fully deprotonated and 20% singly protonated, with <1% doubly protonated. At this pH, two peaks are observed that correspond to subunits A and B in the asymmetric EmrE homodimer. Unexpectedly, additional cross peaks are clearly visible at this pH in the spectrum at 45°C (Figs. 3 A and 5 A). These cross peaks (Fig. 5 A, peaks marked with “x”) are visible at pH 8.0 and above and are aligned with the proton and nitrogen chemical shifts of the major peaks to form a square pattern (Fig. 11 A). This indicates conformational exchange occurring on a tens per second timescale. We have previously observed similar peak patterns in TROSY-HSQC spectra of EmrE bound to ethyltriphenylphosphonium+ (Morrison and Henzler-Wildman, 2014) as a result of interconversion between open-in and open-out forms at a rate of 25 s−1. We demonstrated that cross peaks appear in the TROSY-HSQC spectrum when rates are fast enough for significant exchange to occur during the 11-ms back-transfer portion of the NMR pulse sequence (Morrison and Henzler-Wildman, 2014). Using this 11-ms effective mixing time, we can estimate a conformational exchange rate of 50 s−1 for drug-free EmrE at pH 8.8, 45°C, using the intensities of the well-resolved Ala10 and Gly9 auto and cross peaks. Using the pKa values determined above, at pH 8.8 EmrE is ∼80% fully deprotonated and 20% monoprotonated, and there is essentially no doubly protonated EmrE. The exchange rate is faster, 75 s−1, at pH 8.4 when EmrE is 60–65% deprotonated. Collectively, the data are consistent with the idea that EmrE can exchange in more than just the doubly protonated state.
Figure 11.
Drug-free EmrE exchanges at high pH. Representative cross peaks (connected to auto peaks by blue squares) for well-resolved residues. (A) Cross peaks are visible in the 1H-15N TROSY-HSQC of WT EmrE in isotropic bicelles at pH 8.4 collected at 45°C (red) as compared with the 1H-15N TROSY-HSQC collected at 25°C (navy). (B and C) TROSY-selected ZZ-exchange experiments show cross peaks at lower temperatures. (B) Regions of a ZZ-exchange spectrum collected on WT EmrE in isotropic bicelles at pH 8.4, 35°C, with a 50-ms mixing time (orange) overlaid with a matching 1H-15N TROSY-HSQC (black). (C) Regions of a ZZ-exchange spectrum collected on WT EmrE in DMPC/DHPC isotropic bicelles at pH 8.2, 25°C, with an 80-ms mixing time (cyan) overlaid with a matching 1H-15N TROSY-HSQC (navy).
At lower temperatures, the conformational exchange rate is reduced and cross peaks are not visible in the TROSY-HSQC spectra at 25°C and high pH (Fig. 3 B). To test whether exchange still occurs under these conditions, we conducted TROSY-selected ZZ-exchange NMR experiments (Rotem and Schuldiner, 2004; Li and Palmer, 2009; Bay and Turner, 2012; Morrison et al., 2012). As a result of the difficulty in maintaining high pH for the multiple days required for ZZ-exchange NMR data collection, these experiments were performed at pH 8.2–8.4. Cross peaks were clearly visible in ZZ-exchange spectra collected at both 35°C (Fig. 11 B) and 25°C (Fig. 11 C), confirming that conformational exchange is still occurring at this pH at lower temperatures. Unfortunately, pH 8.2–8.3 is close to the upper pKa value and EmrE is therefore not in a truly apo state, with approximately equal populations of singly and deprotonated species and <5% doubly protonated.
DISCUSSION
Glu14 defines the single site that is central to the single-site alternating access model for proton-coupled drug efflux by EmrE (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Soskine et al., 2004; Nagai et al., 2008). As a result, protonation and deprotonation of Glu14 is an important step in the transport mechanism. Much effort has been put into determining the pKa of Glu14 previously, but these experimental studies have inferred the pKa of Glu14 through competition experiments between substrates and protons using pH-dependent substrate binding or functional assays (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Yerushalmi et al., 2001; Soskine et al., 2004; Adam et al., 2007). These approaches are widely used to infer pKa values for protonatable residues located within TM helices because of the functional importance of these residues and the challenges of working with membrane proteins.
NMR has become the method of choice for precise pKa determination of ionizable residues in soluble proteins because of its unique ability to directly monitor the protonation and deprotonation of a specific amino acid side chain in the absence of any other process. The NMR titration curves for residues that are close to Glu14 in the hydrophobic substrate-binding region of EmrE unambiguously show two distinct macroscopic pKa values for Glu14 in WT EmrE, reflecting distinct electrostatic effects of two protonation events within the hydrophobic substrate-binding pocket of EmrE. A critical question when considering the mechanism of proton-coupled transport by EmrE is the microscopic origin of the two distinct Glu14 pKa values. Do these two pKa values represent protonation of subunits A and B, reflecting the unique structural environments of each subunit within the asymmetric homodimer? Or do they arise from electrostatic interaction between the two glutamate residues, such that deprotonation of one Glu14 side chain disfavors deprotonation of the second Glu14, causing a further increase in the pKa value for the second deprotonation event? It is likely that both microscopic processes contribute to the observed asymmetry in the pKa values, and it will be difficult to experimentally disentangle the microscopic processes in this system. Nevertheless, the separation of the two macroscopic pKa values in WT EmrE by more than one pH unit suggests that deprotonation is essentially a sequential process regardless of the microscopic origin of the Glu14 pKa asymmetry.
The two macroscopic pKa values determined for WT EmrE are both highly shifted from the typical pKa of the glutamic acid side chain in water and are among the highest values measured (Oxenoid et al., 2004; Chill et al., 2006; Grimsley et al., 2009; Farrell et al., 2010; Platzer et al., 2014). This is not surprising given the hydrophobic environment of Glu14 (Wüthrich and Wagner, 1979; Chen et al., 2007; Pace et al., 2009) and is consistent with the role of Glu14 in coupling drug efflux to proton import at physiological pH values. In fact, pKa values estimated from substrate competition and functional assays of other proton-coupled transporters are similarly shifted. For example, the apparent pKa for LacY sugar binding is ≈10.5 (Smirnova et al., 2008), and NhaA is estimated to have a pKa of ≈8.8 (Mager et al., 2011).
The chemical shifts of residues that are more removed from the substrate-binding site can be fit well with the same two pKa values determined from the residues close to Glu14. However, unlike the residues in the substrate-binding pocket, the chemical shifts of remote residues move in a more linear manner. These residues are influenced primarily by protonation state–dependent conformational changes rather than direct electrostatic effects of protonation. Their smooth titration profiles suggest a steady progression of conformational change with pH, perhaps an increased opening of the substrate-binding pocket to one side of the membrane as deprotonation occurs and Glu14 becomes charged. Measurement of structural restraints will be needed to test this hypothesis and characterize the fully protonated and fully deprotonated structural states of EmrE.
Our results demonstrate that EmrE exists in a complex mixture of protonation states. Furthermore, quantitative interpretation of the data is complicated by protonation state–dependent conformational exchange. This behavior is predicted by the single-site alternating access model and confirmed by the pH-dependent changes in linewidth and number of peaks per residue as a function of pH. Quantitatively separating these structural and dynamic features is a challenge because quantitative analysis with NMR dynamics methods requires a limited number of states: two- or perhaps three-state exchange. With three protonation states and two subunit conformations, at least four to six states must be considered for EmrE near neutral pH. The pKa values of Glu14 and instability of lipids at extreme pH values make it difficult to drive the system into a single protonation state and reduce the problem to two-state conformational exchange.
However, qualitative insight into the different states and conformational exchange rates of EmrE can be obtained from the current data. The limited temperature dependence of the Glu14 pKa between 25 and 45°C is typical for glutamate side-chain protonation (Nagai et al., 2008). This is in striking contrast to the rate of conformational exchange between open-in and open-out EmrE, which is highly temperature dependent. This difference allows one to differentiate between the two processes by comparing spectra collected at different temperatures.
High pH is of particular interest because of the prediction that conformational exchange between open-in and open-out forms should be blocked in truly apo EmrE (no bound proton or drug substrates; Fig. 1 A). At the highest pH values we were able to maintain during the NMR experiments, we estimate that EmrE was 80% fully deprotonated with <1% doubly protonated, and conformational exchange was still occurring at a rate of ≈50 s−1. This rate at pH 8.8 is reduced compared with the estimated 75-s−1 conformational exchange rate at pH 8.4, but it is still significantly faster than the open-in to open-out conformational exchange rate of EmrE bound to most polyaromatic cation substrates (Morrison and Henzler-Wildman, 2014). This is consistent with the drug-bound conformational exchange rate or drug off-rate as the rate-limiting step in the transport cycle, as proposed previously (Morrison and Henzler-Wildman, 2014). In this context, whether truly apo EmrE does or does not interconvert between open-in and open-out structures becomes an academic question, as EmrE will virtually never exist in a fully deprotonated state at physiological pH. However, the existence of conformational exchange between open-in and open-out forms in multiple protonation states raises an interesting question: Can EmrE leak protons (Fig. 1 B)? If so, it may provide an alternative explanation for the enhanced sensitivity of E. coli overexpressing EmrE to high pH conditions (Rotem and Schuldiner, 2004; Bay and Turner, 2012).
The differences we observe in the pH titrations of E14D and WT EmrE also provide some insight into their previously reported functional differences. The Asp14 and Glu14 pKa values fall in the broad ranges previously estimated from functional and substrate competition experiments (Muth and Schuldiner, 2000; Yerushalmi and Schuldiner, 2000b; Yerushalmi et al., 2001; Soskine et al., 2004; Adam et al., 2007). However, the Asp14 pKa values are much less shifted from a typical aspartate side-chain pKa than the Glu14 pKa values are from a typical glutamate side-chain pKa. Widespread chemical-shift differences between WT and E14D suggest that the overall structure is also different in the mutant. The most striking difference is the behavior of E14D-EmrE at low pH. The slow protonation/deprotonation suggests that fully protonated E14D-EmrE is in a conformation where protons are not readily accessible to the active-site residue. This could be a result of local structural changes or a more global occluded state that excludes water from the vicinity of E14D. Limited water accessibility to the core of TM helices is not entirely unexpected. Hydrogen/deuterium exchange of backbone amides is generally much slower in a membrane environment (Oxenoid et al., 2004; Chill et al., 2006). In fact, it is often difficult to acquire 1H/15N TROSY spectra of soluble proteins at pH values much above neutral because of increased H/D exchange of backbone amides at high pH (Wüthrich and Wagner, 1979). The spectra presented here all have excellent sensitivity, even at pH 8.8, and were acquired without increased signal averaging, suggesting limited water accessibility. Strikingly, the high pH spectra are some of the highest quality spectra, with good chemical-shift dispersion and signal-to-noise ratio, presumably because of reduced dynamics as EmrE populates the less dynamic fully deprotonated state.
A broad range of pKa values have been reported previously for Glu14 of EmrE based on pH-dependent substrate binding and functional assays. Our detection of two distinct macroscopic pKa values for EmrE Glu14, which span the previously reported range, helps explain the inconsistent values obtained in previous studies. The site-specific measurement of protonation states reveals two distinct protonation events that likely occur sequentially. The fact that these protonation events are coupled to both global structural changes as well as the open-in/open-out exchange process implies that the pKa values are central to the coupling mechanism that drives EmrE transport.
Supplementary Material
Acknowledgments
We thank Bryan Balthazor for preliminary studies of EmrE protonation states and Geoff Chang for the EmrE expression plasmid.
This work was supported by National Institutes of Health (grant 1R01GM095839) and National Science Foundation graduate research fellowships to E.A. Morrison and A.E. Robinson (DGE-1143954).
The authors declare no competing financial interests.
Merritt C. Maduke served as editor.
Footnotes
Abbreviations used in this paper:
- DHPC
- 1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DLPC
- 1,2-dilauroyl-sn-glycero-3-phosphocholine
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- Glu14
- glutamate 14
- HSQC
- heteronuclear single quantum coherence
- NMR
- nuclear magnetic resonance
- TM
- transmembrane
- TPP+
- tetraphenylphosphonium
- TROSY
- transverse relaxation optimized spectroscopy
References
- Adam Y., Tayer N., Rotem D., Schreiber G., and Schuldiner S.. 2007. The fast release of sticky protons: Kinetics of substrate binding and proton release in a multidrug transporter. Proc. Natl. Acad. Sci. USA. 104:17989–17994. 10.1073/pnas.0704425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay D.C.D., and Turner R.J.R.. 2012. Small multidrug resistance protein EmrE reduces host pH and osmotic tolerance to metabolic quaternary cation osmoprotectants. J. Bacteriol. 194:5941–5948. 10.1128/JB.00666-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda C.A., Fitch C.A., Majumdar A., Khangulov V., Schlessman J.L., and García-Moreno B.E.. 2009. Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins. 77:570–588. 10.1002/prot.22470 [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Pornillos O., Lieu S., Ma C., Chen A.P., and Chang G.. 2007. X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. USA. 104:18999–19004. 10.1073/pnas.0709387104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chill J.H., Louis J.M., Miller C., and Bax A.. 2006. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 15:684–698. 10.1110/ps.051954706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., and Bax A.. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- Farrell D., Miranda E.S., Webb H., Georgi N., Crowley P.B., McIntosh L.P., and Nielsen J.E.. 2010. Titration_DB: Storage and analysis of NMR-monitored protein pH titration curves. Proteins. 78:843–857. 10.1002/prot.22611 [DOI] [PubMed] [Google Scholar]
- Grimsley G.R., Scholtz J.M., and Pace C.N.. 2009. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 18:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Schmidt-Rohr K., and Hong M.. 2012. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J. Am. Chem. Soc. 134:3703–3713. 10.1021/ja2081185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Fu R., Nishimura K., Zhang L., Zhou H.X., Busath D.D., Vijayvergiya V., and Cross T.A.. 2006. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. USA. 103:6865–6870. 10.1073/pnas.0601944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner I., Basting D., Meyer B., Haase W., Manolikas T., Kaiser C., Karas M., and Glaubitz C.. 2008. The key residue for substrate transport (Glu14) in the EmrE dimer is asymmetric. J. Biol. Chem. 283:3281–3288. 10.1074/jbc.M707899200 [DOI] [PubMed] [Google Scholar]
- Li Y., and Palmer A.G. III. 2009. TROSY-selected ZZ-exchange experiment for characterizing slow chemical exchange in large proteins. J. Biomol. NMR. 45:357–360. 10.1007/s10858-009-9385-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindman S., Linse S., Mulder F.A.A.F., and André I.. 2006. Electrostatic contributions to residue-specific protonation equilibria and proton binding capacitance for a small protein. Biochemistry. 45:13993–14002. 10.1021/bi061555v [DOI] [PubMed] [Google Scholar]
- Mager T., Rimon A., Padan E., and Fendler K.. 2011. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: An electrophysiological study. J. Biol. Chem. 286:23570–23581. 10.1074/jbc.M111.230235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh L.P.L., Naito D., Baturin S.J.S., Okon M., Joshi M.D.M., and Nielsen J.E.J.. 2011. Dissecting electrostatic interactions in Bacillus circulans xylanase through NMR-monitored pH titrations. J. Biomol. NMR. 51:5–19. 10.1007/s10858-011-9537-x [DOI] [PubMed] [Google Scholar]
- Morrison E.A., and Henzler-Wildman K.A.. 2012. Reconstitution of integral membrane proteins into isotropic bicelles with improved sample stability and expanded lipid composition profile. Biochim. Biophys. Acta. 1818:814–820. 10.1016/j.bbamem.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E.A., and Henzler-Wildman K.A.. 2014. Transported substrate determines exchange rate in the multidrug resistance transporter EmrE. J. Biol. Chem. 289:6825–6836. 10.1074/jbc.M113.535328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E.A., DeKoster G.T., Dutta S., Vafabakhsh R., Clarkson M.W., Bahl A., Kern D., Ha T., and Henzler-Wildman K.A.. 2012. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 481:45–50. 10.1038/nature10703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth T.R., and Schuldiner S.. 2000. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 19:234–240. 10.1093/emboj/19.2.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H., Kuwabara K., and Carta G.. 2008. Temperature dependence of the dissociation constants of several amino acids. J. Chem. Eng. Data. 53:619–627. 10.1021/je700067a [DOI] [Google Scholar]
- Nielsen J.E. 2008. Analyzing protein NMR pH-titration curves. Annual Reports in Computational Chemistry. Wheeler R.A. and Spellmeyer D.C., Elsevier, Amsterdam: 89–106. [Google Scholar]
- Oxenoid K., Kim H.J., Jacob J., Sönnichsen F.D., and Sanders C.R.. 2004. NMR assignments for a helical 40 kDa membrane protein. J. Am. Chem. Soc. 126:5048–5049. 10.1021/ja049916m [DOI] [PubMed] [Google Scholar]
- Pace C.N., Grimsley G.R., and Scholtz J.M.. 2009. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J. Biol. Chem. 284:13285–13289. 10.1074/jbc.R800080200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellecchia M., Iwai H., Szyperski T., and Wüthrich K.. 1997. The 2D NMR experiments H(C)CO2 and HCCO2 for assignment and pH titration of carboxylate groups in uniformly 15N/13C-labeled proteins. J. Magn. Reson. 124:274–278. 10.1006/jmre.1996.1058 [DOI] [PubMed] [Google Scholar]
- Platzer G., Okon M., and McIntosh L.P.. 2014. pH-dependent random coil 1H, 13C, and 15N chemical shifts of the ionizable amino acids: a guide for protein pKa measurements. J. Biomol. NMR. 60:109–129. 10.1007/s10858-014-9862-y [DOI] [PubMed] [Google Scholar]
- Reijenga J., van Hoof A., van Loon A., and Teunissen B.. 2013. Development of methods for the determination of pKa values. Anal. Chem. Insights. 8:53–71. 10.4137/ACI.S12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem D., and Schuldiner S.. 2004. EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J. Biol. Chem. 279:48787–48793. 10.1074/jbc.M408187200 [DOI] [PubMed] [Google Scholar]
- Shrager R.I., Cohen J.S., Heller S.R., Sachs D.H., and Schechter A.N.. 1972. Nuclear magnetic resonance titration curves of histidine ring protons. II. Biochemistry. 11:541–547. 10.1021/bi00754a010 [DOI] [PubMed] [Google Scholar]
- Smirnova I.N., Kasho V., and Kaback H.R.. 2008. Protonation and sugar binding to LacY. Proc. Natl. Acad. Sci. USA. 105:8896–8901. 10.1073/pnas.0803577105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.O., Smith C.S., and Bormann B.J.. 1996. Strong hydrogen bonding interactions involving a buried glutamic acid in the transmembrane sequence of the neu/erbB-2 receptor. Nat. Struct. Biol. 3:252–258. 10.1038/nsb0396-252 [DOI] [PubMed] [Google Scholar]
- Soskine M., Adam Y., and Schuldiner S.. 2004. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J. Biol. Chem. 279:9951–9955. 10.1074/jbc.M312853200 [DOI] [PubMed] [Google Scholar]
- Tomlinson J.H., Green V.L., Baker P.J., and Williamson M.P.. 2010. Structural origins of pH-dependent chemical shifts in the B1 domain of protein G. Proteins. 78:3000–3016. 10.1002/prot.22825 [DOI] [PubMed] [Google Scholar]
- Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinás M., Ulrich E.L., Markley J.L., Ionides J., and Laue E.D.. 2005. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 59:687–696. 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Webb H., Tynan-Connolly B.M., Lee G.M., Farrell D., O’Meara F., Søndergaard C.R., Teilum K., Hewage C., McIntosh L.P., and Nielsen J.E.. 2011. Remeasuring HEWL pKa values by NMR spectroscopy: Methods, analysis, accuracy, and implications for theoretical pKa calculations. Proteins. 79:685–702. 10.1002/prot.22886 [DOI] [PubMed] [Google Scholar]
- Weinglass A.B., Soskine M., Vazquez-Ibar J.-L., Whitelegge J.P., Faull K.F., Kaback H.R., and Schuldiner S.. 2005. Exploring the role of a unique carboxyl residue in EmrE by mass spectrometry. J. Biol. Chem. 280:7487–7492. 10.1074/jbc.M413555200 [DOI] [PubMed] [Google Scholar]
- Wüthrich K., and Wagner G.. 1979. Nuclear magnetic resonance of labile protons in the basic pancreatic trypsin inhibitor. J. Mol. Biol. 130:1–18. 10.1016/0022-2836(79)90548-5 [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., and Schuldiner S.. 2000a. A model for coupling of H(+) and substrate fluxes based on “time-sharing” of a common binding site. Biochemistry. 39:14711–14719. 10.1021/bi001892i [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., and Schuldiner S.. 2000b. An essential glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 275:5264–5269. 10.1074/jbc.275.8.5264 [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., Mordoch S.S., and Schuldiner S.. 2001. A single carboxyl mutant of the multidrug transporter EmrE is fully functional. J. Biol. Chem. 276:12744–12748. 10.1074/jbc.M010979200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.