Abstract
The wall proteome and the secretome of the fungal pathogen Candida albicans help it to thrive in multiple niches of the human body. Mass spectrometry has allowed researchers to study the dynamics of both subproteomes. Here, we discuss some major responses of the secretome to host-related environmental conditions. Three β-1,3-glucan-modifying enzymes, Mp65, Sun41, and Tos1, are consistently found in large amounts in culture supernatants, suggesting that they are needed for construction and expansion of the cell wall β-1,3-glucan layer and thus correlate with growth and might serve as diagnostic biomarkers. The genes ENG1, CHT3, and SCW11, which encode an endoglucanase, the major chitinase, and a β-1,3-glucan-modifying enzyme, respectively, are periodically expressed and peak in M/G1. The corresponding protein abundances in the medium correlate with the degree of cell separation during single-yeast-cell, pseudohyphal, and hyphal growth. We also discuss the observation that cells treated with fluconazole, or other agents causing cell surface stress, form pseudohyphal aggregates. Fluconazole-treated cells secrete abundant amounts of the transglucosylase Phr1, which is involved in the accumulation of β-1,3-glucan in biofilms, raising the question whether this is a general response to cell surface stress. Other abundant secretome proteins also contribute to biofilm formation, emphasizing the important role of secretome proteins in this mode of growth. Finally, we discuss the relevance of these observations to therapeutic intervention. Together, these data illustrate that C. albicans actively adapts its secretome to environmental conditions, thus promoting its survival in widely divergent niches of the human body.
INTRODUCTION
The fungal pathogen Candida albicans is a highly specialized inhabitant of warm-blooded animals (mammals and birds). It preferentially colonizes mucosal surfaces and the skin but can also invade deeper-lying tissues and cause systemic infections that are difficult to treat and frequently lethal (1). To survive under the challenging and divergent conditions associated with the various mucosal surfaces in the human body, C. albicans disposes of a wide arsenal of virulence traits that help it to cope with antimicrobial peptides, the complement system, engulfment by macrophages, antibodies, hypoxic conditions, iron restriction, etc. A fascinating trait is its ability to switch reversibly between various growth forms, including among others the single-cell yeast form, which is especially suitable for dispersion of the fungus; the hyphal form, which facilitates adhesion to host tissues and promotes invasive growth and escape from engulfment by immune cells; and an intermediate, pseudohyphal growth form. C. albicans also forms biofilms (surface-associated microbial communities), which clinically speaking represent a highly relevant mode of growth and in which yeast, pseudohyphal, and hyphal cells cooccur and become encapsulated by substantial amounts of extracellular, macromolecular material. Biofilm formation on abiotic surfaces of medical devices and prostheses and on teeth has therefore been extensively studied (2, 3). The first contacts between C. albicans and host cells occur predominantly at the cell surface, and this presumably explains why the external protein coat of C. albicans cell walls consists of a wide variety of glycoproteins with specialized functions, many of which are under tight control, thus promoting survival under diverse stress conditions (4, 5). Equally important, C. albicans secretes a variety of glycoproteins that help to forage for nutrients by degrading host proteins, lipids, and glycogen, while others acquire iron and zinc ions and provide protection against antimicrobial peptides. Other glycoproteins help to form and strengthen biofilms and to accumulate extracellular matrix material. Together, we designate these secreted proteins as the secretome sensu stricto (see below). The introduction of mass spectrometry in protein research has made it possible to study the protein assortment of entire cells or tissues and also well-defined subsets of proteins (subproteomes such as the cell wall proteome and the secretome), not only qualitatively but also quantitatively. This review discusses recent mass spectrometric explorations of the dynamics of the secretome of C. albicans depending, for example, on growth form and pH or in response to cell surface stress. For complementary reviews, the reader is referred to reference 4, which includes an extensive section about secretome proteins, and to a more recent review (6). In addition, the Candida albicans PeptideAtlas and the Candida Genome Database are recommended for detailed information about mass spectrometrically identified peptides (7, 8).
CLASSIFICATION OF MAJOR SECRETOME PROTEINS (SENSU STRICTO) OF C. ALBICANS
In this review, we will restrict ourselves to the secretome in the narrow sense of the word (secretome sensu stricto), that is, we will discuss only those proteins that possess an N-terminal signal peptide for entering the classical secretory pathway and lack internal transmembrane sequences. The major advantages of analyzing this protein category separately are that it is physiologically well defined, is limited in size, and is not affected by accidentally released proteins. This facilitates statistical analysis of the data (9), simplifies discussion of the results, and leads to physiologically relevant conclusions and testable hypotheses. For more information about the secretome in the wide sense of the word (secretome sensu lato), which includes a number of known cytosolic proteins, we refer to references 6 and 10.
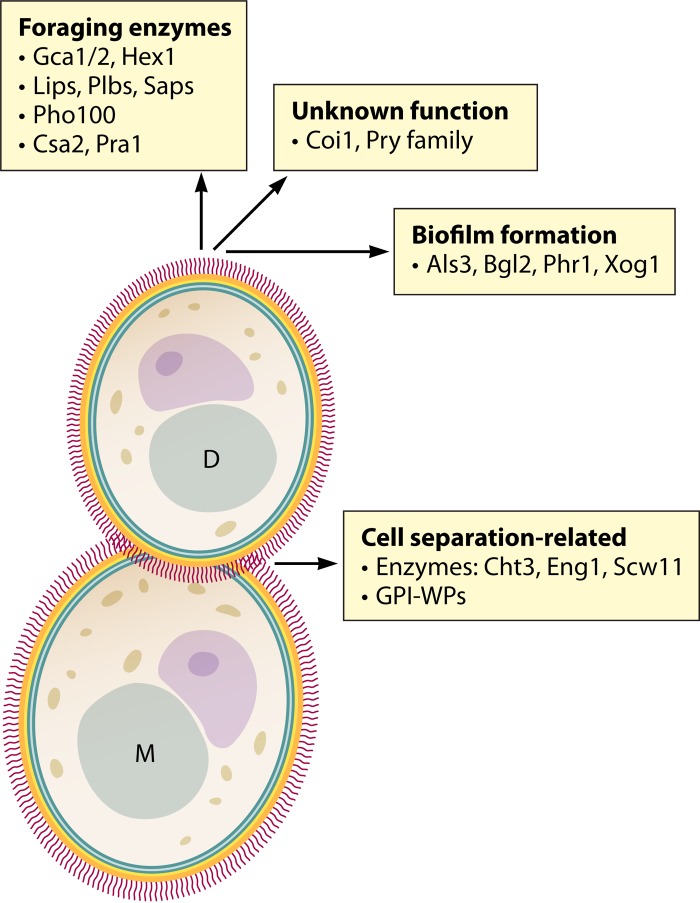
The experimentally identified secretome proteins (sensu stricto) (currently, about 70 [9–15]) can be classified into two major groups: nonglycosylphosphatidylinositol (non-GPI) proteins, which lack a C-terminal signal sequence for the addition of a GPI anchor in the endoplasmic reticulum, and GPI proteins (Fig. 1). To avoid an excessively long list, this review focuses on the more abundant secretome proteins that are observed under more than one growth condition and/or have a known or predicted function.
FIG 1.
Major features of the secretome (sensu stricto) of C. albicans. The wall proteins are represented as short line segments perpendicular to the cell surface. M, mother cell; D, daughter cell; GPI-WP, GPI-modified wall protein. The role of Csa2 in heme binding is speculative (6, 40, 41). Note that Als3 and Phr1 are possibly directly released from the cell wall by Sap9/10 activity (47).
NON-GPI PROTEINS
Modification of cell wall polysaccharides.
Eleven enzymes (distributed over six families of glycosylhydrolases [GHs]) are involved in glycan chain elongation and branching and in glycan degradation (for example, during cell separation and emergence of a new bud or hyphal branch). Throughout this paper, we will follow the GH family classification according to the CAZy (for carbohydrate-active enzymes) database (16).
(i) β-1,3-Glucan-modifying enzymes.
β-1,3-Glucan-modifying enzymes include Xog1 (GH5); Tos1 (GH16); Bgl2, Mp65, and Scw11 (GH17); Eng1 (GH81); Sim1 and Sun41 (GH132); and Dag7 (Barwin-like endoglucanase domain, PF03330) and GH (unspecified) (8).
(ii) Chitin-degrading enzymes.
Chitin-degrading enzymes include Cht1 and Cht3 (GH18); they carry out degradation of the primary septum between mother and daughter cells, thus initiating cell separation. Note that the GH18 family member Cht2 is a GPI protein.
Three of the 11 corresponding genes (CHT3, which encodes the major chitinase; ENG1, which encodes an endo-β-1,3-glucanase; and SCW11, which encodes a β-1,3-glucan-modifying enzyme) are periodic genes with maximal expression in the M/G1 period of the cell cycle and are target genes of the transcription factor Ace2 (17–20), consistent with a role in cell separation. Agar-grown colonies, mucosal biofilms, and biofilms formed on abiotic surfaces produce abundant amounts of extracellular matrix material (3, 21–23). The transglucosylase Bgl2 and the exoglucanase Xog1 (together with the GPI-modified, putative β-1,3-glucan-elongating enzyme Phr1) seem to be directly involved in formation and modification of extracellular matrix material in biofilms (24).
Nutrient acquisition. (i) Glucoamylases.
Glucoamylases include Gca1 and Gca2 (GH31); the predicted substrates are glycogen and starch (4, 16, 25, 26). Interestingly, maltose, which is a degradation product of both polysaccharides, is a carbon source known to promote hyphal growth (27). It has been suggested previously that Gca1 and Gca2 are directly involved in promoting matrix production in biofilms by enzymatic release of soluble β-1,3-glucan fragments from insoluble β-1,3-glucan chains (28). However, Gca1 and Gca2 possess the hallmarks of an α-glucosidase/glucoamylase, cleaving alpha-1,4-glucosidic linkages (8), and hence, it seems unlikely that they might cleave beta-1,3-glucosidic linkages, which have a spatial structure highly different from that of alpha-1,4-glucosidic linkages.
(ii) N-Acetylhexosaminidase.
HEX1 is a glucose-repressed gene (29). Hex1 (GH20) is found in the periplasm and in the culture supernatant (29). It is possibly involved in the release of GlcNAc residues from host tissues for use as a carbon or nitrogen source (29). Note that GlcNAc also acts as a signaling molecule and induces and maintains hyphal growth in glucose-derepressed cells (30–33).
(iii) Lipid degradation by Lips and Plbs.
Lipids are degraded by lipases (Lips) including Lip1 to Lip6 and Lip8 to Lip10 and by phospholipases (Plbs) including Plb1 and Plb2, reviewed in references 4 and 34.
(iv) Protein degradation by aspartyl proteases.
Protein degradation by aspartyl proteases includes, for example, degradation of mucins and host immune proteins (4, 34–37), by aspartyl proteases such as Sap1 to Sap8. Note that Sap9 and Sap10 are cell surface-associated GPI proteins (38).
(v) Metal ion acquisition.
Metal ion acquisition includes acquisition of zinc by Pra1 (39) and, probably, acquisition of heme (iron) by Csa2 (6, 12, 40, 41). Intriguingly, Pra1 is also involved in immune evasion (42, 43).
Pry family.
The Pry family (11) consists of five members: Pry1, Rbe1, Rbt4, and two uncharacterized open reading frames (ORFs) (19.6200 and 19.2336). Rbe1 and Rbt4 have been often identified in culture supernatants (10–14). Interestingly, whereas Rbe1 is much more abundant in yeast culture supernatants, Rbt4 is much more abundant in hyphal culture supernatants (11–13) (see also Table 2). The virulence of an Δrbe1 Δrbt4 double deletion strain in the mouse model for systemic infection is strongly diminished (11), but the precise function of the Pry family proteins is still unknown.
TABLE 2.
Yeast state- and hyphal state-enriched secretome proteins
| Change and protein | Apparent abundance (%)a |
Fold change | |
|---|---|---|---|
| Yeast | Hyphal | ||
| Yeast to hyphal change | |||
| Cht3b | 8.6 | 1.9 | 4.5 |
| Scw11b | 8.6 | 1.9 | 4.5 |
| Xog1 | 8.4 | 1.2 | 7.1 |
| Sim1 | 7.8 | 2.6 | 3.0 |
| Eng1b | 7.3 | NDd | >28 |
| Bgl2 | 3.6 | 0.5 | 7.5 |
| Rbe1 | 3.0 | ND | >12 |
| Cht1 | 2.5 | ND | >9 |
| Dag7 | 2.3 | ND | >9 |
| Hyphal to yeast change | |||
| Sap6 | ND | 13.1 | >62 |
| Rbt4 | 1.8 | 9.7 | 5.7 |
| Als3c | ND | 9.3 | >44 |
| Sap4 | ND | 8.8 | >42 |
Coi1.
Coi1 is a small protein of 191 amino acids that is relatively abundant and is consistently found in the medium under all growth conditions tested (10–14). Homologous proteins are found in only a limited number of Candida spp.—Candida dubliniensis, Candida orthopsilosis, Candida parapsilosis, and Candida tropicalis—and in Lodderomyces elongisporus (8). Its function is unknown.
Signaling protein Msb2.
The signaling protein Msb2 is located in the plasma membrane and involved in signaling through activation of the Cek1 mitogen-activated protein (MAP) kinase (44). Although Msb2 has an internal transmembrane sequence, it is included in this classification and in the secretome sensu stricto, because it has a large extracellular, highly O-glycosylated domain that is shed into the medium and consistently identified in culture supernatants (10–14, 45). Interestingly, it also serves as a broad-range protectant against antimicrobial peptides (45, 46).
GPI PROTEINS
GPI proteins are targeted to the plasma membrane or become covalently linked to the β-glucan layer of the cell wall through their GPI anchor or are found at both locations. However, most GPI proteins are also identified in culture supernatants (10–14). There are several (possible) explanations for their presence.
Target proteins of Sap9 and Sap10.
Some wall-bound GPI proteins are released by the surface-bound aspartyl proteases Sap9 and Sap10, such as Cht2 (47). Other candidates for active and controlled release, such as the adhesion protein Als3 and the transglucosylase Phr1, will be discussed below.
GPI proteins from the neck region.
In single-cell yeast cultures, wall-bound GPI proteins are released from the neck region during cell separation. The wall between mother cell and growing bud is continuous, and complete cell separation therefore requires not only degradation of the primary septum by chitinase activity but also degradation of the lateral wall in the neck region.
Accidental release.
Especially in shaken cultures, wall-bound GPI proteins might be released during emergence of a new bud or hyphal branch, which requires localized cell wall softening, or during periods of isotropic growth, which requires insertion of new cell wall polysaccharides and wall proteins into the existing wall.
Wall protein precursors may be washed out into the medium before they become covalently linked to the glucan-chitin network, especially in shaken cultures.
DYNAMICS OF THE SECRETOME OF C. ALBICANS
In this section, the term “apparent abundance” is introduced. It is defined as the number of spectral counts per protein divided by the total number of spectral counts of all secretome proteins (sensu stricto) and expressed as a percentage (48). This is a semiquantitative measure that allows comparison of the individual contributions of the secretome proteins and, importantly, allows estimating and comparing the fold changes of individual secretome proteins upon changes in environmental conditions, including conditions that induce growth as single yeast, pseudohyphal, or hyphal cells. We prefer the use of apparent abundances to that of normalized spectral abundance factors (NSAFs [9, 10, 49]). In the latter approach, the number of spectral counts per protein is normalized for protein length, which for nonglycosylated proteins results in more accurate estimates of protein abundance. However, secretome proteins sensu stricto frequently contain long, heavily O-glycosylated sequences, which rarely result in detectable tryptic peptides and thus lead to serious underestimations of protein abundances (for example, about 10-fold in the case of Msb2 [7, 8]) and decreased accuracy. A rough estimate of the number of secretome proteins per cell present in yeast culture supernatants, based on the data in references 12 and 50 and assuming an average protein mass of 40 kDa, is about 4 × 105 to 5 × 105.
Three prominent secretome proteins.
The three β-glucan-modifying enzymes Mp65 (GH17), Sun41 (GH132), and Tos1 (GH16) belong to the most prominent (detectable) secretome proteins, both in single-cell yeast culture supernatants and hyphal culture supernatants and during pseudohypha-like growth induced by fluconazole (Table 1). Similar values for their apparent abundances have been obtained under diverse growth conditions (6, 9, 10, 12–14). Conceivably, they are involved in various ways in the construction and remodeling of the β-1,3-glucan layer in the cell wall during growth (8, 16, 51–55).Their combined apparent abundance accounts for one-fourth to one-third of all secretome proteins. Consistent with this, the gene sequences of Mp65, Sun41, and Tos1 have a relatively high codon bias index (MP65, 0.71; SUN41, 0.64; TOS1, 0.59 [8]), suggesting that these genes are strongly expressed. Surprisingly, although Mp65 lacks a C-terminal GPI anchor to connect it covalently to the β-glucan network, it is usually also found in (hot-SDS-extracted) cell walls (10, 13, 14, 30, 37, 40). Scw11 and Sim1 belong to the same families as Mp65 and Sun41, respectively, and both show high apparent abundances in single-cell yeast cultures (Tables 2 to 4). However, in hyphal cultures and to a lesser extent also in pseudohyphal-growth cultures, their apparent abundances are considerably lower, suggesting that Scw11 and Sim1 might play a direct role in cell separation.
TABLE 1.
Three prominent secretome (sensu stricto) proteins of C. albicans grown under various conditions
TABLE 4.
Main features of secretomes of low-pH-grown cultures
| Change and protein | Apparent abundance (%) at pHa: |
Fold change | |
|---|---|---|---|
| 4 | 7.4 | ||
| pH 4 to pH 7.4 change | |||
| Utr2b | 8.0 | 3.2 | 2.5 |
| Bgl2 | 7.4 | 2.8 | 2.6 |
| Plb4.5b | 6.0 | ND | >32 |
| Pir1 | 3.6 | 1.1 | 3.2 |
Data based on reference 14. ND, not detected.
GPI proteins are in bold.
Yeast state-enriched secretome proteins: yeast versus hyphal cultures.
In single-cell yeast cultures, the daughter cells become separated from the mother cell, a process that requires chitinase activity to degrade the primary septum formed during cytokinesis and β-1,3-endoglucanase activity to degrade the β-1,3-glucan layer in the lateral wall of the neck region. Conceivably, also transglucosylase activity is needed for repair activity. This results in much higher apparent abundances of the cell separation enzymes Cht3 and Eng1 and the potential repair enzyme Scw11 in the culture solution of unicellular budding yeast than in hyphal cells (Table 2). Although the apparent abundances of Bgl2, Cht1, Dag1, Rbe1, Sim1, and Xog1in the culture solution of hyphal cultures are also strongly reduced, the corresponding genes do not seem to be periodically expressed (8, 17).
Hyphal state-enriched secretome proteins: hyphal versus yeast cultures.
Many host-related chemical and physical conditions, such as a neutral pH, a temperature of 37°C, low oxygen levels and high CO2 concentrations, the presence of GlcNAc (derivatives) or serum, iron restriction, and low glucose concentrations, promote hyphal growth (30, 56–58). The natural inducer N-acetylglucosamine (GlcNAc) offers several advantages. Under glucose-derepressing conditions, which occur in many niches in the human body, GlcNAc and also GlcNAc derivatives strongly initiate and sustain hyphal growth (31, 32, 59). GlcNAc is, for example, found in the polysaccharide hyaluronan, which consists of alternating residues of glucuronic acid and GlcNAc and is an abundant component of the extracellular matrix of epithelial and connective tissues (60) and is a known inducer of hyphal growth (59). An additional advantage of using GlcNAc to induce hyphal growth is that it, in contrast to bovine serum, allows (mass spectrometric) analysis of the hyphal secretome (12). GlcNAc-induced hyphal cultures contain four additional proteins in the culture medium that become about as equally prominent as Mp65, Sun41, and Tos1 (Tables 1 and 2), namely, the aspartyl proteases Sap4 and Sap6; the Pry family protein Rbt4, whose function is unknown; and the GPI-modified adhesion protein Als3 (Tables 1 and 2). This is consistent with the strongly increased expression levels of the corresponding genes in GlcNAc-induced hyphal cultures (31; see also reference 61) for serum-induced hyphal cultures. Of interest, Als3 is a hypha-specific adhesive GPI protein that is targeted to the cell wall and required for biofilm formation (62); it is also known to promote the formation of mixed fungal-bacterial biofilms through its N-terminal adhesion domain (63, 64). This raises the question of whether biofilm matrix material might become enriched with released Als3 and whether it might promote the cohesion of the extracellular matrix material. It is unknown whether Als3 is passively released from the cell wall or is released through a regulated, enzymatic process, possibly involving the GPI proteins Sap9 and/or Sap10 (38, 47). The secreted aspartyl proteases Sap4 and Sap6 are strongly associated with the yeast-to-hypha transition (65). Note that the presence of an ammonium salt as a rich nitrogen source in the culture medium at neutral pH does not seem to interfere with the accumulation of Sap4 and Sap6 at high levels in the medium (Table 2).
Major changes in apparent abundance in the secretome of fluconazole-treated cells.
Pseudohyphal (or pseudohypha-like) growth can be induced in various ways, for example, by treating single-cell yeast cultures with sublethal concentrations of the antifungal compound fluconazole (14). Fluconazole blocks an Erg11-mediated demethylation step of the planar ring structure during ergosterol synthesis, and this results in the formation of structurally suboptimal sterols and increased fluidity of the plasma membrane (66). The cells respond by suppression of cell separation and by increased phosphorylation of the MAP kinase Mkc1 (14), which mediates the cell wall integrity pathway. Pseudohypha-like cell aggregates are formed, which separate into single cells when treated with exogenous chitinase (14). These effects are not specific for fluconazole but also occur when the cells are treated with the detergent SDS, grown at the harmful temperature of 42°C, or treated with the cell wall construction-perturbing compound calcofluor white or Congo red (14, 51) (Fig. 2). This suggests that many forms of plasma membrane stress or cell wall stress, together here referred to as cell surface stress, will cause pseudohypha-like growth and constitutively activate the cell wall integrity pathway. Indeed, cell separation suppression is a common phenotype among all genetic mutants that have to cope with a weakened cell wall resulting from defective N- or O-glycosylation of secretory glycoproteins (67–70) or lack proteins involved in cell wall formation, such as Bgl2 (71); Ecm33, which is a GPI protein required for cell wall integrity (72); the secretome enzyme Sun41 (51, 52); and the GPI-modified and cell surface-located aspartyl proteases Sap9 and Sap10 (38), as well as Pir1, a putative cross-linker of β-glucan chains (73), and this list could be easily extended. Interestingly, CHT3 transcription is markedly downregulated in micafungin-treated cells (74), suggesting that this inhibitor of β-1,3-glucan synthesis might also induce cell separation suppression. Similar observations have been described by O'Meara and coworkers, who treated wild-type cultures with serum in combination with various antifungal drugs at sublethal concentrations and found that under these conditions hyphal growth was suppressed and, instead, pseudohyphal cell clusters were formed (see Fig. 2c in reference 75). A relevant implication of these observations is that “cell separation suppression” as a phenotype does not necessarily prove that the mutated gene is directly involved in cell separation.
FIG 2.
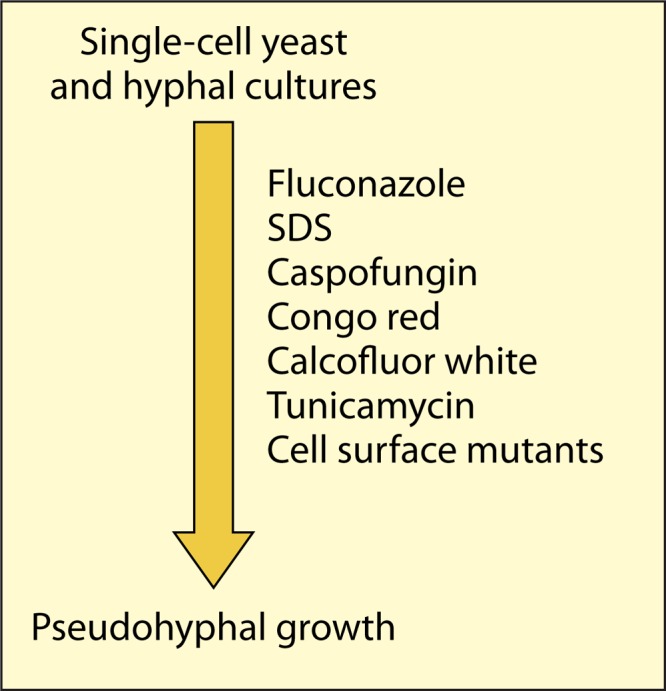
Cell surface stress induces pseudohyphal growth. For further discussion, see the text and references 14 and 75.
Consistent with the partial suppression of cell separation in fluconazole-treated cells, resulting in pseudohypha-like growth, the major chitinase Cht3 (GH18), the endo-β-1,3-glucanase Eng1 (GH81), and the β-1,3-glucan-modifying enzyme Scw11 are all decreased in the supernatants of fluconazole-treated cultures, although less so than in hyphal cultures (Tables 2 and 3). Other chitin- and β-1,3-glucan-modifying enzymes, namely, Cht1 (GH18) and Sim1 (GH132), are also decreased in fluconazole-treated and hyphal cells. How exactly suppression of cell separation is regulated in response to cell surface stress is currently unknown. As CHT3, ENG1, and SCW11 are known target genes of the transcription factor Ace2 (18–20), a regulator of cell separation that controls the expression of M/G1-specific genes, it seems likely that fluconazole treatment and many other forms of cell surface stress lead to suppression of Ace2 activity in cell separation. Possibly, cell separation is suppressed through phosphorylation of the transcription factor Efg1, which in turn represses Ace2 target genes and promotes pseudohypha-like growth (76).
TABLE 3.
Main features of the secretomes of fluconazole-supplemented cultures
Another striking observation is the high apparent abundance of the GPI protein Phr1 in the medium of fluconazole-treated cells, whereas it is not detectable in the medium of single-cell yeast cultures and hyphal cultures. This observation is consistent with the finding that the antifungal compounds ketoconazole and caspofungin stimulate the expression of PHR1 (77). Phr1 (GH72) is a pH-responsive transglucosylase and probably involved in expansion of the β-1,3-glucan layer (78, 79). It also plays an important role in the accumulation of β-1,3-glucan in the extracellular matrix of biofilms, possibly by extending the β-1,3-glucan chains there and thus increasing the cohesiveness of the biofilm (24). This raises the interesting question of whether biofilm-associated Phr1in fluconazole-treated cells might contribute to protection against fluconazole. It is currently unknown how Phr1 is released from the cell surface. However, Phr1 contains two consecutive lysine residues (K451 and K452), suggesting that it might be a substrate of Sap9/10 (47).
Finally, the culture medium of fluconazole-treated yeast cultures contains several low-abundance secretory proteins that so far have not been observed under other growth conditions (13). This does not necessarily prove that this secretome response is specific for fluconazole-treated cells, because, as we have seen above, many other conditions lead to cell surface stress, and this might be in itself enough to trigger the release of members of this subset of proteins.
Secretome proteins increased at low pH.
Acidic conditions are found on the skin (80); in the vagina, due to formation of lactic acid by vaginal epithelial cells and by lactobacilli (81); and in dental plaque, where they are caused by the formation of lactic acid by Streptococcus mutans (82). Two GPI-modified wall proteins (Plb4.5 and Utr2) and the β-1,3-glucan-linked wall protein Pir1 become much more abundant in the culture medium upon lowering the environmental pH, either because at acidic pH their incorporation into the cell wall becomes less efficient or because they are actively released from the wall (Table 4). The apparent abundance of the non-GPI, β-1,3-glucan-modifying protein Bgl2 (GH17) also shows a considerable increase. As this protein contributes to β-1,3-glucan accumulation in biofilms (24), this might mean that at acidic pHs biofilms produce more extracellular matrix.
Lactate-induced secretome proteins.
Although at many infection sites glucose levels are low and, consequently, C. albicans cells are in a glucose-derepressed state, depending instead on alternative carbon sources for growth, glucose-grown cells are often used for research. The results obtained in this way can therefore not always be directly extrapolated to glucose-derepressed cells (83). For example, the levels of three secretome proteins, glucoamylase(s) Gca1/2 (GH31), the aspartyl protease Sap7, and the exo-β-1,3-glucanase Xog1 (GH15), strongly increase when lactate, a nonfermentable carbon source and a much poorer carbon source than glucose, is used to support growth (9, 84). On the other end of the spectrum, there are also a number of secretome proteins whose apparent abundances strongly decrease in lactate-grown cultures (9). The induction of hyphal growth by GlcNAc as discussed above is already blocked at a glucose concentration of 20 mM (33), and this represents another striking example of the importance of selecting the appropriate carbon source and/or glucose concentration in Candida research.
CONCLUDING REMARKS
Similarly to the wall proteome, the secretome of C. albicans operates at the fungus-host interface, and pronounced changes in abundance of individual proteins occur in distinct host niches. For example, the switch from single-cell yeast growth to hyphal growth results in the abundant secretion of the aspartyl proteases Sap4 and Sap6, which significantly contribute to virulence (85). As expected, the pH of the host niche also strongly affects the composition of the secretome. The ability to form biofilms, in which yeast, pseudohyphal, and hyphal cells are found, with the concurrent accumulation of extracellular matrix material and the often strongly lowered susceptibility to antifungal agents, is another important virulence trait. Several secretome proteins, such as Als3, Bgl2, Mp65, Phr1, Xog1, and Sun41, are believed to be involved in biofilm formation (8). In view of their high apparent abundances under widely diverging growth conditions, the secretome proteins Mp65, Sun41, and Tos1 seem attractive candidates for diagnostic purposes. In addition, as Candida infections are usually associated with invasive, hyphal growth, and as Als3, Rbt4, Sap4, and Sap6 become abundant in the supernatants of hyphal cultures, these proteins also are potential diagnostic candidates. Two secretome proteins, Als3 (actually, a recombinant protein that covers the N-terminal immunoglobulin-like domain of Als3) and Sap2, are currently being actively pursued for vaccine development (86–88). Als3 is an abundant, multifunctional GPI-modified cell wall protein (86, 89, 90) and as such is a highly attractive vaccine candidate. The finding that Als3 also becomes an abundant secretome protein in hyphal cultures (12) with a proposed role in biofilm formation further increases its attractiveness. It has been argued that a multivalent vaccine might be more effective to combat the various types of Candida infections (91). Conceivably, a recombinant protein consisting of two or three of the most immunogenic epitopes from various cell wall proteins (92) and from secretome proteins such as the aspartyl proteases Sap4 and Sap6 (12, 85); the three secretome proteins involved in the formation of biofilm matrix beta-glucan (Bgl2, Phr1, and Xog1) (24); Sun41 and Sim1, which form a two-member family and are synthetically lethal (52); and the Pry proteins Rbe1 and Rbt4 (11), each separated from the other by a nonimmunogenic linker sequence, might be an option. Similar approaches might be considered for other pathogenic fungi. Finally, many regulatory pathways have been shown to be involved in controlling the abundance of individual secretome proteins, but more-systematic studies of how the secretome as a whole is regulated are still scarce.
ACKNOWLEDGMENTS
We thank Alice Sorgo and Clemens Heilmann for their stimulating involvement in the research presented here.
REFERENCES
- 1.Moran G, Coleman D, Sullivan D. 2012. An introduction to the medically important Candida species, p 11–25. In Calderone RA, Clancy CJ (ed), Candida and candidiasis, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 2.Bonhomme J, d'Enfert C. 2013. Candida albicans biofilms: building a heterogeneous, drug-tolerant environment. Curr Opin Microbiol 16:398–403. doi: 10.1016/j.mib.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Finkel JS, Mitchell AP. 2012. Biofilm formation in Candida albicans, p 299–315. In Calderone RA, Clancy CJ (ed), Candida and candidiasis, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 4.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klis FM, Sosinska GJ, de Groot PW, Brul S. 2009. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res 9:1013–1028. doi: 10.1111/j.1567-1364.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 6.Sorgo AG, Heilmann CJ, Brul S, de Koster CG, Klis FM. 2013. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol Lett 338:10–17. doi: 10.1111/1574-6968.12049. [DOI] [PubMed] [Google Scholar]
- 7.Vialas V, Sun Z, Loureiro y Penha CV, Carrascal M, Abián J, Monteoliva L, Deutsch EW, Aebersold R, Moritz RL, Gil C. 2014. A Candida albicans PeptideAtlas. J Proteomics 97:62–68. doi: 10.1016/j.jprot.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. 2012. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ene IV, Heilmann CJ, Sorgo AG, Walker LA, de Koster CG, Munro CA, Klis FM, Brown AJ. 2012. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics 12:3164–3179. doi: 10.1002/pmic.201200228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil-Bona A, Llama-Palacios A, Parra CM, Vivanco F, Nombela C, Monteoliva L, Gil C. 2015. Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J Proteome Res 14:142–153. doi: 10.1021/pr5007944. [DOI] [PubMed] [Google Scholar]
- 11.Röhm M, Lindemann E, Hiller E, Ermert D, Lemuth K, Trkulja D, Sogukpinar O, Brunner H, Rupp S, Urban CF, Sohn K. 2013. A family of secreted pathogenesis-related proteins in Candida albicans. Mol Microbiol 87:132–1351. doi: 10.1111/mmi.12087. [DOI] [PubMed] [Google Scholar]
- 12.Sorgo AG, Heilmann CJ, Dekker HL, Brul S, de Koster CG, Klis FM. 2010. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27:661–672. doi: 10.1002/yea.1775. [DOI] [PubMed] [Google Scholar]
- 13.Sorgo AG, Heilmann CJ, Dekker HL, Bekker M, Brul S, de Koster CG, de Koning LJ, Klis FM. 2011. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot Cell 10:1071–1081. doi: 10.1128/EC.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann CJ, Sorgo AG, Mohammadi S, Sosinska GJ, de Koster CG, Brul S, de Koning LJ, Klis FM. 2013. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot Cell 12:254–264. doi: 10.1128/EC.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddi A, Bowman SM, Free SJ. 2009. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet Biol 46:768–781. doi: 10.1016/j.fgb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Côte P, Hogues H, Whiteway M. 2009. Transcriptional analysis of the Candida albicans cell cycle. Mol Biol Cell 20:3363–3373. doi: 10.1091/mbc.E09-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot Cell 5:2001–2013. doi: 10.1128/EC.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol 53:969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 20.Esteban PF, Ríos I, García R, Dueñas E, Plá J, Sánchez M, de Aldana CR, Del Rey F. 2005. Characterization of the CaENG1 gene encoding an endo-1,3-beta-glucanase involved in cell separation in Candida albicans. Curr Microbiol 51:385–392. doi: 10.1007/s00284-005-0066-2. [DOI] [PubMed] [Google Scholar]
- 21.Joshi KR, Wheeler EE, Gavin JB. 1973. Scanning electron microscopy of colonies of six species of Candida. J Bacteriol 115:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol 11:30–36. doi: 10.1016/S0966-842X(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 23.Harriott MM, Lilly EA, Rodriguez TE, Fidel PL Jr, Noverr MC. 2010. Candida albicans forms biofilms on the vaginal mucosa. Microbiology 156:3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturtevant J, Dixon F, Wadsworth E, Latge JP, Zhao XJ, Calderone R. 1999. Identification and cloning of GCA1, a gene that encodes a cell surface glucoamylase from Candida albicans. Med Mycol 37:357–366. doi: 10.1046/j.1365-280X.1999.00244.x. [DOI] [PubMed] [Google Scholar]
- 26.Dennerstein GJ, Ellis DH. 2001. Oestrogen, glycogen and vaginal candidiasis. Aust N Z J Obstet Gynaecol 41:326–328. doi: 10.1111/j.1479-828X.2001.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd MG, Sullivan PA. 1976. The production and growth characteristics of yeast and mycelial forms of Candida albicans in continuous culture. J Gen Microbiol 93:361–370. doi: 10.1099/00221287-93-2-361. [DOI] [PubMed] [Google Scholar]
- 28.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niimi K, Niimi M, Shepherd MG, Cannon RD. 1997. Regulation of N-acetylglucosaminidase production in Candida albicans. Arch Microbiol 168:464–472. doi: 10.1007/s002030050523. [DOI] [PubMed] [Google Scholar]
- 30.Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, de Koster CG, de Koning LJ, Klis FM. 2011. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157:2297–2307. doi: 10.1099/mic.0.049395-0. [DOI] [PubMed] [Google Scholar]
- 31.Naseem S, Araya E, Konopka JB. 2015. Hyphal growth in Candida albicans does not require induction of hyphal-specific gene expression. Mol Biol Cell 26:1174–1187. doi: 10.1091/mbc.E14-08-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naseem S, Konopka JB. 2015. N-Acetylglucosamine regulates virulence properties in microbial pathogens. PLoS Pathog 11:e1004947. doi: 10.1371/journal.ppat.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shepherd MG, Poulter RT, Sullivan PA. 1985. Candida albicans: biology, genetics, and pathogenicity. Annu Rev Microbiol 39:579–614. doi: 10.1146/annurev.mi.39.100185.003051. [DOI] [PubMed] [Google Scholar]
- 34.Schaller M, Borelli C, Korting HC, Hube B. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- 35.Schaller M, Bein M, Korting HC, Baur S, Hamm G, Monod M, Beinhauer S, Hube B. 2003. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect Immun 71:3227–3234. doi: 10.1128/IAI.71.6.3227-3234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C. 2009. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol 47:465–475. doi: 10.1016/j.molimm.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Sosinska GJ, de Koning LJ, de Groot PW, Manders EM, Dekker HL, Hellingwerf KJ, de Koster CG, Klis FM. 2011. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157:136–146. doi: 10.1099/mic.0.044206-0. [DOI] [PubMed] [Google Scholar]
- 38.Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, Maccallum D, Odds FC, Schäfer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem 281:688–694. doi: 10.1074/jbc.M509297200. [DOI] [PubMed] [Google Scholar]
- 39.Citiulo F, Jacobsen ID, Miramón P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D. 2012. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog 8:e1002777. doi: 10.1371/journal.ppat.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorgo AG, Brul S, de Koster CG, de Koning LJ, Klis FM. 2013. Iron restriction-induced adaptations in the wall proteome of Candida albicans. Microbiology 159:1673–1682. doi: 10.1099/mic.0.065599-0. [DOI] [PubMed] [Google Scholar]
- 41.Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, Turano P, Becker J, Lewinson O, Kornitzer D. 2014. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog 10:e1004407. doi: 10.1371/journal.ppat.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo S, Blom AM, Rupp S, Hipler UC, Hube B, Skerka C, Zipfel PF. 2011. The pH-regulated antigen 1 of Candida albicans binds the human complement inhibitor C4b-binding protein and mediates fungal complement evasion. J Biol Chem 286:8021–8029. doi: 10.1074/jbc.M110.130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zipfel PF, Skerka C, Kupka D, Luo S. 2011. Immune escape of the human facultative pathogenic yeast Candida albicans: the many faces of the Candida Pra1 protein. Int J Med Microbiol 301:423–430. doi: 10.1016/j.ijmm.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Román E, Cottier F, Ernst JF, Pla J. 2009. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot Cell 8:1235–1249. doi: 10.1128/EC.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szafranski-Schneider E, Swidergall M, Cottier F, Tielker D, Román E, Pla J, Ernst JF. 2012. Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog 8:e1002501. doi: 10.1371/journal.ppat.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swidergall M, Ernst AM, Ernst JF. 2013. Candida albicans mucin Msb2 is a broad-range protectant against antimicrobial peptides. Antimicrob Agents Chemother 57:3917–3922. doi: 10.1128/AAC.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schild L, Heyken A, de Groot PW, Hiller E, Mock M, de Koster C, Horn U, Rupp S, Hube B. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell 10:98–109. doi: 10.1128/EC.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Sadygov RG, Yates JR III. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 49.McIlwain S, Mathews M, Bereman MS, Rubel EW, MacCoss MJ, Noble WS. 2012. Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinformatics 13:308. doi: 10.1186/1471-2105-13-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klis FM, de Koster CG, Brul S. 2014. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot Cell 13:2–9. doi: 10.1128/EC.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiller E, Heine S, Brunner H, Rupp S. 2007. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot Cell 6:2056–2065. doi: 10.1128/EC.00285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Firon A, Aubert S, Iraqui I, Guadagnini S, Goyard S, Prévost MC, Janbon G, d'Enfert C. 2007. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol Microbiol 66:1256–1275. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- 53.Sandini S, Stringaro A, Arancia S, Colone M, Mondello F, Murtas S, Girolamo A, Mastrangelo N, De Bernardis F. 2011. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol 11:106. doi: 10.1186/1471-2180-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gastebois A, Aimanianda V, Bachellier-Bassi S, Nesseir A, Firon A, Beauvais A, Schmitt C, England P, Beau R, Prévost MC, d'Enfert C, Latgé JP, Mouyna I. 2013. SUN proteins belong to a novel family of β-(1,3)-glucan-modifying enzymes involved in fungal morphogenesis. J Biol Chem 288:13387–13396. doi: 10.1074/jbc.M112.440172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steczkiewicz K, Knizewski L, Rychlewski L, Ginalski K. 2010. TOS1 is circularly permuted 1,3-beta-glucanase. Cell Cycle 9:201–204. doi: 10.4161/cc.9.1.10510. [DOI] [PubMed] [Google Scholar]
- 56.Sudbery P, Gow N, Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol 12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 57.Thompson DS, Carlisle PL, Kadosh D. 2011. Coevolution of morphology and virulence in Candida species. Eukaryot Cell 10:1173–1182. doi: 10.1128/EC.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hameed S, Prasad T, Banerjee D, Chandra A, Mukhopadhyay CK, Goswami SK, Lattif AA, Chandra J, Mukherjee PK, Ghannoum MA, Prasad R. 2008. Iron deprivation induces EFG1-mediated hyphal development in Candida albicans without affecting biofilm formation. FEMS Yeast Res 8:744–755. doi: 10.1111/j.1567-1364.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- 59.Shepherd MG, Sullivan PA. 1984. The control of morphogenesis in Candida albicans. J Dent Res 63:435–440. doi: 10.1177/00220345840630031501. [DOI] [PubMed] [Google Scholar]
- 60.Laurent TC, Fraser JR. 1992. Hyaluronan. FASEB J 6:2397–2404. [PubMed] [Google Scholar]
- 61.Hube B, Monod M, Schofield DA, Brown AJ, Gow NA. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol 14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 62.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. 2015. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology 161:18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abe F, Usui K, Hiraki T. 2009. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48:8494–8504. doi: 10.1021/bi900578y. [DOI] [PubMed] [Google Scholar]
- 67.Munro CA, Bates S, Buurman ET, Hughes HB, Maccallum DM, Bertram G, Atrih A, Ferguson MA, Bain JM, Brand A, Hamilton S, Westwater C, Thomson LM, Brown AJ, Odds FC, Gow NA. 2005. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J Biol Chem 280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bates S, Hughes HB, Munro CA, Thomas WP, MacCallum DM, Bertram G, Atrih A, Ferguson MA, Brown AJ, Odds FC, Gow NA. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem 281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- 69.Southard SB, Specht CA, Mishra C, Chen-Weiner J, Robbins PW. 1999. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J Bacteriol 181:7439–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst JF. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J Biol Chem 273:20837–20846. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- 71.Sarthy AV, McGonigal T, Coen M, Frost DJ, Meulbroek JA, Goldman RC. 1997. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-beta-glucosyltransferase. Microbiology 143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 72.Martinez-Lopez R, Monteoliva L, Diez-Orejas R, Nombela C, Gil C. 2004. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology 150:3341–3354. doi: 10.1099/mic.0.27320-0. [DOI] [PubMed] [Google Scholar]
- 73.Martínez AI, Castillo L, Garcerá A, Elorza MV, Valentín E, Sentandreu R. 2004. Role of Pir1 in the construction of the Candida albicans cell wall. Microbiology 150:3151–3161. doi: 10.1099/mic.0.27220-0. [DOI] [PubMed] [Google Scholar]
- 74.Kaneko Y, Ohno H, Kohno S, Miyazaki Y. 2010. Micafungin alters the expression of genes related to cell wall integrity in Candida albicans biofilms. Jpn J Infect Dis 63:355–357. [PubMed] [Google Scholar]
- 75.O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. 2015. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 6:6741. doi: 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang A, Raniga PP, Lane S, Lu Y, Liu H. 2009. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol Cell Biol 29:4406–4416. doi: 10.1128/MCB.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu TT, Lee RE, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. 2005. Genome wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother 49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Bernardis F, Mühlschlegel FA, Cassone A, Fonzi WA. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun 66:3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mouyna I, Fontaine T, Vai M, Monod M, Fonzi WA, Diaquin M, Popolo L, Hartland RP, Latgé JP. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J Biol Chem 275:14882–14889. doi: 10.1074/jbc.275.20.14882. [DOI] [PubMed] [Google Scholar]
- 80.Zlotogorski A. 1987. Distribution of skin surface pH on the forehead and cheek of adults. Arch Dermatol Res 279:398–401. doi: 10.1007/BF00412626. [DOI] [PubMed] [Google Scholar]
- 81.Linhares IM, Summers PR, Larsen B, Giraldo PC, Witkin SS. 2011. Contemporary perspectives on vaginal pH and lactobacilli. Am J Obstet Gynecol 204:120.e1–5. doi: 10.1016/j.ajog.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 82.Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. 2013. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog 9:e1003616. doi: 10.1371/journal.ppat.1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ene IV, Cheng SC, Netea MG, Brown AJ. 2013. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun 81:238–248. doi: 10.1128/IAI.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NA, Brown AJ. 2012. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol 14:1319–1335. doi: 10.1111/j.1462-5822.2012.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanglard D, Hube B, Monod M, Odds FC, Gow NA. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun 65:3539–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ibrahim AS, Luo G, Gebremariam T, Lee H, Schmidt CS, Hennessey JP Jr, French SW, Yeaman MR, Filler SG, Edwards JE Jr. 2013. NDV-3 protects mice from vulvovaginal candidiasis through T- and B-cell immune response. Vaccine 31:5549–5556. doi: 10.1016/j.vaccine.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sandini S, LaValle R, Deaglio S, Malavasi F, Cassone A, DeBernardis F. 2011. A highly immunogenic recombinant and truncated protein of the secreted aspartic proteases family (rSap2t) of Candida albicans as a mucosal anticandidal vaccine. FEMS Immunol Med Microbiol 62:215–224. doi: 10.1111/j.1574-695X.2011.00802.x. [DOI] [PubMed] [Google Scholar]
- 89.Coleman DA, Oh SH, Zhao X, Zhao H, Hutchins JT, Vernachio JH, Patti JM, Hoyer LL. 2009. Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J Microbiol Methods 78:71–78. doi: 10.1016/j.mimet.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cassone A. 2013. Development of vaccines for Candida albicans: fighting a skilled transformer. Nat Rev Microbiol 11:884–891. doi: 10.1038/nrmicro3156. [DOI] [PubMed] [Google Scholar]
- 92.Klis FM, de Koster CG, Brul S. 2011. A mass spectrometric view of the fungal wall proteome. Future Microbiol 6:941–651. doi: 10.2217/fmb.11.72. [DOI] [PubMed] [Google Scholar]