Abstract
Using confocal microscopy, we observed ring-like organelles, similar in size to nuclei, in the hyphal tip of the filamentous fungus Neurospora crassa. These organelles contained a subset of vacuolar proteins. We hypothesize that they are novel prevacuolar compartments (PVCs). We examined the locations of several vacuolar enzymes and of fluorescent compounds that target the vacuole. Vacuolar membrane proteins, such as the vacuolar ATPase (VMA-1) and the polyphosphate polymerase (VTC-4), were observed in the PVCs. A pigment produced by adenine auxotrophs, used to visualize vacuoles, also accumulated in PVCs. Soluble enzymes of the vacuolar lumen, alkaline phosphatase and carboxypeptidase Y, were not observed in PVCs. The fluorescent molecule Oregon Green 488 carboxylic acid diacetate, succinimidyl ester (carboxy-DFFDA) accumulated in vacuoles and in a subset of PVCs, suggesting maturation of PVCs from the tip to distal regions. Three of the nine Rab GTPases in N. crassa, RAB-2, RAB-4, and RAB-7, localized to the PVCs. RAB-2 and RAB-4, which have similar amino acid sequences, are present in filamentous fungi but not in yeasts, and no function has previously been reported for these Rab GTPases in fungi. PVCs are highly pleomorphic, producing tubular projections that subsequently become detached. Dynein and dynactin formed globular clusters enclosed inside the lumen of PVCs. The size, structure, dynamic behavior, and protein composition of the PVCs appear to be significantly different from those of the well-studied prevacuolar compartment of yeasts.
INTRODUCTION
The vacuolar compartments in cells of fungi and plants have many of the same functions as the lysosomal compartments of animal cells (1–5). The defining characteristics of vacuoles and lysosomes are (i) the presence of a variety of hydrolytic enzymes such as proteases and phosphatases that help to degrade and recycle macromolecules and (ii) the maintenance of an acidic internal pH. An electrochemical gradient for protons is generated across vacuolar/lysosomal membranes by the vacuolar ATPase, a large complex enzyme (1, 6).
We have been investigating the structure and function of the vacuolar compartments in filamentous fungi, using Neurospora crassa as our model organism. In fungi, vacuoles have multiple functions in addition to the degradation of macromolecules. They are involved in the regulation of arginine and ornithine metabolism, osmoregulation, and cytosolic ion and pH homeostasis. Fungal vacuoles contain high concentrations of basic amino acids, calcium, and polyphosphate (1, 4, 5).
The vacuolar compartment in filamentous fungi has a complex structure and is highly dynamic (3, 7, 8). Many vacuoles have internal contents with a different refractive index than that of the cytosol, allowing them to be visualized with Nomarski optics. These vacuoles, prominent in older parts of the hypha, are spherical, ranging in size from <0.5 μm to >10 μm. Other vacuoles, prominent in the first hyphal segment, take the form of a dense network of interconnected tubules (9, 10). For N. crassa, previous work showed that the first hyphal segment can be described as having four regions containing different distributions of organelles (11, 12). Region I, extending 2 to 3 μm behind the apex, contains the Spitzenkörper, a structured collection of vesicles that delivers material to the growing hyphal tip. Region II, 20 to 30 μm beyond region I, contains small vesicles and mitochondria but few nuclei or vacuoles. Nuclei are added to the mix of organelles in region III, 30 to 40 μm beyond region II. Region IV, which extends to the first septum, is characterized by having an increased density of mitochondria, nuclei, and vesicles, and it is in this region that the tubular vacuolar network appears.
Several years ago, we examined the structure and distribution of organelles in N. crassa by using enzymes tagged with green fluorescent protein (GFP) and red fluorescent protein (RFP) as markers (13). Subunits of the vacuolar ATPase (VMA-1 and VMA-5) and a vacuolar calcium transporter (CAX) were abundant in structures that had not been previously described. These structures were roughly spherical, 1 to 3 μm in diameter, and located only at the boundary of regions III and IV (∼50 to 200 μm from the hyphal tip). Unlike other spherical vacuoles, the structures were not visible with Nomarski optics. They appeared to contain only a subset of vacuolar proteins. PEP-12, a vacuolar SNARE protein (misidentified as VAM-3 in reference 13), and the calcium transporters NCA-2 and NCA-3 were not observed in the new organelle. In that initial report, we speculated that the new organelle could be a specialized type of vacuole that controlled calcium levels at the hyphal tip. Observations presented in this report have led us to hypothesize that the new organelle is involved in the formation of the tubular vacuolar network. Therefore, we tentatively call it a prevacuolar compartment (PVC).
The biogenesis of the vacuole has been studied extensively in yeast (3, 14, 15). Vacuolar proteins are initially synthesized by ribosomes attached to the endoplasmic reticulum (ER) and transported via small vesicles to the Golgi complex. At least three pathways have been described for the subsequent transport of Golgi-derived vesicles to the vacuole/lysosome. Some proteins, such as alkaline phosphatase (product of PHO8 in Saccharomyces cerevisiae), travel directly to the vacuole (16, 17). Other proteins, such as carboxypeptidase Y (CPY) and the vacuolar ATPase, are transported first to an intermediate compartment called the late endosome/multivesicular body (MVB) (18). The MVB is so named because it is a membrane-bound compartment that contains vesicles in its interior. MVB proteins travel to and fuse with existing vacuoles. In the third pathway, proteins from the Golgi complex travel via two intermediate compartments, the early endosomes and the late endosomes/MVBs (19, 20). In addition to the proteins that come from the Golgi complex, many proteins of the plasma membrane are also transported to the vacuole via a series of transport vesicles, including the late endosome/MVB. The complex set of proteins that controls the identity of the compartments involved in protein transport includes several Rab GTPases (21, 22), which we investigate in this report.
For filamentous fungi, much less is known about the biogenesis of vacuoles. The structure and biogenesis of the vacuole have been examined in Aspergillus species, where deletion strains lacking homologs of proteins predicted to play a role in forming vacuoles in S. cerevisiae were generated. vpsA is the Aspergillus nidulans homolog of S. cerevisiae VPS1, which encodes a dynamin-like protein involved in vesicle budding from the Golgi complex and endosomes (23, 24). Aovps24 encodes the Aspergillus oryzae homolog of S. cerevisiae VPS24, involved in vesicle transport from endosomes to the MVB (25, 26). avaA is the A. nidulans homolog of S. cerevisiae YPT7 (Rab7 in animal cells) and is required for vesicle fusion with the vacuole (27, 28). Deletion of any of these three genes in Aspergillus caused fragmentation of the spherical vacuoles. A detailed study of early and late endosomes in A. nidulans has shown that, as in S. cerevisiae, homologs of Rab5 facilitate the transport of cargo to the early endosome, while Rab7 is involved in traffic from the early endosome to the late endosome and then to the vacuole (29). These studies of Aspergillus indicate that the transport of proteins to the vacuole in filamentous fungi is similar to that observed in S. cerevisiae. However, significant differences have been reported (reviewed in reference 1), and the genomes of N. crassa and other filamentous fungi contain homologs of at least two Rab GTPases that are found in animal cells but not in S. cerevisiae (30–32). In this report, we provide evidence that a subset of vacuolar proteins and Rab GTPases are components of a novel prevacuolar compartment in N. crassa.
MATERIALS AND METHODS
Construction of plasmids and Neurospora crassa strains.
We used four plasmids to generate proteins with fluorescent tags: pMF272 for enhanced GFP (eGFP) at the C terminus (33); pCCG::N-GFP for eGFP at the N terminus (34); pMF334 for tdimer2, a variant of dsRED, at the N terminus (35); and pTSL48b for mCherry at the N terminus (gift from T. Starr and L. Glass, University of California, Berkeley). The genes of interest (see Table S1 in the supplemental material) were amplified by PCR, using DNA from wild-type strain 74A and using either the Pfu Turbo DNA polymerase (Agilent Technologies) or Phusion polymerase (Life Technologies). The primers were designed with restriction endonuclease sites to facilitate insertion into the multiple-cloning-site region of the plasmids. DNA amplified from the PCRs was initially inserted into the vector pJET1.2 (Fermentas Life Sciences), which facilitates cloning of PCR products with blunt ends. The pJET1.2 plasmid containing the insert was digested with the two restriction endonucleases that recognized the sites in the primers. In this way, a DNA fragment with a high likelihood that each end had been cut with the correct enzyme was obtained. This fragment was ligated with a plasmid digested with the same pair of restriction endonucleases. All plasmids listed in Table S1 in the supplemental material are available from the Fungal Genetics Stock Center, Kansas State University, Manhattan, KS (36). To incorporate the tagged genes into N. crassa, we transformed the his-3 mus-51 mat A strain by electroporation as previously described (13). Three strains from previous studies were used, the GFP–YPT-1 (51), PEP-12–dsRED (13), and eGFP-dynactin-P150 (55) strains.
Microscopy.
N. crassa strains were inoculated with conidia on 100-mm petri plates containing Vogel's minimal medium (VMM) (39) with 2% sucrose plus 2% agar and grown for 16 to 18 h at 30°C. An agar block of ∼1 by 2 cm was cut from the leading edge of the colony and mounted onto a glass coverslip as described previously (9). To visualize GFP-tagged and RFP-tagged proteins in the same hypha, we prepared heterokaryons. Conidia from each strain were suspended in water and counted. Typically, 4 μl or 12 μl of each strain (10,000 conidia per μl) was coinoculated at ratios of 1:1, 1:3, and 3:1. For most samples, the best images were obtained with a 1:1 ratio. As a control experiment, heterokaryons were made by using wild-type strain 74A and the VMA-1–dsRED A strain. As described in Results, for this experiment, the ratio of conidia varied from 0.8% to 100% VMA-1–dsRED.
Confocal laser scanning microscopy was performed by using a Leica TCS SP5 system with a Leica DMI6000 inverted microscope. GFP images were obtained by excitation at 488 nm with emission collected at 500 to 600 nm. RFP images were obtained with excitation at 543 nm and emission at 555 to 700 nm. Lasers were set with a scan speed of 100 to 200 Hz, and the camera resolution was typically 1,024 by 1,024 pixels. The microscope objective was 63×, oil, and images were typically digitally magnified 4 times. To obtain two-color images, the sample was scanned sequentially line by line, and emission was collected at 500 to 535 nm for GFP and at 570 to 640 nm for RFP. In one experiment, the intensity of fluorescence was quantitated by measuring the mean gray value. In each hypha, four adjacent 100-μm2 regions of the tubular vacuolar network were scanned, and the values were averaged.
For experiments with the ad-3B strain, mycelia were grown on Vogel's medium with 2% sucrose and 0.1 mM adenine. In some experiments, organelles were visualized by using Oregon Green 488 carboxylic acid diacetate (carboxy-DFFDA) (Life Technologies). A 30-μl drop of a 3 μM solution was placed onto a coverslip, and an inverted agar block with mycelia was placed onto the coverslip and immediately examined with a microscope. For both the ad-3B strain and the carboxy-DFFDA experiments, the microscope settings were the same as those for GFP-tagged strains.
RESULTS
Structure of the vacuole in N. crassa hyphae.
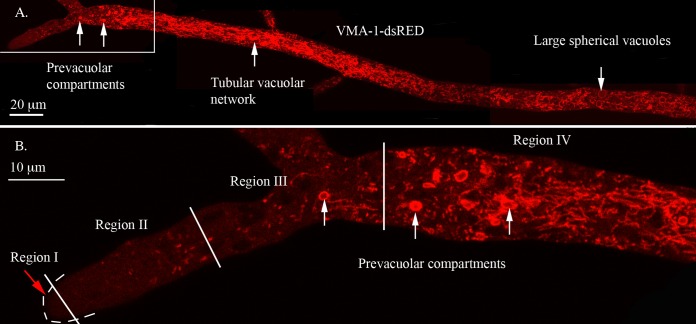
Vacuolar membranes were visualized in a strain expressing RFP-tagged subunit A of the vacuolar ATPase (VMA-1–dsRED). The vacuoles were highly diverse in size and shape (Fig. 1A). Regions I and II had few visible vacuoles. Region III had small, roughly spherical organelles (typically 2 to 3 μm in diameter) and tubular elements, which then merged into region IV containing the tubular vacuolar network. Small organelles and tubules persisted in more distal parts of the hypha, and large spherical vacuoles were observed in hyphal segments distal to the first septum. The round organelles in region III (Fig. 1B) resembled spherical vacuoles in more distal regions of the hypha, but they also differed significantly. Unlike most spherical vacuoles, they were not visible with Nomarski optics, indicating a different internal content. In addition, as evidenced below, they were highly dynamic and pleomorphic (see Movie S1 in the supplemental material). Most importantly, these organelles were composed of only a subset of the proteins found in the spherical vacuoles and the tubular vacuolar network. We propose that these organelles are prevacuolar compartments (PVCs).
FIG 1.
Visualization of the vacuolar ATPase in different regions of the hypha. The hypha was transformed with VMA-1–dsRED. (A) Composite image made from four overlapping photos of the hypha. The structures seen in different regions are described in the text. (B) Enlargement of the boxed area of the hyphal tip in panel A. Regions I, II, III, and IV are separated by white lines, with white arrows pointing to examples of PVCs.
PVC morphology and distribution are not affected by changes in the levels of expression of tagged proteins.
The PVCs were initially observed in hyphae expressing vacuolar ATPase or a vacuolar calcium transporter (CAX) tagged with RFP or GFP (13). We used previously developed vectors containing the ccg-1 promoter in front of the fusion protein (13, 33, 35, 40). These vectors have been widely used because they have allowed visualization of many proteins, including some that could not be seen when expressed under the control of their endogenous promoter. The ccg-1 promoter was first isolated as part of a glucose-repressible gene (41) and subsequently reisolated as a clock-controlled gene, ccg-1 (42). The level of expression of the ccg-1 transcript is elevated after 1 h when cells are deprived of glucose or are grown on xylose (41). The level of expression of ccg-1-controlled genes in cells grown overnight on Vogel's medium has not been rigorously examined. This medium typically contains glucose or sucrose, sugars that repress the expression of ccg-1-controlled genes. One way to control the level of expression of a gene in N. crassa is to generate heterokaryons with different nuclear ratios. To test whether the size and distribution of PVCs varied with the level of expression of the tagged protein, we inoculated agar plates (Vogel's medium with 2% sucrose) with a mixture of wild-type and VMA-1–dsRED conidia of the same mating type. Germlings quickly fuse and form heterokaryons in which a single hyphal compartment can have as many as 100 nuclei (43). We used conidial mixtures in which 0.8% to 100% of the conidia were from the VMA-1–dsRED strain. The amount of red fluorescence was quantified by measuring the pixel intensity in regions of the hyphae with the tubular vacuolar network. The strength of the red fluorescence signal was proportional to the percentage of VMA-1–dsRED conidia (see Fig. S1A in the supplemental material). By digitally enhancing the signal, it was possible to visualize the organelles in the heterokaryons with few VMA-1–dsRED nuclei. The size and distribution of the PVCs were the same in all samples, even in those with the smallest proportion of VMA-1–dsRED (Fig. 1B). In another experiment, the VMA-1–dsRED strain was grown on medium with 2% xylose. The level of red fluorescence was ∼4-fold higher than when grown on the usual sucrose medium, and the size and distribution of the PVCs were not different (data not shown). Thus, the expression of VMA-1–dsRED could be varied by >100-fold without causing an observable difference in the size and distribution of the PVCs.
Visualization of PVCs in an adenine-requiring auxotroph.
We recently discovered an alternative way to visualize the vacuolar compartments without using GFP or RFP. It has long been known that ADE2 mutant strains of S. cerevisiae accumulate a red fluorescent pigment in vacuoles (44). Mutant strains of ad-3B, the N. crassa homolog of ADE2, also accumulate this pigment (45). Using confocal microscopy, we observed fluorescence in large spherical vacuoles, in the tubular vacuolar network, and also in the PVCs (Fig. 2; see also Movie S2 in the supplemental material). It was interesting that the pigment filled the interior of the large vacuoles but only formed a ring in the PVCs.
FIG 2.
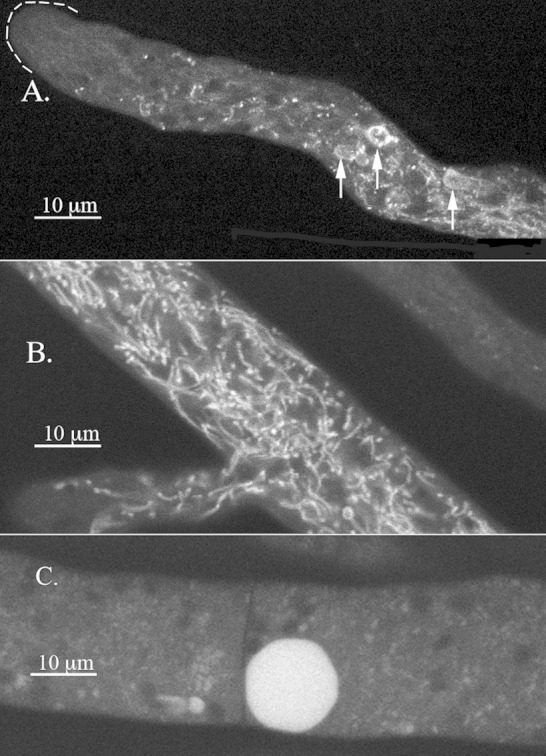
Visualization of the pigment in the adenine auxotroph ad-3B strain grown on medium with 0.1 mM adenine. (A) Hyphal tip region with arrows pointing to the PVCs. (B) Region of the hypha ∼200 μm from the tip where the tubular vacuolar network appears. (C) Region of the hypha several millimeters from the tip with a large spherical vacuole.
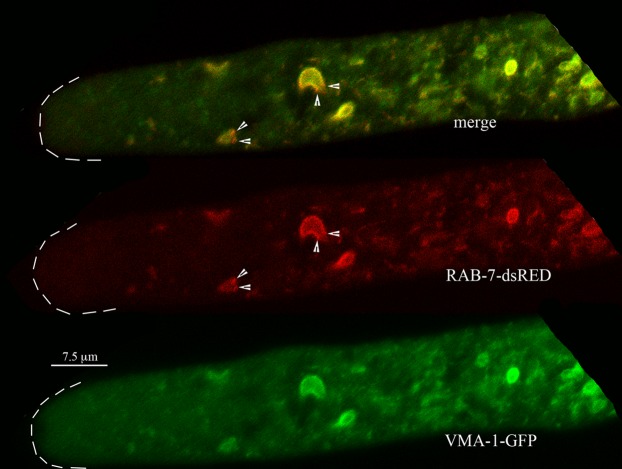
Colocalization of RAB-7 and VMA-1.
If the PVC is indeed a vacuolar compartment, we might expect RAB-7, a GTPase involved in targeting proteins to the vacuole (29), to localize to PVCs. The fluorescence from RAB-7–dsRED almost completely overlapped the fluorescence from VMA-1–GFP (Fig. 3). The only subtle difference observed was that in regions closest to the hyphal tip, small red puncta corresponding to RAB-7 were seen on the PVCs (Fig. 3). In observations of 10 different hyphae, 26 of 39 PVCs (67%) had these small red puncta. Many of the PVCs were somewhat similar in size and shape to nuclei. As we reported previously, the nuclear envelope can be observed with proteins of the endoplasmic reticulum, such as the calcium ATPase NCA-1 (13). Cells coexpressing NCA-1–GFP and RAB-7–dsRED showed that these proteins are clearly located in different compartments (Fig. 4).
FIG 3.
Localization of Rab-7 GTPase (RAB-7) compared to the localization of the vacuolar ATPase (VMA-1) at the hyphal tip. Some small red puncta, marked by arrowheads, occur at the edges of the PVCs.
FIG 4.
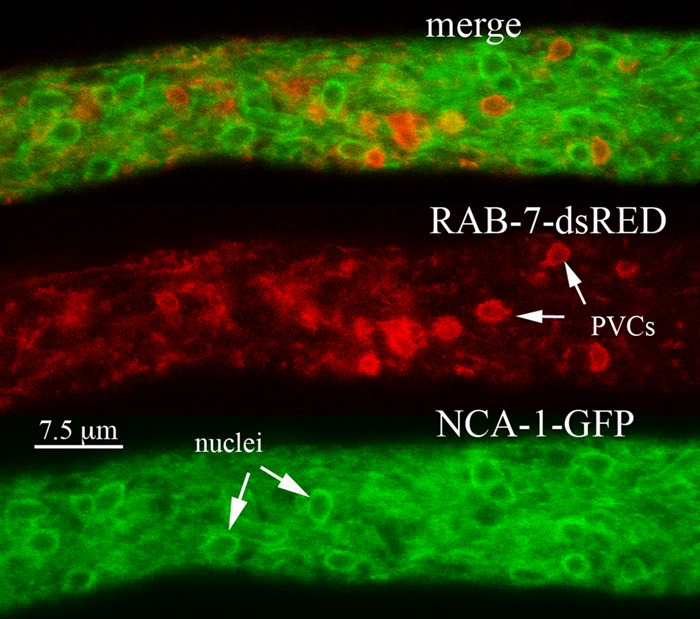
Localization of the nuclear envelope and endoplasmic reticulum with NCA-1–GFP compared with the localization of RAB-7–dsRED. The region of the hypha shown is ∼100 μm from the hyphal tip.
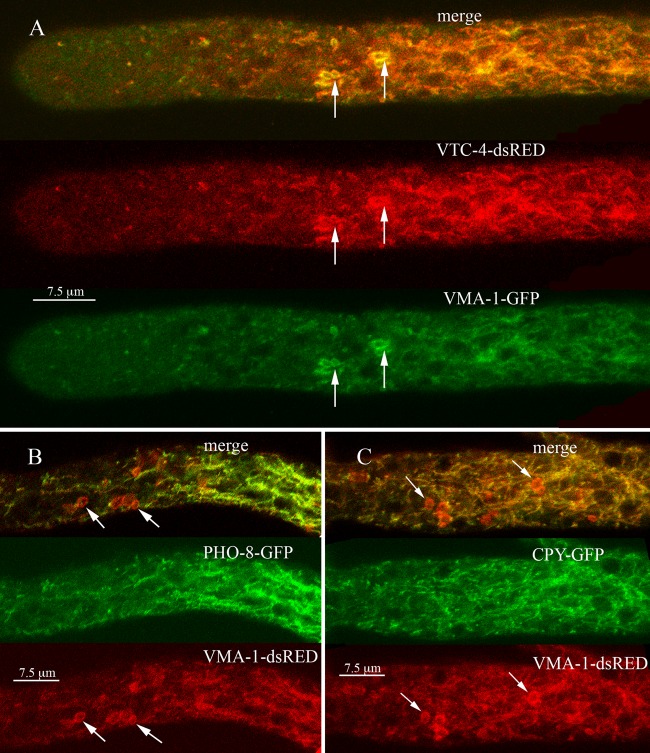
Localization of vacuolar proteins VTC-4, PHO-8, and CPY.
We examined the cell locations of several other proteins predicted to be in the vacuole. The vtc-4 gene encodes an integral membrane subunit of polyphosphate polymerase (46, 47). When we looked at the localization of VTC-4–dsRED and VMA-1–GFP, we found both tagged proteins in the PVCs and the tubular vacuolar network (Fig. 5A) as well as in the large spherical vacuoles (data not shown). The trafficking of the vacuolar alkaline phosphatase encoded by the PHO8 gene has been studied in detail in S. cerevisiae (17, 48). We tagged the N. crassa homolog of PHO8 and examined its cellular location. PHO-8–GFP was observed in large spherical vacuoles (not shown) and in the tubular vacuolar network but not in PVCs (Fig. 5B). Carboxypeptidase Y, a soluble protein, has also been used to investigate protein trafficking to the vacuole (49). In N. crassa, we observed CPY-GFP fluorescence in large spherical vacuoles and in the tubular vacuolar network but not in PVCs (Fig. 5C).
FIG 5.
(A) Localization of the vacuolar polyphosphate polymerase (VTC-4) compared to the localization of the vacuolar ATPase (VMA-1). (B) Localization of the vacuolar alkaline phosphatase (PHO-8) compared to the localization of the vacuolar ATPase (VMA-1). (C) Localization of carboxypeptidase Y (CPY) compared to the localization of the vacuolar ATPase (VMA-1). In each panel, arrows point to examples of PVCs.
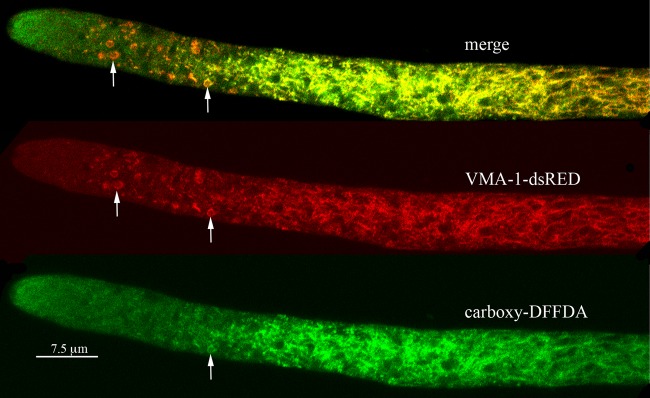
Visualization of vacuoles with carboxy-DFFDA.
Another good way to visualize vacuolar compartments in N. crassa is to use the molecule carboxy-DFFDA, which becomes fluorescent when cleaved by an esterase in the lumen of the vacuole (9). The fluorescent molecule accumulated in the tubular vacuolar network (Fig. 6). In hyphae that also expressed VMA-1–dsRED, carboxy-DFFDA and the red-tagged protein were in the same tubular network. However, we consistently observed a gradient of accumulation of carboxy-DFFDA in the PVCs. The PVCs nearest the hyphal tip had little carboxy-DFFDA, while some PVCs furthest from the tip had both red and green fluorescence. As observed for the pigment produced by ad-3B strains, the green fluorescence was consistently seen in the periphery of the PVC and not in the central space.
FIG 6.
Visualization of carboxy-DFFDA a soluble dye that localizes to vacuoles, compared to the localization of the vacuolar ATPase (VMA-1). The VMA-1–dsRED strain was incubated with carboxy-DFFDA as described in Materials and Methods. Arrows point to examples of PVCs.
Localization of Rab GTPases.
Rab GTPases are an important part of the cellular machinery by which proteins are sorted to the correct organelles (21, 37, 50). As described above, we observed that RAB-7, predicted to be located in vacuoles, was also localized to the PVCs. We tested eight other Rab proteins identified in N. crassa. The nomenclature that we used and the relationship to Rab proteins of other organisms are shown in Table 1. Rab-1 and -11 are involved in the early steps of protein targeting, associating with the ER, Golgi complex, and early endosomes. We previously reported that in N. crassa, these Rabs are seen in different populations of vesicles at the Spitzenkörper (51). RAB-1 (YPT-1) and RAB-11 (YPT-31) did not appear to associate with the PVCs (see Fig. S2A and S2B in the supplemental material). Two orthologs of Rab-5 have been identified in N. crassa (52). These orthologs are predicted to associate with vesicles that carry proteins between endosomes and the plasma membrane. Neither Rab-5A nor Rab-5B appeared to associate with the PVCs (see Fig. S2C and S2D in the supplemental material).
TABLE 1.
Genes used in this study
| N. crassa gene | Product | Locus tag | S. cerevisiae homolog |
|---|---|---|---|
| vma-1 | V-ATPase, subunit A | NCU01207 | VMA1 |
| vma-5 | V-ATPase, subunit C | NCU09897 | VMA5 |
| cpy | Carboxypeptidase Y | NCU00477 | CPY1 |
| pho-8 | Alkaline phosphatase | NCU08997 | PHO8 |
| vtc-4 | Polyphosphate polymerase | NCU08110 | VTC4 |
| ypt-1/rab1 | Rab GTPase 1 | NCU08477 | YPT1 |
| rab-2 | Rab GTPase 2 | NCU01647 | None |
| rab-4 | Rab GTPase 4 | NCU08477 | None |
| rab-5A | Rab GTPase 5A | NCU00895 | YPT51/52/53 |
| rab-5B | Rab GTPase 5B | NCU06410 | YPT51/52/53 |
| rab-6 | Rab GTPase 6 | NCU05234 | YPT6 |
| rab-7 | Rab GTPase 7 | NCU06410 | YPT7 |
| rab-8 | Rab GTPase 8 | NCU06404 | SEC4 |
| rab-11 | Rab GTPase 11 | NCU01523 | YPT31/32 |
| tlg-1 | t-SNARE, endosomes | NCU01199 | TLG1 |
| atg-8 | Autophagosome protein | NCU01545 | ATG8 |
| pep-12 | t-SNARE, vacuole | NCU06777 | PEP12/VAM3 |
| ro-3 | Dynactin subunit P150 | NCU03483 | NIP100/PAC13 |
Rab-6 is predicted to be associated with the Golgi complex and possibly late endosomes. The RAB-6–mCherry strain gave a weak signal, largely in the cytosol, and did not appear to be located in the PVCs (see Fig. S3A in the supplemental material). Rab-8 is involved in the transport of proteins from the Golgi complex to the plasma membrane. No PVCs were visible in the RAB-8–mCherry strain; only small vesicles and tubules were visible. However, in a heterokaryon with both RAB-8–mCherry and VMA-1–GFP, the RAB-8 protein appeared to be associated with PVCs although not mixed with VMA-1–GFP (see Fig. S3B in the supplemental material). The two proteins appeared to form a chimeric structure with distinct red and green regions. Also visible were tubular elements with distinct red and green regions.
N. crassa and other filamentous fungi contain at least two Rab proteins, RAB-2 and RAB-4, that are present in mammalian cells but not in S. cerevisiae (30–32). The function of these proteins in fungi has not been reported. We were surprised to find that both of these proteins localized to the PVCs. In N. crassa, we observed a complex pattern of localization for RAB-2. Virtually all PVCs contained some RAB-2–mCherry, but the ratio of RAB-2–mCherry to VMA-1–GFP varied with distance from the tip (compare PVCs marked with arrows in Fig. 7). Closest to the tip were structures that appeared all red, further away were PVCs with little red, and in between were PVCs that were roughly equally red and green. Other structures appeared as chimeras with red and green regions (Fig. 7, arrowheads). In mammalian cells, Rab-4 has been observed on early endosomes. In N. crassa, RAB-4–mCherry colocalized strongly with VMA-1–GFP (Fig. 8). Like RAB-7, in the merged images, we also observed some tiny red-only particles associated with PVCs (Fig. 8, arrowheads). Observation of 10 different hyphae showed that 17 of 36 PVCs (47%) had these small red puncta.
FIG 7.
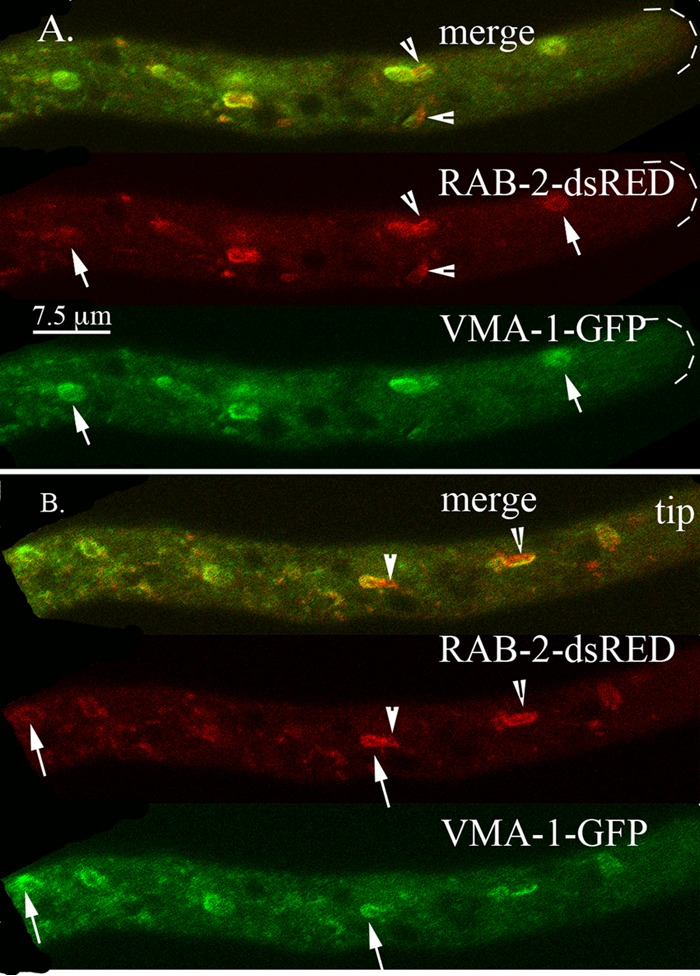
Localization of the Rab-2 GTPase (RAB-2) compared to the localization of the vacuolar ATPase (VMA-1). Arrows point to examples of PVCs. Arrowheads point to structures that contain both RAB-2–dsRED and VMA-1–GFP. Panels A and B show examples of different hyphae from the same heterokaryon.
FIG 8.
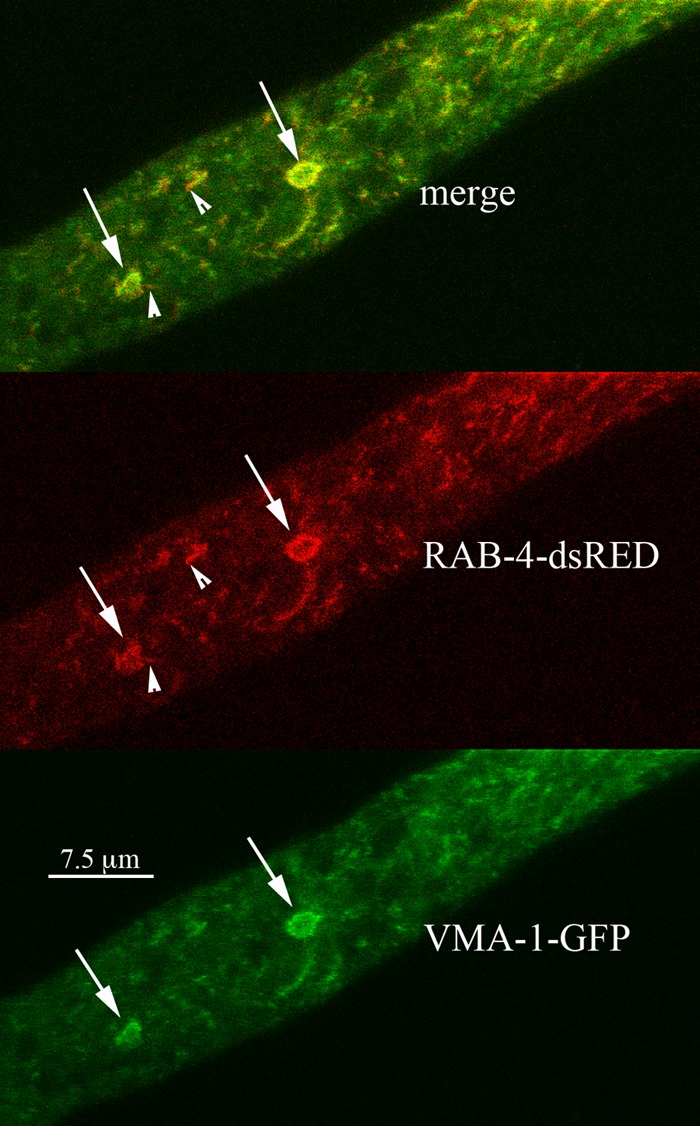
Localization of the Rab-4 GTPase (RAB-4) compared to the localization of the vacuolar ATPase (VMA-1). Some small red puncta, marked by arrowheads, were observed at the edges of the PVCs.
Localization of TLG-1 and ATG-8.
Another protein reported to be associated with the early endosomes/late-Golgi complex is the t-SNARE encoded by TLG1 (53). In N. crassa, TLG-1–dsRED strongly localized at the Spitzenkörper, like RAB-11 (see Fig. S4A in the supplemental material). It was also seen as small puncta near the tip and in the tubular vacuolar network. TLG-1–dsRED did not appear to localize to the PVCs.
Some features of the PVCs are reminiscent of the features of autophagosomes. Both can have irregular shapes, and the autophagosomes are formed with a double-lipid bilayer, a feature that could be shared with the PVCs. The product of the ATG8 gene has been shown to be a marker of autophagosomes (54). In N. crassa, we observed ATG-8–dsRED in small puncta near the hyphal tip and in the tubular vacuolar network (see Fig. S4B in the supplemental material). It did not appear to be associated with PVCs.
Dynamic behavior of PVCs.
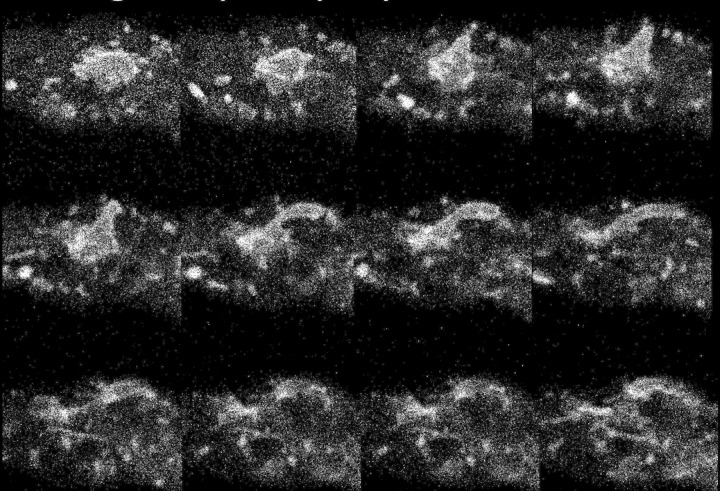
An important characteristic of the PVCs is the rapid change in their size and shape, as shown in the four movies in the supplemental material. This dynamic behavior was observed with proteins tagged with fluorescent markers (e.g., VMA-1–GFP or RAB-7–mCherry) and with the ad-3B strain, which accumulates a red pigment in the vacuole. We frequently observed a blebbing phenomenon in which a roughly spherical PVC would extend a tubular projection that subsequently became detached. Figure 9 shows frames from Movie S3 in the supplemental material. The blebbing was fast, typically complete within 10 to 60 s. However, because these structures can move out of the focal plane, it was not possible to determine their fate definitively. In roughly 1 to 2% of hyphae, with both tagged proteins and the ad-3B strain, we observed unusually large and bright PVCs (see Movie S4 in the supplemental material). Blister-like distortions formed on the surface of these large PVCs and appeared to be the site at which spherical and tubular projections formed. We noted that the boundary envelope forming these large PVCs was nearly 1 μm thick, much larger than would be expected for a single lipid bilayer.
FIG 9.
Changes in the shape of PVCs visualized with RAB-7–dsRED. The figure shows successive frames taken at 5.4-s intervals from a movie (see Movie S3 in the supplemental material).
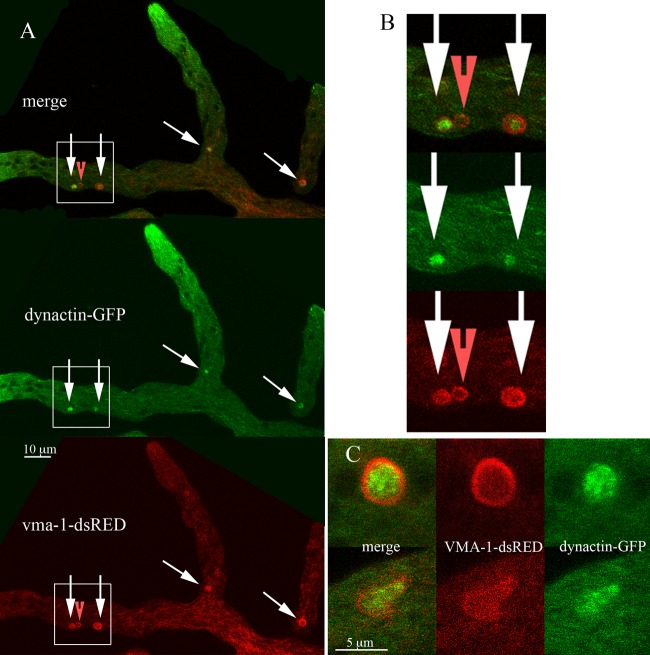
In our observations, we did not see vesicles or other organelles such as mitochondria within PVCs (data not shown). However, an unrelated investigation of the cytoskeleton in N. crassa led to the serendipitous observation that dynein and dynactin were associated with the PVCs (55). As shown in Fig. 10A, dynactin-GFP was observed on filaments at the hyphal tip (presumably attached to microtubules) and also as roughly spherical globules in the region of the hyphae where PVCs were observed. VMA-1–dsRED appeared to surround and enclose the dynactin-GFP. However, as shown in Fig. 10A and B, not all PVCs appeared to contain dynactin-GFP. In other experiments, “empty” PVCs were consistently seen. At a higher magnification, it appeared that the dynactin-GFP was inside the spherical PVCs (Fig. 10B and C). We were fortunate to catch a few PVCs in the process of blebbing, one of which is shown in Fig. 10C. Dynactin-GFP had the altered shape of the PVC and appeared to be enclosed within.
FIG 10.
Localization of dynactin compared to the localization of the vacuolar ATPase (VMA-1). (A) Low-magnification image of the heterokaryon. Arrows point to the PVCs. The red arrowhead points to a PVC that does not appear to contain dynactin-GFP. (B) Boxed regions of panel A at a higher magnification. (C) Two PVCs from other hyphae at a higher magnification.
DISCUSSION
Several years ago, while using confocal microscopy to identify organelles in N. crassa, we observed a cellular compartment that appeared to be different from anything described in the research literature (13). The information in this report allows us to hypothesize a function for this unidentified organelle. We propose that it is a prevacuolar compartment that is involved in the formation of the tubular vacuolar network in N. crassa.
PVCs are enriched in a subset of vacuolar proteins (Table 2). These include the vacuolar ATPase, a calcium transporter, the polyphosphate polymerase, and the Rab-7 GTPase. The PVCs either lack or have small amounts of other vacuolar proteins such as carboxypeptidase Y and alkaline phosphatase. In previous reports, vacuoles have been shown to contain significant amounts of proteins that function in the plasma membrane, such as the plasma membrane proton pump (PMA-1) and two plasma membrane calcium pumps (NCA-2 and NCA-3) (13, 38). These plasma membrane proteins were not observed in the PVCs. Tagged marker proteins for other organelles—nuclei, the ER, the Golgi complex, peroxisomes, and mitochondria—were not observed in vacuoles or PVCs.
TABLE 2.
Proteins localized by confocal microscopy
| Protein | Presence of protein in: |
|
|---|---|---|
| PVC | Vacuole | |
| Plasma membrane ATPase (PMA-1) | ✓ | |
| Calcium ATPase (NCA-2) | ✓ | |
| Calcium ATPase (NCA-3) | ✓ | |
| t-SNARE (PEP-12) | ✓ | |
| Carboxypeptidase Y (CPY) | ✓ | |
| Alkaline phosphatase (PHO-8) | ✓ | |
| V-ATPase subunit A (VMA-1) | ✓ | ✓ |
| V-ATPase subunit C (VMA-5) | ✓ | ✓ |
| Calcium/H+ transporter (CAX) | ✓ | ✓ |
| Polyphosphate polymerase (VTC-4) | ✓ | ✓ |
| RAB-2 GTPase | ✓ | ✓ |
| RAB-4 GTPase | ✓ | ✓ |
| RAB-7 GTPase | ✓ | ✓ |
PVCs can be visualized without the use of fluorescently tagged proteins. Adenine auxotrophs of N. crassa, such as the ad-3B strain, accumulate a red pigment in vacuoles when grown with low concentrations of adenine (45). For S. cerevisiae, it has been shown that these adenine auxotrophs accumulate an intermediate in the synthesis of adenine. This compound is conjugated to glutathione in the cytosol and then moved into the vacuole by at least two specific transport proteins in the vacuolar membrane (56). The envelope that forms the N. crassa PVC apparently contains at least one such transporter, because PVCs were easily observed in ad-3B strains grown with low concentrations of adenine. Of interest, the red pigment filled the lumen of spherical vacuoles but did not fill the lumen of round PVCs.
Observation of the localization of another soluble vacuolar marker, carboxy-DFFDA, was informative. This molecule becomes fluorescent when activated by esterases within the vacuole. As shown previously, it is an excellent dye for visualizing the tubular vacuolar network and spherical vacuoles (9, 57). Some PVCs, but not all, fluoresced in the presence of carboxy-DFFDA. As with the ad-3B strains, fluorescence was seen at the periphery of the PVCs (i.e., as a ring). Most PVCs, those nearest the hyphal tip, did not fluoresce, but the most distal ones did. A possible explanation is that PVCs acquire other vacuolar proteins, such as the esterases that hydrolyze carboxy-DFFDA, with the most “mature” PVCs in the most distal position. The idea that the composition of PVCs varies with their distance from the hyphal tip is also suggested by the localization of RAB-2–dsRED. This protein is associated with PVCs at the hyphal tip but is less abundant in the most distal PVCs. S. cerevisiae has no homolog of RAB-2. Although present in filamentous fungi, its function in these organisms has not been reported.
Our observations of the N. crassa PVC are consistent with the hypothesis that this compartment has a function analogous to that of the late endosome or the multivesicular body in S. cerevisiae. Analysis of the cellular location of RAB proteins supports this idea. RAB proteins that function in earlier steps in the protein targeting pathway, RAB-1, -5A, -5B, -6, and -11, do not appear to colocalize with PVCs. RAB-7, a well-documented component of late endosomes and vacuoles/lysosomes in yeast and animal cells, colocalizes with other PVC proteins.
We did not know what to expect with Rab-2 and -4 because these proteins are not present in most yeasts and, to our knowledge, have not been investigated in any fungus (30–32). RAB-2 appeared to associate with PVCs, although partly in a chimeric fashion. Hyphae expressing RAB-2–dsRED and VMA-1–GFP had PVCs with distinct red and green regions. The localization of RAB-4 was virtually the same as that of RAB-7 or VMA-1. It was clearly a component of PVCs. An investigation of the evolution of fungal rab genes reported difficulty in identifying the orthologues of this pair of fungal RAB GTPases (32). The protein encoded by NCU00889, named RAB-4, is likely an ortholog of mammalian RAB-4, but we suspect that NCU01647, named RAB-2, is not an ortholog of mammalian RAB-2. When we aligned the sequences of N. crassa and mouse Rab proteins, we found that N. crassa RAB-2 and RAB-4 are more closely related to each other and to mouse RAB4 than they are to any of the other Rab proteins. Our observations suggest that RAB-4, like RAB-7, has a role in targeting proteins to the vacuole. RAB-2 may have a similar role, but it appears to be partially absent from the PVCs that are most distal from the hyphal tip.
The association of RAB-7 with N. crassa PVCs is consistent with these compartments functioning as a multivesicular body (MVB). However, the size, shape, and protein components of the N. crassa PVC are different from those of the well-studied MVB in S. cerevisiae in several ways. The MVB is a much smaller organelle, <0.5 μm in diameter (14). N. crassa PVCs are highly variable in shape, with many about the size of nuclei, ∼3 μm. We have not seen vesicles within the PVCs, but small vesicles that would be observable in electron micrographs may not be resolvable with a confocal microscope. In S. cerevisiae, alkaline phosphatase and carboxypeptidase Y take different routes to the vacuole, the latter going via the MVB. In N. crassa, neither of these proteins localized to the PVC. A well-characterized component of the MVB of S. cerevisiae is the t-SNARE Pep12p, which interacts with another t-SNARE with a very similar sequence, Vam3p (58). In fact, overexpression of Pep12p can suppress the phenotype of ΔVam3p and vice versa (59, 60). The genomes of N. crassa and other filamentous fungi encode only one protein with a similar sequence, which has led to an inconsistent naming of this gene. Kitamoto's group, working with Aspergillus oryzae, named the gene Aovam3 (61). We used the N. crassa ortholog (NCU06777) and named it vam-3. Previous to our initial publication (13) and that of Kitamoto et al., Gupta and Brent Heath analyzed the genomes of multiple fungi and concluded that VAM3 was almost universally absent; they named the gene pep-12, the name that we have subsequently used (31, 62). Kitamoto et al. reported that AoVam3-GFP localized to vacuoles and to small punctate structures adjacent to large spherical vacuoles (8). Unlike the vacuoles, the small punctate structures did not stain with the molecule CMAC (7-amino-4-chloromethylcoumarin), suggesting that these puncta could be prevacuolar compartments. Our observations with N. crassa were different: PEP-12 was seen only in the tubular vacuolar network and in large vacuoles but not in the PVCs (13). For proteins like PEP-12, carboxypeptidase, and alkaline phosphatase, we cannot conclude absolutely that they are not present in the PVC, but they are not visible with a confocal microscope, whereas other proteins, like VMA-1 and RAB-7, appear to be more abundant in PVCs than in the vacuole.
Perhaps the most striking features of the N. crassa PVC are its large size and dynamic structure. All the tagged proteins that mark the PVC and the red pigment of the ad-3B strain are seen in tubules and rings of various sizes. Although roughly spherical, these structures sometimes appear to have an open side. A split membrane was occasionally observed with very large PVCs (see Movie S4 in the supplemental material). The images suggest that PVCs can be folded or distorted, an observation that merits further investigation. The movies in the supplemental material show that PVCs can change shape rapidly, with round structures blebbing to produce long tubules. These dynamic shape changes occur in a very small region of the hypha, near the apical end of the tubular vacuolar network. We found one report of similar organelles. Hyphal tips of the basidiomycete Phanerochaete velutina, when stained with carboxy-DFFDA, were observed to have dynamic, ring-like structures in the tip region just in front of the tubular vacuolar network (57). The authors of that report suggested that these structures were either a distinct subcompartment of the vacuole or some other organelle.
Our observations generate many questions. For example, we have not yet looked at the role of molecular motors and the cytoskeleton in positioning and shaping the PVCs. The fact that dynein and dynactin are associated with the PVCs is consistent with the findings in other organisms that these proteins associate with effectors of RAB-7 (29, 63). At this stage of the investigation, our data allow us to formulate a testable hypothesis. The hypothesis is that the N. crassa PVC functions like a late endosome/multivesicular body. A subset of vacuolar proteins, especially integral membrane proteins, is delivered to the PVC. Subsequent reorganization of the PVC envelope produces tubular elements that either fuse to or mature into elements of the tubular vacuolar network.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ben Abrams, Facilities Manager of the UCSC Life Sciences Microscopy Center, for his assistance and advice.
This study was supported by Public Health Service grant GM058903 from the Institute of General Medicine to B.J.B.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00128-15.
REFERENCES
- 1.Bowman EJ, Bowman BJ. 2010. Vacuoles in filamentous fungi, p 179–190. In Borkovich K, Ebbole DJ (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC. [Google Scholar]
- 2.Klionsky DJ, Herman PK, Emr SD. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 54:266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards A, Veses V, Gow NAR. 2010. Vacuole dynamics in fungi. Fungal Biol Rev 24:93–105. doi: 10.1016/j.fbr.2010.04.002. [DOI] [Google Scholar]
- 4.Veses V, Richards A, Gow NA. 2008. Vacuoles and fungal biology. Curr Opin Microbiol 11:503–510. doi: 10.1016/j.mib.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Li SC, Kane PM. 2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta 1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 7.Cole L, Orlovich DA, Ashford AE. 1998. Structure, function, and motility of vacuoles in filamentous fungi. Fungal Genet Biol 24:86–100. doi: 10.1006/fgbi.1998.1051. [DOI] [PubMed] [Google Scholar]
- 8.Shoji JY, Arioka M, Kitamoto K. 2006. Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryot Cell 5:411–421. doi: 10.1128/EC.5.2.411-421.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickey PC, Swift SR, Roca MG, Read ND. 2004. Live-cell imaging of filamentous fungi using vital fluorescent dyes and confocal microscopy. Methods Microbiol 34:63–87. doi: 10.1016/S0580-9517(04)34003-1. [DOI] [Google Scholar]
- 10.Shepherd VA, Orlovich DA, Ashford AE. 1993. A dynamic continuum of pleiomorphic tubules in filamentous fungi. J Cell Sci 104:495–507. [Google Scholar]
- 11.Araujo-Palomares CL, Castro-Longoria E, Riquelme M. 2007. Ontogeny of the Spitzenkorper in germlings of Neurospora crassa. Fungal Genet Biol 44:492–503. doi: 10.1016/j.fgb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Riquelme M, Roberson RW, McDaniel DP, Bartnicki-Garcia S. 2002. The effects of ropy-1 mutation on cytoplasmic organization and intracellular motility in mature hyphae of Neurospora crassa. Fungal Genet Biol 37:171–179. doi: 10.1016/S1087-1845(02)00506-6. [DOI] [PubMed] [Google Scholar]
- 13.Bowman BJ, Draskovic M, Freitag M, Bowman EJ. 2009. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell 8:1845–1855. doi: 10.1128/EC.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowers K, Stevens TH. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Wickner W. 2010. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol 26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 16.Cowles CR, Snyder WB, Burd CG, Emr SD. 1997. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J 16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piper RC, Bryant NJ, Stevens TH. 1997. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol 138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. 1994. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell 77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 19.Sipos G, Brickner JH, Brace EJ, Chen L, Rambourg A, Kepes F, Fuller RS. 2004. Soi3p/Rav1p functions at the early endosome to regulate endocytic trafficking to the vacuole and localization of trans-Golgi network transmembrane proteins. Mol Biol Cell 15:3196–3209. doi: 10.1091/mbc.E03-10-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha SA, Torabinejad J, DeWald DB, Wenk MR, Lucast L, De Camilli P, Newitt RA, Aebersold R, Nothwehr SF. 2003. The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol Biol Cell 14:1319–1333. doi: 10.1091/mbc.E02-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 22.Pfeffer SR. 2013. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol 25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekena K, Vater CA, Raymond CK, Stevens TH. 1993. The VPS1 protein is a dynamin-like GTPase required for sorting proteins to the yeast vacuole. Ciba Found Symp 176:198–211, discussion 211–194. [DOI] [PubMed] [Google Scholar]
- 24.Tarutani Y, Ohsumi K, Arioka M, Nakajima H, Kitamoto K. 2001. Cloning and characterization of Aspergillus nidulans vpsA gene which is involved in vacuolar biogenesis. Gene 268:23–30. doi: 10.1016/S0378-1119(01)00418-8. [DOI] [PubMed] [Google Scholar]
- 25.Raiborg C, Rusten TE, Stenmark H. 2003. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol 15:446–455. doi: 10.1016/S0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 26.Tatsumi A, Shoji JY, Kikuma T, Arioka M, Kitamoto K. 2007. Aggregation of endosomal-vacuolar compartments in the Aovps24-deleted strain in the filamentous fungus Aspergillus oryzae. Biochem Biophys Res Commun 362:474–479. doi: 10.1016/j.bbrc.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 27.Schimmoller F, Riezman H. 1993. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J Cell Sci 106(Part 3):823–830. [DOI] [PubMed] [Google Scholar]
- 28.Ohsumi K, Arioka M, Nakajima H, Kitamoto K. 2002. Cloning and characterization of a gene (avaA) from Aspergillus nidulans encoding a small GTPase involved in vacuolar biogenesis. Gene 291:77–84. doi: 10.1016/S0378-1119(02)00626-1. [DOI] [PubMed] [Google Scholar]
- 29.Abenza JF, Galindo A, Pinar M, Pantazopoulou A, de los Rios V, Penalva MA. 2012. Endosomal maturation by Rab conversion in Aspergillus nidulans is coupled to dynein-mediated basipetal movement. Mol Biol Cell 23:1889–1901. doi: 10.1091/mbc.E11-11-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, Zelter A, Kalkman ER, O'Rourke R, Bowring F, Yeadon J, Ishii C, Suzuki K, Sakai W, Pratt R. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta GD, Brent Heath I. 2002. Predicting the distribution, conservation, and functions of SNAREs and related proteins in fungi. Fungal Genet Biol 36:1–21. doi: 10.1016/S1087-1845(02)00017-8. [DOI] [PubMed] [Google Scholar]
- 32.Pereira-Leal JB. 2008. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9:27–38. doi: 10.1111/j.1600-0854.2007.00667.x. [DOI] [PubMed] [Google Scholar]
- 33.Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol 41:897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Honda S, Selker EU. 2009. Tools for fungal proteomics: multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182:11–23. doi: 10.1534/genetics.108.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freitag M, Selker EU. 2005. Expression and visualization of red fluorescent protein (RFP) in Neurospora crassa. Fungal Genet Newsl 52:14–17. [Google Scholar]
- 36.McCluskey K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv Appl Microbiol 52:245–262. doi: 10.1016/S0065-2164(03)01010-4. [DOI] [PubMed] [Google Scholar]
- 37.Hutagalung AH, Novick PJ. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fajardo-Somera RA, Bowman B, Riquelme M. 2013. The plasma membrane proton pump PMA-1 is incorporated into distal parts of the hyphae independently of the Spitzenkorper in Neurospora crassa. Eukaryot Cell 12:1097–1105. doi: 10.1128/EC.00328-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis RH. 2000. Neurospora. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 40.Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. 2004. HP1 is essential for DNA methylation in Neurospora. Mol Cell 13:427–434. doi: 10.1016/S1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- 41.McNally MT, Free SJ. 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Curr Genet 14:545–551. doi: 10.1007/BF00434079. [DOI] [PubMed] [Google Scholar]
- 42.Loros JJ, Dunlap JC. 1991. Neurospora crassa clock-controlled genes are regulated at the level of transcription. Mol Cell Biol 11:558–563. doi: 10.1128/MCB.11.1.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roper M, Ellison C, Taylor JW, Glass NL. 2011. Nuclear and genome dynamics in multinucleate ascomycete fungi. Curr Biol 21:R786–R793. doi: 10.1016/j.cub.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisman LS, Bacallao R, Wickner W. 1987. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J Cell Biol 105:1539–1547. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Serres FJ, Kolmark HG. 1958. A direct method for determination of forward-mutation rates in Neurospora crassa. Nature 182:1249–1250. doi: 10.1038/1821249a0. [DOI] [PubMed] [Google Scholar]
- 46.Boyce KJ, Kretschmer M, Kronstad JW. 2006. The vtc4 gene influences polyphosphate storage, morphogenesis, and virulence in the maize pathogen Ustilago maydis. Eukaryot Cell 5:1399–1409. doi: 10.1128/EC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, Ladurner AG, Herrmann C, Scheffzek K, Mayer A. 2009. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324:513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 48.Klionsky DJ, Emr SD. 1989. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J 8:2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryant NJ, Stevens TH. 1998. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev 62:230–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, Pantazopoulou A. 2012. Searching for gold beyond mitosis: mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist 2:2–14. doi: 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Leon E, Bowman B, Seidel C, Fischer R, Novick P, Riquelme M. 2015. The Rab GTPase YPT-1 associates with Golgi cisternae and Spitzenkorper microvesicles in Neurospora crassa. Mol Microbiol 95:472–490. doi: 10.1111/mmi.12878. [DOI] [PubMed] [Google Scholar]
- 52.Seidel C, Moreno-Velasquez SD, Riquelme M, Fischer R. 2013. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth. Eukaryot Cell 12:1020–1032. doi: 10.1128/EC.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holthuis JC, Nichols BJ, Pelham HR. 1998. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol Biol Cell 9:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klionsky DJ, Cuervo AM, Seglen PO. 2007. Methods for monitoring autophagy from yeast to human. Autophagy 3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 55.Sivagurunathan S, Schnittker RR, Nandini S, Plamann MD, King SJ. 2012. A mouse neurodegenerative dynein heavy chain mutation alters dynein motility and localization in Neurospora crassa. Cytoskeleton (Hoboken) 69:613–624. doi: 10.1002/cm.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma KG, Kaur R, Bachhawat AK. 2003. The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch Microbiol 180:108–117. doi: 10.1007/s00203-003-0566-z. [DOI] [PubMed] [Google Scholar]
- 57.Zhuang X, Tlalka M, Davies DS, Allaway WG, Watkinson SC, Ashford AE. 2009. Spitzenkorper, vacuoles, ring-like structures, and mitochondria of Phanerochaete velutina hyphal tips visualized with carboxy-DFFDA, CMAC and DiOC6(3). Mycol Res 113:417–431. doi: 10.1016/j.mycres.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 58.Gerrard SR, Levi BP, Stevens TH. 2000. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic 1:259–269. doi: 10.1034/j.1600-0854.2000.010308.x. [DOI] [PubMed] [Google Scholar]
- 59.Darsow T, Rieder SE, Emr SD. 1997. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol 138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerrard SR, Mecklem AB, Stevens TH. 2000. The yeast endosomal t-SNARE, Pep12p, functions in the absence of its transmembrane domain. Traffic 1:45–55. doi: 10.1034/j.1600-0854.2000.010108.x. [DOI] [PubMed] [Google Scholar]
- 61.Kuratsu M, Taura A, Shoji JY, Kikuchi S, Arioka M, Kitamoto K. 2007. Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae. Fungal Genet Biol 44:1310–1323. doi: 10.1016/j.fgb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 62.Bowman BJ, Abreu S, Johl JK, Bowman EJ. 2012. The pmr gene, encoding a Ca2+-ATPase, is required for calcium and manganese homeostasis and normal development of hyphae and conidia in Neurospora crassa. Eukaryot Cell 11:1362–1370. doi: 10.1128/EC.00105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T, Ming Z, Xiaochun W, Hong W. 2011. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal 23:516–521. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.