Abstract
Scope
The microorganisms that make up kefir grains are well known for lactose fermentation, but the extent to which they hydrolyze and consume milk proteins remains poorly understood. Peptidomics technologies were used to examine the proteolytic activity of kefir grains on bovine milk proteins.
Methods and results
Gel electrophoresis revealed substantial digestion of milk proteins by kefir grains, with mass spectrometric analysis showing the release of 609 protein fragments and alteration of the abundance of >1,500 peptides that derived from 27 milk proteins. Kefir contained 25 peptides identified from the literature as having biological activity, including those with antihypertensive, antimicrobial, immunomodulatory, opioid and anti-oxidative functions. 16S rRNA and shotgun metagenomic sequencing identified the principle taxa in the culture as Lactobacillus species.
Conclusion
The model kefir sample contained thousands of protein fragments released in part by kefir microorganisms and in part by native milk proteases.
Keywords: Functional peptide, Kefir, Milk, Peptidomics, Protease
1 Introduction
Food fermentation is one of the oldest methods of extending shelf-life, and more than 3,500 fermented foods are known (Farnworth, 2003). Kefir, an acidic, fermented milk beverage that originated thousands of years ago in the Caucasus Mountains, is still consumed worldwide. To produce it, kefir grains—a complex of polysaccharides, proteins, symbiotic lactic acid bacteria (e.g. Lactobacillus, Lactococcus, Leuconostoc and Streptococcus) and yeast (e.g. Saccharomyces, Candida, Kluyveromyces, Debaryomyces and Torulaspora)—are incubated with heat-treated milk under aerobic conditions (Angulo, Lopez, & Lema, 1993; Leite, Miguel, Peixoto, Rosado, Silva, & Paschoalin, 2013; S.-Y. Wang, Chen, Lo, Chiang, Chen, Liu, et al., 2012). The fermentation of milk lactose by these microorganisms results in acidification of the product, which prevents the growth of spoilage organisms. Typically, kefir grains are inoculated at 2–8% concentration and allowed to incubate for 18–24 h at 20–25 °C (Otles & Cagindi, 2003). After incubation, the kefir is allowed to mature further at 4 °C for 20–24 hours (Otles & Cagindi, 2003).
Though the lactic acid bacteria in kefir are known to hydrolyze casein, which is critical for texture and flavor development (Kunji, Mierau, Hagting, Poolman, & Konings, 1996), the extent to which caseins and other milk proteins are hydrolyzed by kefir microorganisms remains unclear. This study employed mass spectrometry-based peptidomics and gel electrophoresis to examine the peptides released from bovine milk proteins by the kefir microorganisms. To determine whether peptides in kefir were the result of kefir microorganism activity, were naturally occurring peptides, or were released by native milk proteases during incubation, this study compared peptides in heat- and kefir-treated raw milk with those in unincubated raw milk and in heat-treated incubated milk without kefir.
Milk peptides released during kefir fermentation were examined for homology with milk peptides known to have antimicrobial, antihypertensive, immunomodulatory, opioid and prebiotic properties (Clare & Swaisgood, 2000). Determining which functional peptides are released can lead to exploration of possible peptide-induced health benefits from kefir consumption.
2 Materials and methods
2.1 Samples
Fresh milk was collected from a pool of six healthy Holstein cows at the University of California, Davis (USA) as described previously (D. Dallas, Guerrero, Parker, Garay, Bhandari, Lebrilla, et al., 2013). Before attaching the milking pumps, all four teats were washed with water and then dipped in an antiseptic solution (Chlorhexidine Active Mastitis Prevention) with 0.5% chlorhexidine gluconate as the active ingredient. The milk was immediately frozen at −30 °C until use. Kefir grains were purchased from Fusion Teas (McKinney, Texas, USA) and were preserved in pasteurized milk at 4 °C.
2.2 Sample preparation
After thawing and gently mixing, the freshly collected, frozen raw milk was apportioned into twelve 1-mL subsamples (3 subsamples for each of the 4 study groups). Three subsamples were frozen at −20 °C to serve as the untreated control (Raw Milk, RM). The nine remaining subsamples were heated at 93 °C for 7 min using a thermomixer (Thermo Mixer C, Eppendorf, Hamburg, Germany), cooled in an ice bath for 20 min and brought to room temperature. This heat treatment was selected based on previous literature on kefir production (Otles & Cagindi, 2003). To have a concentration of 4.15% kefir grains, 41.5 mg of kefir grains were added to three of the nine samples (Kefir, group K). Prior to collection, the kefir grain supply was thoroughly mixed to ensure a representative sample was collected. The nine samples were incubated on a thermomixer at 23 °C for 24 h at 800 rpm and then matured at 4 °C for 24 h. During the incubation and maturation steps, the three sample vials with kefir grains (group K) were kept open to match the aerobic conditions typical for kefir production. To control for any environmental contamination by airborne microorganisms, three of the six vials of heat-treated milk without kefir grains were closed (heat-treated milk with closed vials, group HMc), whereas the remaining three were open (heat-treated milk with open tubes, group HMo). The three vials of raw milk (group RM) were defrosted and all 12 subsamples were centrifuged at 16,000 ×g at 4.5 °C for 10 min to separate and remove the milk fat and 500 μL of delipidated milk were collected from each vial.
2.2.1 Trichloroacetic acid protein precipitation
Skim milk proteins in each of the twelve subsamples were precipitated by addition of trichloroacetic acid (200 g/L TCA, EMD Millipore, Darmstadt, Germany) in a 1:1 v/v ratio. After centrifugation at 4,000 ×g, at 20 °C for 10 min, 850 μL of the supernatant from each of the twelve subsamples were collected for extraction of peptides.
2.2.2 Extraction of peptides with C18 microplate
Sugars, salt and TCA were removed from the supernatants using a C18 solid-phase extraction microplate procedure (D. C. Dallas, Smink, Robinson, Tian, Guerrero, Parker, et al., 2015) with no modifications. Peptide fractions were dried by centrifugal evaporation (miVac Quattro, Genevac, Ipswich, UK) at 44 °C and preserved at −20 °C.
2.3 Sample analysis
2.3.1 Mass spectrometry-based peptide analysis
LC separation was performed on a Waters Nano Acquity UHPLC (Waters Corporation) with a Proxeon nanospray source. The peptides were reconstituted in 2% ACN, 0.1% TFA. Two micrograms of each sample were loaded onto the column based on measured absorbance at 280 nm. Peptides were first loaded onto the trap column (a 100 μm × 25 mm Magic C18 100Å 5U reverse-phase column) for online desalting and then onto a 75 μm × 150 mm Magic C18 200Å 3U reverse-phase column (Waters, Milford, MA) for analytical separation. Peptides were eluted using a gradient of 0.1% formic acid (A) and 100% acetonitrile (B) with a flow rate of 300 nL/min. The 60-min gradient was designed as follows: 5–35% B over 50 min, 35–80% B over 3 min, 80% B for 1 min, 80–5% B over 1 min and then held at 5% B for 5 min. Each sample injection was followed by a 30 min column wash.
Mass spectra were collected on a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) in a data-dependent mode with one MS precursor scan followed by 15 MS/MS scans. A dynamic exclusion of 20 s was used. MS spectra were acquired with a resolution of 70,000 and a target of 1 × 106 ions or a maximum injection time of 30 ms. MS/MS spectra were acquired with a resolution of 17,500 and a target of 5 × 104 ions or a maximum injection time of 50 ms. Peptide fragmentation was performed using higher-energy collision dissociation with a normalized collision energy value of 27. Unassigned charge states as well as ions > +6 were excluded from MS/MS fragmentation.
2.3.2 Spectral analysis and peptide identification
Spectra were analyzed by database searching in X!Tandem as described previously (D. Dallas, Guerrero, Khaldi, Castillo, Martin, Smilowitz, et al., 2013; D. Dallas, Guerrero, Parker, et al., 2013), with minor modifications. No complete (required) modifications or potential modifications were allowed. Spectra were searched against a bovine milk library compiled from previous bovine milk proteome literature. The data were deposited to the ProteomeXchange with identifier PXD001826.
2.3.3 Peptide peak area determination
An in-house curated bovine milk protein library was imported in .fasta file format into Skyline (Schilling, Rardin, MacLean, Zawadzka, Frewen, Cusack, et al., 2012). A library of identified peptides was uploaded from the .xml outputs of the X!Tandem program for each sample to create the spectral library. After applying all settings, the spectral library was searched against the raw data files (.raw) to extract the peaks for each peptide in each sample.
The settings for the extraction were as follows. Precursor mass was calculated based on the monoisotopic ion. Allowed precursor charges were 1–6. Ion types were set as precursor only. The ion match tolerance was set to 0.5 m/z. The instrument acquisition window was set between 300 and 1,600 m/z with a tolerance of 0.055 m/z. For MS1 filtering, isotope peaks included by count were employed. The precursor mass analyzer was set to “Orbitrap.” The resolving power was set to 60,000 at 400 m/z. The precursor isotopic import filter was set to a count of 3 (M, M+1, M +2). MS/MS filtering was set to none. Retention time filtering was set to 1 min of the MS/MS identifications.
After data import, all peaks were manually inspected for proper peak picking of the MS1-filtered peptides. Only peaks with ≤3 ppm mass error and an idotp score of ≥80 were retained. Peaks were selected based on mass error and retention time proximity to the identified peptide retention time. Peaks that did not match these criteria were deleted. Peaks too close to the noise level to be visually discernable were excluded.
After manual inspection, the data were exported to a .csv file. To reassign the protein name to peptide sequence and collapse multiple charge states into a single compound, the file was processed through an in-house script.
2.3.4 Functional peptide search
Identified peptide sequences from the samples were searched against a library of known functional milk peptides from the literature (Hayes, Stanton, Fitzgerald, & Ross, 2007; Maruyama, Nakagomi, Tomizuka, & Suzuki, 1985; Minervini, Algaron, Rizzello, Fox, Monnet, & Gobbetti, 2003; Recio & Visser, 1999). Peptides in the samples that completely encompassed a known functional peptide were counted for the bioactive peptide table, and only peptides with 100% homology to known functional peptides were reported.
2.3.5 Electrophoresis gel
Another set of samples for the four experimental groups were created in triplicate. After maturation, the fat was removed by centrifugation at 16,000 ×g at 4.5 °C for 10 min. The upper fat layer was removed with a pipette tip and the mix of skim and pelleted proteins for each sample were collected and frozen at −20 °C. The centrifugally pelleted proteins were intentionally collected as part of the sample for gel electrophoresis to avoid potential loss of casein micelles. In addition, five commercial kefir products were purchased and examined in the gel as a comparison.
To determine whether milk proteins were hydrolyzed extensively by the kefir microbiota, electrophoresis was performed using a 12% polyacrylamide gel (Kesenkas, Yerlikaya, & Ozer, 2013). The quantity of proteins in each sample was determined with the A280 method. Twenty microliters of each sample were mixed with 6.6 μL of 4x Laemmli loading buffer (277.8 mM Tris-HCl, pH 6.8, 4.4% lithium dodecyl sulfate, 44.4% (w/v) glycerol, 0.02% bromophenol blue (Bio-Rad)) and 2.75 μL of 1 M DTT, and heated at 95 °C for 5 min. Each sample (50 μg) was loaded into the gel using a Mini Protean II apparatus (Bio-Rad). 200 V were applied to the gel for 35 min at room temperature. The gel was fixed in three baths of nanopure water for 15 min each, stained with BioSafe Coomassie G 250 solution (Bio-Rad) and destained in nanopure water overnight. Sample bands were compared with 10 blue-stained recombinant protein bands (molecular weight standards) from 10 to 250 kDa (Bio-Rad).
2.4 Statistical analysis
All statistical analyses were conducted in R software, version 3.1.2. Peptides with p ≤0.05 were considered significant.
2.4.1 Number of unique peptides across treatment group
The identified peptides were grouped by milk treatment (raw milk, RM; milk with heat treatment and fermented by kefir grains, K; heat-treated milk with tubes opened, HMo; heat-treated milk with tubes closed, HMc). The number of peptides in each sample was modeled as a function of treatment using the generalized linear model with quasi-Poisson distribution. For pairwise comparisons, the Tukey’s familywise comparison was employed.
2.4.2 Total peptide abundance across sample groups
To model the overall mean abundance of peptides, a linear regression model was applied. Analysis of variance (ANOVA) was used to compare the groups with this model.
2.4.3 Individual peptide peak area across sample groups
To compare the peak areas of individual peptides, all missing values were transformed to “1” for log transformation. Peptides absent from >75% of the samples (i.e. >9 samples) were omitted. Peak areas were transformed into natural log scale. To examine the main effect of the different treatments, a linear regression model was built for each peptide’s abundance across the four groups. Peptides that had a significant treatment effect were analyzed via post-hoc testing of all pairwise comparisons between the different groups with Tukey’s honest significant difference (HSD) method.
2.4.4 Number of peptides per protein across sample groups
To model the number of unique peptides for each protein as a function of treatment (sample groups), the generalized linear model with quasi-Poisson distribution was applied. For proteins with significant differences, Tukey’s HSD methods were employed for all pairwise comparisons between groups. No proteins were removed from this analysis as none was absent in >75% of samples.
2.4.5 Peptide abundance (summed peptide intensities) per protein across sample groups
Peptide intensities across groups on the protein level were compared by one-way ANOVA. The Tukey’s HSD test was employed to determine which proteins differed in total peptide abundance across treatment groups. Proteins included in this comparison were the most abundant by peptide count and intensity.
2.5 Sequence alignment visualization
Peptide sequences identified were aligned to the protein sequences and their intensities were summed to visualize the origins of peptides from each protein across the sample groups, as described previously (A. Guerrero, Dallas, Contreras, Chee, Parker, Sun, et al., 2014).
2.6 Microbial analysis
Total genomic DNA was extracted from 150 mg kefir grain samples (performed in duplicate) using the ZR Fecal DNA Miniprep Kit (Zymo Research, Irvine, CA). DNA library construction was carried out as previously described with the exception that the PCR reaction conditions were an initial 94°C for 3 min, followed by 25 cycles of 94°C for 45 sec, 50°C for 60 sec, and 72°C for 90 sec with a final extension at 72°C for 10 min (Bokulich, Thorngate, Richardson, & Mills, 2014). The DNA library was submitted to the UC Davis Genome Center DNA Technologies Core for sequencing on an Illumina MiSeq instrument. The resulting sequencing data was analyzed with QIIME software package 1.8.0, which was used for quality filtering and demultiplexing (Caporaso, Lauber, Walters, Berg-Lyons, Lozupone, Turnbaugh, et al., 2011). Operational taxonomic units were assigned using UCLUST based on 97% pairwise identity (Edgar, 2010) and taxonomic classification was based on the Ribosomal Database Project classifier against a representative subset of the greengenes 16S rRNA database (gg_otus_13_8 release) (DeSantis, Hugenholtz, Larsen, Rojas, Brodie, Keller, et al., 2006; Q. Wang, Garrity, Tiedje, & Cole, 2007).
For metagenomics analysis, genomic DNA from kefir was prepared for 200bp single reads on an Ion Torrent Proton, which yielded 11,572,313 sequences with a mean sequence length of 148 bp after the removal of DNA mapped as originating from the bovine genome, quality filtering, and analysis using the MG-RAST server (http://metagenomics.anl.gov), where the data are now publicly available. To estimate the total fungal population, quantitative PCR was carried out on the kefir grains. Primers ITS1f (5′ – CTTGGTCATTTAGAGGAAGTAA – 3′) and ITS2 (5′ - GCTGCGTTCTTCATCGATGC – 3′) were used to amplify the ITS region (Smith & Peay, 2014). QPCR was performed on each sample in triplicate in 20 μL reaction mixtures including 2 μL of the DNA template, 1 μL of each 4 μM primer, and 10 μL of 2x SYBR® Premix Ex Taq™ II (Tli RNase H Plus), ROX Plus (Takara Clontech, Mountainview, CA). Fungal cell concentrations were determined by comparison to a standard curve of serially diluted Saccharomyces cerevisiae of known concentration.
3 Results
3.1 pH changes with fermentation
The samples with kefir grains added (K) showed a decrease in pH from an initial mean of 6.55 (SD = 0.007) to a final mean of 4.54 (SD = 0.050) after 48 h of incubation and maturation. No pH change was observed for the samples without kefir grains (the raw milk with no incubation group (RM) and the heat-treated milk groups without kefir with tubes opened (HMo) and closed (HMc) during incubation). The pH commonly decreases during kefir fermentation due to the production of lactic acid (Otles & Cagindi, 2003; Zhou, Liu, Jiang, & Dong, 2009).
3.2 Protein analysis by gel electrophoresis
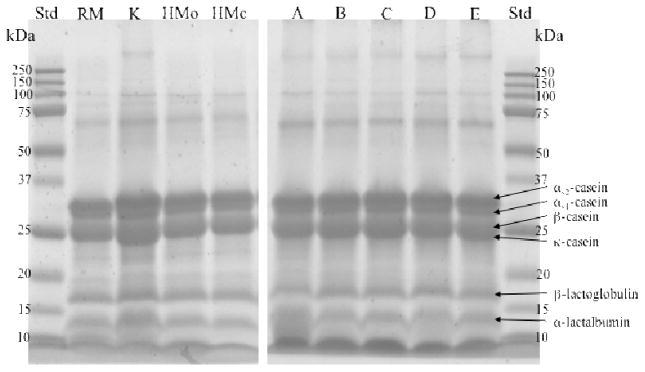
The electrophoresis gel (Fig. 1) shows that all the milk samples, including those with kefir grains (the raw milk with no incubation group (RM), the kefir group (K) and the heat-treated milk groups without kefir with tubes opened (HMo) and closed (HMc) during incubation) had the same protein profiles. Milk proteins, including αs1-casein, αs2-casein, β-casein, κ-casein, β-lactoglobulin and α-lactalbumin, were clearly visible for all groups. The five commercial kefir products (A, B, C, D and E) show the same protein profile as the RM, K, HMo and HMc groups.
Figure 1.
Electrophoresis gel (15% polyacrylamide gel) of the proteins in the raw milk group (RM), the kefir group (K), and the heat-treated milk groups without kefir with tubes opened (HMo) and closed (HMc) during incubation. Lanes A, B, C, D and E are 5 commercial kefir products for comparison.
3.3 Number of peptides identified
The peptidomic technique developed here allows for the most in-depth profile of the naturally occurring milk peptides and those released by kefir microorganisms to date. Overall, 2,689 peptides were identified in the four treatment groups (RM, K, HMo, HMc). All identified peptide sequences were deposited along with the X!Tandem result files and MS/MS raw data in the ProteomeXchange with identifier PXD001826.
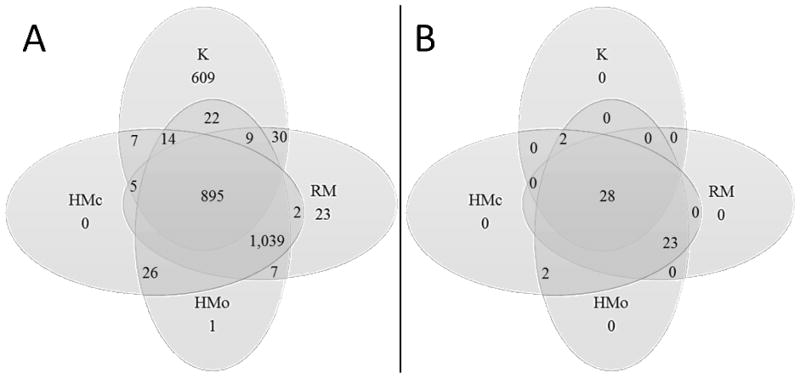
The K samples contained 1,591 peptides. To determine which of these peptides were naturally occurring in milk (D. Dallas, Guerrero, Khaldi, et al., 2013; D. Dallas, Guerrero, Parker, et al., 2013), the K samples were compared with the RM group. To determine which peptides in the K samples were released by native milk enzymes during incubation, the K samples were compared with the two control groups HMo and HMc. Of the peptides identified in kefir, 609 were not present in the three control groups (Fig. 2). These peptides represented, by count, 38.3% of the total peptides identified in kefir (20.3% by abundance). These newly released peptides derived from 20 proteins, including β-casein (38.3% by count, 48.7% by abundance), αs1-casein (14.1% by count, 6.2% by abundance), polymeric immunoglobulin receptor (13.8% by count, 28.1% by abundance), κ-casein (9.2% by count, 3.5% by abundance), glycosylation-dependent cell adhesion molecule 1 (8.1% by count, 4.3% by abundance) and αs2-casein (4.8% by count and 4% by abundance).
Figure 2.
A: Venn diagram of peptide numbers that were the same and unique in each group (Raw milk group, RM; kefir group, K; heat-treated milk groups with vials open, HMo, and with vials closed, HMc). A peptide present in at least one of a triplicate’s samples was considered present. B: Venn diagram of protein numbers that released peptides in the K, RM, HMo and HMc groups. A protein present in at least one of a triplicate’s samples was considered present.
The HMo and the HMc samples contained 63 and 47 peptides, respectively, that were absent from the RM samples. Removing duplicates, the 70 peptides present in the combined list of HMo and HMc samples but absent from RM samples derived from 17 proteins, including β-casein (18 peptides by count), polymeric immunoglobulin receptor (9 peptides), αs1-casein (9 peptides), αs2-casein (7 peptides), κ-casein (5 peptides), glycosylation-dependent cell adhesion molecule 1 (4 peptides), lactoferrin (3 peptides), osteopontin (3 peptides) and other proteins with less than 3 peptides.
One thousand thirty-nine peptides from 23 proteins were present in all three controls (RM, HMo and HMc groups) but absent from the K samples. These 23 proteins included xanthine dehydrogenase/oxidase, actin cytoplasmic 1, platelet glycoprotein 4, synaptosomal-associated protein and staphylococcal nuclease domain-containing protein 1.
3.4 Number of unique peptides in each treatment group
On average, the K samples contained fewer peptides (1,421 ± 42, SD) than the three control group samples (HMo, 1,965 ± 8; HMc, 1,917 ± 46; and RM, 1,942 ± 21). Based on the generalized linear model, the number of unique peptides in the K group was different from each of the three control groups (p <1 × 10−4).
3.5 Total peptide abundance across sample groups
ANOVA based on the linear regression model indicated no significant treatment effect (difference among the groups) based on total peptide abundance (p = 0.6466). The minimum average was 3.10 × 1012 ion counts and the maximum average was 3.91 × 1012 ion counts.
3.6 Individual peptide peak area across sample groups
Around 1,500 peptides were significantly different in intensity between the K group and each of the three control groups (Table 1). The intensities of 58.9% of these peptides were increased in the K group and 37.4% were decreased. HMo compared with RM and HMc compared with RM had fewer significantly different peak areas than K compared with RM (Table 1). Comparison of HMo and HMc revealed the least number of significantly different peptide abundances.
Table 1.
Comparison of statistically significant changes between treatment groups for peptides, proteins by peptide count and proteins by total peptide abundance.
| Comparison | Number of peptides | Number of proteins (by count) | Number of proteins (by peptide abundance) |
|---|---|---|---|
|
| |||
| Global | 1,689 | 51 | 53 |
| K vs. RM | 1,531 | 40 | 45 |
| K vs. HMc | 1,515 | 45 | 45 |
| K vs. HMo | 1,572 | 45 | 49 |
| RM vs. HMc | 217 | 3 | 11 |
| RM vs. HMo | 201 | 4 | 7 |
| HMc vs. HMo | 97 | 1 | 4 |
Column 2: Number of peptides significantly different (p ≤0.05) in abundance in treatment groups. Column 3: Number of proteins significantly different (p ≤0.05) in unique peptide count between treatment groups. Column 4: Number of proteins significantly different (p ≤0.05) in peptide abundance between treatment groups. The raw milk group, RM; the kefir group, K; the heat-treated milk groups with vials open, HMo, and with vials closed, HMc.
3.7 Number of peptides per protein across sample groups
Of the milk proteins, 40–45 were significantly different in peptide count between the K group and the three control groups (RM, HMo and HMc) (Table 1). Only a few proteins differed significantly by count between the HMo and RM samples (three proteins), and between the HMc and RM samples (four proteins). Only one protein differed significantly in peptide count between the HMo and HMc groups.
3.8 Peptide abundance (summed peptide intensities) per protein across sample groups
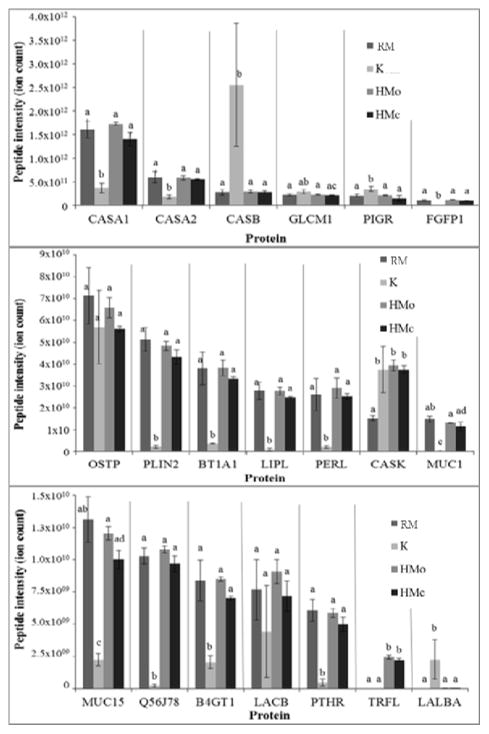
Identified peptides were derived from 55 milk protein precursors. The peptide intensity means for each protein were significantly different across the four treatment groups for 53 out of the 55 proteins identified. Peptides from β-casein, polymeric immunoglobulin receptor and α-lactalbumin were significantly more abundant in K than in the control groups (Fig. 3 and Supplemental Table 1). Peptides derived from αs1-casein, αs2-casein, fibroblast growth factor-binding protein 1, perilipin, butyrophilin, lipoprotein lipase, lactoperoxidase, mucin 1, mucin 15, serum amyloid A protein, β-1,4-galactosyltransferase 1 and parathyroid hormone 1 receptor were significantly less abundant in the K group than in the other groups (Fig. 3 and Supplemental Table 1). As was true for peptide counts, osteopontin (p = 0.319) and β-lactoglobulin (p = 0.161) were the only protein precursors from those selected that were not altered significantly across the groups in terms of peptide abundance means. The p-values of the peptide abundances grouped by protein are shown in the Supplemental Table 1. Forty-five to forty-nine proteins (up to 81% of the total number of proteins identified) in the K group differed significantly in peptide abundance compared with the three control groups (Table 1). Seven and eleven proteins differed by peptide abundance between the RM group and the HMc and HMo groups, respectively. Only four proteins differed between the two heat-treated controls. Proteolytic maps for selected proteins, showing the regions from which the peptides for each treatment group originated in terms of total ion abundance, were created with PepEx software (Supplemental Figs. 1–7) (A. Guerrero, et al., 2014; Parker, 2014). These maps demonstrate that the extent to which specific protein regions were represented by identified peptides changed upon fermentation with kefir microorganisms, even in the cases of β-lactoglobulin and osteopontin (Supplemental Fig. 2), which had no overall statistical difference in peptide abundance.
Figure 3.
Peptide abundance for each protein by treatment group (K, RM, HMo and HMc; see Fig. 1 for abbreviations). The letters (a, b, c) indicate whether the peptide abundances between the different groups were significantly different (p <0.05) for each protein. Values are mean ± SEM. αs1-casein, CASA1; αs2-casein, CASA2; β-casein, CASB; glycosylation-dependent cell adhesion molecule 1, GLCM1; polymeric immunoglobulin receptor, PIGR; fibroblast growth factor-binding protein 1, FGFP1; osteopontin, OSTP; perilipin-2, PLIN2; butyrophilin subfamily 1 member A1, BT1A1; lipoprotein lipase, LIPL; lactoperoxidase, PERL; κ-casein, CASK; mucin-1, MUC1; mucin-15, MUC15; serum amyloid A protein, Q56J78; β-1,4-galactosyltransferase 1, B4GT1; β-lactoglobulin, LACB; parathyroid hormone, PTHR; lactoferrin, TRFL; and α-lactalbumin, LALBA.
3.9 Comparison of identified peptides with the functional peptide database
Peptides in all samples were compared for sequence matches with an in-house database of known functional peptides. Across the categories, 29 peptides had exactly the same sequence as the library peptides (i.e. sequence similarity = 100%) (Table 2). These peptides represented, by count, 0.95% of peptides in samples without kefir grains and 1.76% of samples with kefir grains. Sixteen of these 29 peptides were present in samples from all four groups. All 29 peptides derived from αs1-, αs2-, β- and κ-casein. Five of these peptides were antibacterial, 15 were antihypertensive, 2 were immunomodulatory, 5 were opioid, one a strong anti-oxidant and one (casecidin-17) antimicrobial, antihypertensive, immunomodulatory and a strong anti-oxidant. Of these 29 peptides, the mean abundances of 20 were significantly different between the K group and the control groups (p < 0.05). The peptide abundances of β-casein f.177–183 (β-casokinin-7, antihypertensive), β-casein f.114–121 (casohypotensin, antihypertensive), β-casein f.193–209 (casecidin-17, antimicrobial), β-casein f.114–119 (β-neocasomorphin-6, opioid), β-casein f.166–175 (antihypertensive) and β-casein f.192–209 (immunomodulatory) were significantly higher in the K group than the control groups. Seven functional peptides were present only in the K group: β-casein f.193–207 (casecidin-15, antimicrobial), β-casein f.63–68 (immunomodulatory), β-casein f.183–190 (antihypertensive), β-casein f.60–72 (Pro8-β-casomorphin-13, opioid), β-casein f.60–68 (Pro8-β-casomorphin-9, opioid), β-casein f.73–82 (antihypertensive) and β-casein f.59–68 (V-β-casomorphin-9, anti-oxidative). However, the peptide abundance of αs1-casein f.95–105 (antihypertensive), αs1-casein f.30–37 (antimicrobial), β-casein f.124–133 (antihypertensive), αs1-casein f.25–32 (antihypertensive) and αs1-casein f.24–31(antihypertensive) were significantly lower in the K group than the control groups. Two peptides in the three controls were absent from in the kefir group: αs1-casein f.30–38 (caseicin B, antimicrobial) and αs1-casein f.201–212 (antihypertensive).
Table 2.
Peptides that contained a sequence 100% homologous to a known functional peptide.
| Peptide | Sequence | Function | Ref | RM Mean abundance |
K mean abundance |
HMo mean abundance |
HMc mean abundance |
ANOVA p- value* |
presence in group |
|---|---|---|---|---|---|---|---|---|---|
| αs2-casein f.189–197 | FALPQYLK | antihypertensive | [47] | 3.19 ×108 | 1.91 ×109 | 3.48 ×108 | 3.96 ×108 | 5.25 ×10−2 | RM/K/HMo/HMc |
| αs2-casein f.204–212 | AMKPWIQPK | antihypertensive | [48] | 1.23 ×109 | 7.01 ×108 | 2.21 ×109 | 1.82 ×109 | 6.2 ×10−2 | RM/K/HMo/HMc |
| β-casein f.214–224 | GPVRGPFPIIV | antihypertensive | [49] | 1.25 ×1010 | 9.41 ×109 | 9.13 ×109 | 1.46 ×1010 | 2.61 ×10−1 | RM/K/HMo/HMc |
| β-casokinin-7 | AVPYPQR | antihypertensive | [24] | 5.19 ×108 | 5.40 ×109 | 5.63 ×108 | 5.74 ×108 | 1.03 ×10 −3 | RM/K/HMo/HMc |
| β-casomorphin-7 | YPFPGPI | opioid agonist | [50] | 3.15 ×107 | 3.59 ×107 | 2.36 ×107 | 2.77 ×107 | 8.33 ×10−1 | RM/K/HMo/HMc |
| Casecidin-15 | YQEPVLGPVRGPFPI | antimicrobial | [51] | 0 | 2.80 ×109 | 0 | 0 | 4.54 ×10−3 | K |
| Casecidin-17 | YQEPVLGPVRGPFPIIV | Antimicrobial, antithrombotic, immune-modulatory, anti-oxidant | [52] | 8.60 ×107 | 4.38 ×109 | 2.31 ×108 | 6.64 ×107 | 2.71 ×10−5 | RM/K/HMo/HMc |
| Caseicin B | EVFGKEKVN | antimicrobial | [53] | 1.00 ×108 | 0 | 1.81 ×108 | 1.37 ×108 | 5.37 ×10−4 | RM/HMo/HMc |
| Caseicin C | SDIPNPIGSENSEK | antimicrobial | [53] | 8.96 ×108 | 1.13 ×109 | 9.44 ×108 | 9.51 ×108 | 4.40 ×10−1 | RM/K/HMo/HMc |
| Casohypotensin | YPVEPFTE | antihypertensive | [54] | 1.40 ×107 | 2.81 ×108 | 1.32 ×107 | 1.14 ×107 | 4.33 ×10−9 | RM/K/HMo/HMc |
| β-casein f.183–190 | RDMPIQAF | antihypertensive | [55] | 0 | 5.44 ×109 | 0 | 0 | 1.75 ×10−2 | K |
| β-neocasomorphin-6 | YPVEPF | opioid | [56] | 1.47 ×108 | 7.77 ×108 | 0 | 0 | 1.51 ×10−2 | RM/K |
| Casoplatelin | MAIPPKKNQDK | antithrombotic | [57] | 0 | 2.15 ×107 | 0 | 0 | 4.41 ×10−1 | K |
| Casoxin-A | YPSYGLN | opioid antagonist | [58] | 0 | 8.89 ×107 | 0 | 0 | 2.43 ×10−1 | K |
| Isracidin | RPKHPIKHQGLPQEVLNENLLRF | antimicrobial | [59] | 0 | 0 | 3.99 ×107 | 0 | 4.41 ×10−1 | HMo |
| Pro-8-β-casomorphin 9 | YPFPGPIPN | opioid | [56] | 0 | 4.18 ×109 | 0 | 0 | 2.21 ×10−3 | K |
| Pro-8-β-casomorphin-13 | YPFPGPIPNSLPQ | opioid | [56] | 0 | 4.63 ×107 | 0 | 0 | 3.49 ×10−3 | K |
| V-β-casomorphin-9 | VYPFPGPIPN | strong anti-oxidative | [60] | 0 | 9.24 ×109 | 0 | 0 | 3.34 ×10−2 | K |
| αs1-casein f.30–37 | VLNENLLR | antimicrobial | [53] | 3.34 ×108 | 2.67 ×107 | 3.37 ×108 | 3.50 ×108 | 2.29 ×10−5 | RM/K/HMo/HMc |
| β-casein f.166–175 | SQSKVLPVPQ | antihypertensive | [61] | 6.96 ×108 | 4.14 ×1010 | 7.54 ×108 | 6.52 ×108 | 1.3 ×10−2 | RM/K/HMo/HMc |
| β-casein f.124–133 | MPFPKYPVEP | antihypertensive | [61] | 9.75 ×108 | 7.32 ×107 | 1.05 ×109 | 8,16 ×108 | 1.58 ×10−3 | RM/K/HMo/HMc |
| αs1-casein f.201–212 | IGSENSEKTTMP | antihypertensive | [61] | 2.07 ×109 | 0 | 2.94 ×109 | 2.17 ×109 | 2.49 ×10−3 | RM/HMo/HMc |
| β-casein f.63–68 | PGPIPN | immune-modulatory | [62] | 0 | 2.63 ×107 | 0 | 0 | 3.71 ×10−4 | K |
| β-casein f.192–209 | LYQEPVLGPVRGPFPIIV | immune-modulatory | [63] | 7.39 ×106 | 1.02 ×109 | 3.50 ×108 | 1.70 ×108 | 4.91 ×10−5 | RM/K/HMo/HMc |
| β-casein f.73–82 | NIPPLTQTPV | antihypertensive | [64] | 0 | 1.53 ×1010 | 1.42 ×107 | 0 | 3.73 ×10−2 | K/HMo |
| β-casein f.47–52 | DKIHPF | antihypertensive | [64] | 3.29 ×107 | 3.42 ×108 | 1.61 ×108 | 7.07 ×107 | 1.44 ×10−1 | RM/K/HMo/HMc |
| αs1-casein f.25–32 | VAPFPEVF | antihypertensive | [65] | 9.95 ×109 | 1.99 ×109 | 8.22 ×109 | 9.47 ×109 | 1.62 ×10−4 | RM/K/HMo/HMc |
| αs1-casein f.24–31 | FVAPFPEV | antihypertensive | [65] | 1.60 ×108 | 2.24 ×107 | 1.60 ×108 | 2.17 ×108 | 1.68 ×10−4 | RM/K/HMo/HMc |
Each treatment group (the raw milk group, RM; the kefir group, K; the heat-treated milk groups with vials open, HMo, and with vials closed, HMc) consists of three replicates.
P values were calculated by one-way ANOVA. Differences were considered significant when p <0.05.
3.10 Microbial ecology and metagenomic sequencing
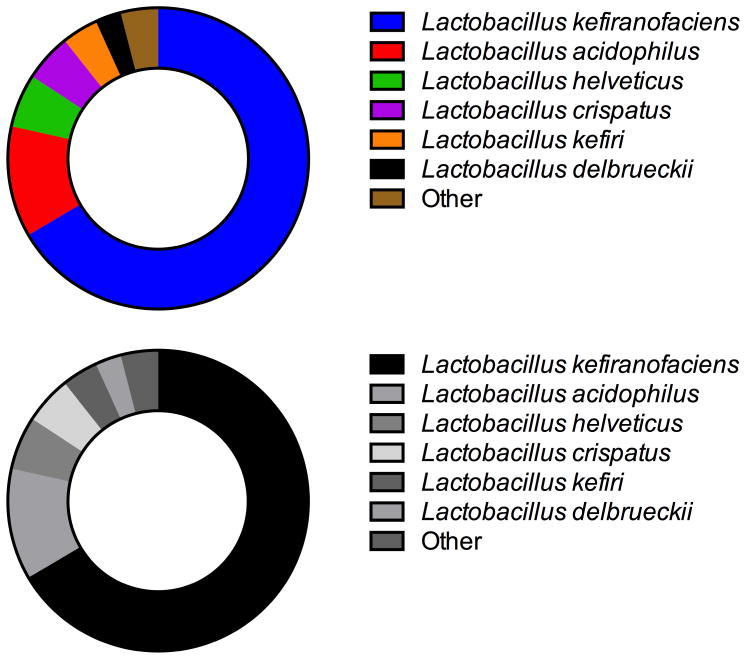
16S rRNA marker gene sequencing of the V4 region demonstrated that 99.9% of all bacteria in the kefir grain were members of the Lactobacillaceae family. Fungal qPCR showed 1.18 × 108 cfu of yeast per 150 mg kefir grain sample (2.32 × 107 standard deviation from triplicate analysis). Species-level classifications via shotgun metagenomic sequencing identified six dominant bacterial species of Lactobacillus in the sample via 16S rRNA classification as L. kefiranofaciens, L. acidophilus, L. helveticus, L. crispatus, L. kefiri, and L. delbrueckii, and other species composing minor proportions of the kefir community (Fig. 4). A small proportion (0.05% of those assigned to Lactobacilliaceae) were classified as fungal metagenomic reads. These reads were overwhelmingly mapped to Saccharomycetaceae.
Figure 4.
Relative abundance of taxonomic assignments by species, as assigned by MG-RAST annotation of shotgun 16S rRNA sequencing.
Metagenomic sequencing found that after chaperone-related ATP-dependent intracellular proteases (Clp proteases), the most abundant proteases were HtrA-like serine proteases, which have been well characterized for their activity on milk proteins (Kok & De Vos, 1994). These HtrA-like serine proteases have signal peptides and transmembrane domains and are predicted to be extracellular. These extracellular proteases could be responsible for the milk protein hydrolysis observed. Of the 4,023 reads annotated as possible serine proteases, MG-RAST found that 65% belonged to L. acidophilus (average sequence match at 88.69%), 32% belonged to L. helveticus (average sequence match at 90.10%) and the remaining reads corresponded to other minor taxa, each contributing <1% of total reads. While L. helveticus and L. acidophilus are well known for their ability to hydrolyze milk proteins via extracellular proteases (Altermann, Russell, Azcarate-Peril, Barrangou, Buck, McAuliffe, et al., 2005; Beganović, Kos, Leboš Pavunc, Uroić, Džidara, & Šušković, 2013) it is interesting to consider the individual proteolytic contributions of specific taxa. Future studies investigating these contributions and the peptide products produced by species-specific proteases would be of value.
Though more abundant, the ATP-dependent Clp proteases are intracellular and, therefore, would not be expected to contribute to extracellular milk protein hydrolysis during kefir fermentation per se. These intracellular proteases could, however, contribute to milk protein hydrolysis in the case of cell lysis. Peptidases were also identified but they were annotated as internally localized peptidases.
4 Discussion
4.1 Kefir microorganisms had proteolytic activity on milk proteins and peptides and likely consumed small peptides
Numerous findings indicated that the kefir microorganisms hydrolyzed milk proteins and peptides. 1,039 peptides from 23 proteins present in the control groups were absent from the K samples. These peptides were likely either hydrolyzed further by extracellular kefir microorganism proteases or taken up by the kefir microorganisms. Some kefir microorganisms, including Lactobacillus identified in the kefir grains used in the present study, have extracellular proteolytic capabilities. Many lactic acid bacteria are auxotrophic for several amino acids—that is, they depend on external amino acids for growth (Kunji, Mierau, Hagting, Poolman, & Konings, 1996). Lactobacillus have extracellular cell envelope proteinases that hydrolyze proteins into protein fragments (oligopeptides and amino acids) (Pritchard & Coolbear, 1993). Lactic acid bacteria transporters carry di- and tripeptides and an Opp protein system transports larger peptides into cells (Doeven, Kok, & Poolman, 2005; Savijoki, Ingmer, & Varmanen, 2006) where proteases hydrolyze small peptides to amino acids (Kunji, Mierau, Hagting, Poolman, & Konings, 1996). The action of the extracellular proteinase and peptide transport systems in Lactobacillus can explain the disappearance of large peptides in the K samples that were present in the control groups. This point aligns with the finding that the K samples contained statistically fewer peptides than the control samples. The K samples contained 609 peptides derived from 20 proteins that were not present in the control samples were likely the result of the Lactobacillus extracellular proteinases. The finding that ~1,500 peptides differed in intensity between K and the control groups (58.9% increased, 37.4% decreased) demonstrated that kefir microorganisms hydrolyzed proteins to release more peptides, and either further hydrolyzed or absorbed peptides present in the milk. That 40–45 proteins that differed in peptide count and 45–49 proteins differed in peptide abundance between K and the control groups also indicated that kefir microorganisms hydrolyzed milk proteins. Many protein total peptide intensities and total peptide counts of proteins were altered significantly in the kefir samples: β-casein and α-lactalbumin increased in intensity and count, indicating that kefir organisms released additional peptides from these proteins, and αs1-casein and αs2-casein decreased in intensity and count, indicating that kefir peptides either took these peptides up or hydrolyzed them further.
Despite the clear proteolytic activity shown via peptidomics, the electrophoretic gel image revealed little to no difference in the protein profile between the K samples and the samples without kefir grains (RM, HMo and HMc). These results demonstrate that a more detailed perspective than gel electrophoresis, such as peptidomics, is required to observe the proteolytic activity of kefir grains on milk proteins in the typical fermentation period for kefir production. Longer fermentation periods may have allowed visualization of changes in the protein profile via gel electrophoresis.
The absence of peptides in the peptidomics data from the K samples did not necessarily indicate absorption into the cell by the kefir microorganisms. The database searching approach for peptidomics applied is unlikely to identify peptides <5 amino acids in length. Therefore, peptides that appeared to be absorbed may have been hydrolyzed to peptides too small to be identified with this approach. Despite this caveat, the results clearly indicated that kefir microorganisms’ proteases were active on milk proteins.
While Lactobacillus in the sample contribute to milk protein proteolysis, the identified yeast—Saccharomyces—likely did not hydrolyze milk proteins. While S. cerevisiae can take up free amino acids via amino acid permeases as a source of nitrogen or for direct use in protein generation (Regenberg, Düring-Olsen, Kielland-Brandt, & Holmberg, 1999), previous research shows that S. cerevisiae do not possess extracellular proteolytic activity (Strauss, Jolly, Lambrechts, & Van Rensburg, 2001).
4.2 Milk proteases survived heat treatment and were active during incubation
Multiple pieces of evidence indicated that native milk proteases survived the heat treatment and were active during incubation. Milk contains numerous proteases, protease activators and inhibitors that are active on milk proteins within the mammary gland and after expression (D. Dallas, Guerrero, Khaldi, et al., 2013; D. Dallas, Guerrero, Parker, et al., 2013; D. C. Dallas, Guerrero, Khaldi, Borghese, Bhandari, Underwood, et al., 2014; D. C. Dallas, et al., 2015; Holton, Vijaykumar, Dallas, Guerrero, Borghese, Lebrilla, et al., 2014; Khaldi, Vijayakumar, Dallas, Guerrero, Wickramasinghe, Smilowitz, et al., 2014). Though pasteurization denatures and inhibits the activity of many enzymes, milk plasmin activity increases with pasteurization due to the degradation of inhibitors but not the enzyme (Prado, Sombers, Ismail, & Hayes, 2006). The data from this study demonstrated that the native milk enzymes continued to act after pasteurization. HMo and HMc contained 63 and 47 peptides, respectively, that were absent in the RM samples, indicating their release by native milk enzymes during incubation. These peptides released by the native milk enzymes derived from β-casein, αs1-casein, polymeric immunoglobulin receptor, αs2-casein and κ-casein. The point that native milk proteases survived heat-treatment and continued to be active is substantiated by the many statistical differences between HMo and HMc compared with RM, including the 201 and 217 peptides, respectively, different in abundance, the 3 and 4 proteins, respectively, different by peptide count and the 7 and 11 proteins, respectively, different by peptide abundance. These findings demonstrate that native milk enzymes survived heat-treatment and acted upon the milk proteins during the incubation steps.
4.2.1 Native milk protease activity post heat-treatment was minimal compared with kefir microorganisms’ protease activity
Though the evidence suggests that some native milk enzymes survived the heat-treatment and continued to hydrolyze proteins during incubation, the degree of proteolysis due to native milk enzymes was less than that due to kefir microorganisms. This point is supported by several findings. Compared with the K group, HMo and HMc contained fewer peptides that were absent from RM, fewer peptides that differed in intensity from RM and fewer proteins that differed in peptide count and abundance from RM. Visual inspection of the gel did not reveal evidence of protein hydrolysis when comparing HMc and HMo with RM. Furthermore, proteolytic maps (Supplemental Figs. 1–7) of selected proteins show relatively little variation between the raw milk and heat-treated controls. Therefore, though peptidomic analysis revealed that native milk enzymes continued to function during incubation in the heat-treated milk, this proteolysis was minimal compared with that carried out by the kefir microorganisms’ proteases.
4.3 No effect of atmosphere or contamination was observed
The study design included two heat-treated controls—one with vials kept open during incubation (HMo) and one with vials closed during incubation (HMc). These controls were selected because the K samples, like traditional kefir, were incubated with open vials, and it was important to verify whether proteolysis observed was due to the kefir microorganisms or to contaminating air-borne microbes. HMo with HMc were compared on a variety of metrics and there were very few differences, indicating that keeping the vials open or closed did not affect the final proteolytic outcomes. HMo and HMc were similar in the following ways: HMo compared with HMc had the fewest differences in peak intensity and the fewest proteins different in peptide count (1) and peptide abundance (4). The gel image showed no clear differences between HMo and HMc. All of these points substantiated the claim that incubating the samples in open or closed vials had little, if any, effect on proteolysis.
4.4 Kefir microorganisms’ proteolysis was protein-specific
Though the kefir microorganisms’ proteases hydrolyzed most milk proteins (caseins and larger whey proteins), some proteins, including osteopontin and β-lactoglobulin were relatively unaffected. Osteopontin and β-lactoglobulin were not altered across groups in terms of total peptide abundance. However, the PepEx maps of osteopontin and β-lactoglobulin show that peptide abundance decreased for some regions of the protein sequences and increased in others (Supplemental Fig. 2), indicating that the kefir microorganisms did act upon the proteins to some extent. However, the statistical evidence suggests that this action was relatively mild. The peptidomics data revealed that peptide peak intensity for α-lactalbumin increased in the K samples over that in the RM samples, indicating that some hydrolysis occurred. These data suggest that β-lactoglobulin, α-lactalbumin and osteopontin were more resistant to kefir microorganism hydrolysis than other milk proteins. These findings support previous reports that the kefir microbiota digested β- and α-caseins but failed to hydrolyze β-lactoglobulin, even with extended fermentation time (Ferreira, Pinho, Monteiro, Faria, Cruz, Perreira, et al., 2010).
4.5 Kefir microorganisms released functional peptides
Of 29 functional peptides identified—including antibacterial, antihypertensive, opioid and anti-oxidative functions—26 were present in the K samples that derived from αs1-, αs2-, β- and κ-casein. Six functional peptides were more abundant in K samples than the controls. Seven functional peptides were present only in the K samples, including peptides with antimicrobial, antihypertensive, opioid and anti-oxidative functions. Seven functional peptides were in common with those previously identified (Ebner, Arslan, Fedorova, Hoffmann, Küçükçetin, & Pischetsrieder, 2015), including those with antihypertensive, anti-oxidative, opioid, antimicrobial and immunomodulatory functions. These data demonstrate that kefir microorganisms release functional peptides that could enhance product functionality. These peptides are likely the result of extracellular serine proteases found in L. acidophilus and L. helveticus.
4.6 Comparison with previous works
The number of peptides identified to date is limited. A study of the antihypertensive properties of caprine kefir identified 16 peptides by RP-HPLC-MS/MS (Quirós, Hernández-Ledesma, Ramos, Amigo, & Recio, 2005). The peptide profiles of kefirs made with traditional kefir grains were compared with those made with commercial starter culture using nano-ESI-LTQ-Orbitrap MS, and 97 casein-derived peptides were common to the two varieties, out of 257 total peptides (Ebner, Arslan, Fedorova, Hoffmann, Küçükçetin, & Pischetsrieder, 2015). When compared with 257 peptides identified in raw milk, 237 were unique to kefir. However, the peptides in kefir were not compared with those in heat-treated controls incubated without kefir microorganisms, thus precluding determination of whether peptides were released by native milk proteases or by kefir microorganisms. The present study of the peptide profile of kefir made from traditional kefir grains identified 1,591 peptides, 609 of which were unique to kefir. Therefore, the present study is the most exhaustive analysis to date of the kefir peptidome and the first to include controls that distinguished peptides released by native milk enzymes from those released by kefir microorganisms.
This work reveals higher numbers of peptides present in raw bovine milk (mean 1,942 ± 21) than previously reported. Our previous research identified 159 (D. Dallas, Guerrero, Parker, et al., 2013), 238 (David C Dallas, Weinborn, de Moura Bell, Wang, Parker, Guerrero, et al., 2014) and 234 (Andres Guerrero, Dallas, Contreras, Bhandari, Cánovas, Islas-Trejo, et al., 2015) peptides in non-mastitic bovine milk. The higher number of peptides identified in the present study reflects improvements in mass spectrometry techniques applied. Our previous research employed an Agilent 6520 Q-TOF, whereas a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer was applied in the present study. The Orbitrap instrument used allows for faster spectral acquisition (50 ms maximum allowed MS/MS spectral acquisition time compared to our optimized Q-TOF settings: 1 MS/MS spectra/s) while maintaining adequate ion abundance (a target of 5 × 104 ions), which allowed for higher numbers of molecules fragmented and increased numbers of peptides identified.
4.8 Kefir-induced protein hydrolysis can improve digestibility
The present study demonstrates that kefir microorganisms hydrolyze milk proteins extensively. Beyond releasing functional peptides, this hydrolysis may be beneficial to consumers in that it improves milk protein digestibility. In a comparison of milk with lactic acid bacteria-fermented milks (kefir, yogurt and sour milk) via in vitro pepsin digestibility assays of free amino-nitrogen and by growth rate in rats, milk fermentation improved the digestibility and biological value of the protein component (Vass, Szakaly, & Schmidt, 1983). In a 10-month study (Puri, Mahapatra, Bijlani, Prasad, & Nath, 1994), rats fed yogurt made from fermented spray dried milk maintained body weight with lower caloric intake than rats fed unfermented spray dried milk, pointing to increased feed efficiency of partially digested fermented milk proteins. In other studies, fermentation of milk to yogurt also improved digestibility and feed efficiency in rats (Lee, Friend, & Shahani, 1988), and in vitro digestibility of yogurt was greater than that of non-fermented milk (Breslaw & Kleyn, 1973). The partial digestion of milk proteins that accompanies kefir microorganism fermentation may be advantageous for specific consumer groups with lowered digestive function. The enhanced digestibility of proteins in kefir could limit the production of inflammatory metabolites by putrefactive bacteria in the colon and thus improve gut health.
Supplementary Material
Highlights.
Kefir-induced degradation of milk proteins was examined via peptidomics.
Kefir microorganisms released 609 peptides from milk proteins.
Native milk enzymes degraded proteins to a lesser extent than kefir microorganisms.
Kefir contains functional peptides that may affect consumer health.
Acknowledgments
The authors thank Cora J. Dillard for editing this manuscript and the University of California Genome Center Proteomics Core (especially Brett Phinney, Darren Weber and Michelle Salemi) for the mass spectrometry work. All authors read and approved the final manuscript. The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001826. This project was funded in part by the K99/R00 Pathway to Independence Career Award, Eunice Kennedy Shriver Institute of Child Health & Development of the National Institutes of Health (K99HD079561) (D.C. Dallas). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303, and metagenomic reads are public accessible at MG-RAST (http://metagenomics.anl.gov). SAF is supported by a National Institutes of Health Postdoctoral Fellowship from the National Center for Complementary and Integrative Medicine (F32AT008533). DAM acknowledges NIH awards R01AT007079 and R01AT008759 and support as the Peter J. Shields Endowed Chair in Dairy Food Science.
Abbreviations
- ANOVA
analysis of variance
- HMc
heat-treated milk closed during incubation
- HMo
heat-treated milk opened during incubation
- HSD
honest significant difference
- K
kefir
- RM
raw milk
- TCA
trichloroacetic acid
Footnotes
Conflict of interest statement
The authors have declared no conflict of interest.
Author Contributions
DCD and FC performed the mass spectrometry data analysis, functional library search and wrote the paper. TT and FC performed the statistical analyses. VLS helped with the kefir fermentation and kefir grain DNA extraction. KMK and DAM performed the bacterial sequencing analysis. SAF and DAM analyzed the metagenomic sequencing data. RCR created the PepEx maps and analyzed proteolytic cleavage sites. DCD, FC and DB designed the study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(11):3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo L, Lopez E, Lema C. Microflora present in kefir grains of the Galician region (North-West of Spain) Journal of Dairy Research. 1993;60(2):263–267. doi: 10.1017/s002202990002759x. [DOI] [PubMed] [Google Scholar]
- Beganović J, Kos B, Leboš Pavunc A, Uroić K, Džidara P, Šušković J. Proteolytic activity of probiotic strain Lactobacillus helveticus M92. Anaerobe. 2013;20(0):58–64. doi: 10.1016/j.anaerobe.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslaw ES, Kleyn DH. In vitro digestibility of protein in yogurt at various stages of processing. Journal of Food Science. 1973;38(6):1016–1021. [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DA, Swaisgood HE. Bioactive milk peptides: a prospectus. Journal of Dairy Science. 2000;83(6):1187–1195. doi: 10.3168/jds.S0022-0302(00)74983-6. [DOI] [PubMed] [Google Scholar]
- Dallas D, Guerrero A, Khaldi N, Castillo P, Martin W, Smilowitz J, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. Journal of Proteome Research. 2013;12(5):2295–2304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas D, Guerrero A, Parker E, Garay L, Bhandari A, Lebrilla C, Barile D, German J. Peptidomic profile of milk of Holstein cows at peak lactation. Journal of Agricultural and Food Chemistry. 2013;62(1):58–65. doi: 10.1021/jf4040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Guerrero A, Khaldi N, Borghese R, Bhandari A, Underwood MA, Lebrilla CB, German JB, Barile D. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. Journal of Nutrition. 2014;144(6):815–820. doi: 10.3945/jn.113.185793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Smink CJ, Robinson RC, Tian T, Guerrero A, Parker EA, Smilowitz JT, Hettinga KA, Underwood MA, Lebrilla CB, German JB, Barile D. Endogenous human milk peptide release is greater following preterm birth than term birth. Journal of Nutrition. 2015;145(3):425–433. doi: 10.3945/jn.114.203646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Weinborn V, de Moura Bell JM, Wang M, Parker EA, Guerrero A, Hettinga KA, Lebrilla CB, German JB, Barile D. Comprehensive peptidomic and glycomic evaluation reveals that sweet whey permeate from colostrum is a source of milk protein-derived peptides and oligosaccharides. Food Research International. 2014;63(B):203–209. doi: 10.1016/j.foodres.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeven MK, Kok J, Poolman B. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Molecular microbiology. 2005;57(3):640–649. doi: 10.1111/j.1365-2958.2005.04698.x. [DOI] [PubMed] [Google Scholar]
- Ebner J, Arslan AA, Fedorova M, Hoffmann R, Küçükçetin A, Pischetsrieder M. Peptide profiling of bovine kefir reveals 236 unique peptides released from caseins during its production by starter culture or kefir grains. Journal of proteomics. 2015;117(0):41–57. doi: 10.1016/j.jprot.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Farnworth ER. Handbook of fermented functional foods. Boca Raton, FL: CRC PRESS; 2003. [Google Scholar]
- Ferreira I, Pinho O, Monteiro D, Faria S, Cruz S, Perreira A, Roque A, Tavares P. Short communication: Effect of kefir grains on proteolysis of major milk proteins. Journal of Dairy Science. 2010;93(1):27–31. doi: 10.3168/jds.2009-2501. [DOI] [PubMed] [Google Scholar]
- Guerrero A, Dallas DC, Contreras S, Bhandari A, Cánovas A, Islas-Trejo A, Medrano JF, Parker EA, Wang M, Hettinga K, Chee S, German JB, Barile D, Lebrilla CB. Peptidomic analysis of healthy and subclinically mastitic bovine milk. International Dairy Journal. 2015;46(0):46–52. doi: 10.1016/j.idairyj.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero A, Dallas DC, Contreras S, Chee S, Parker EA, Sun X, Dimapasoc LM, Barile D, German JB, Lebrilla CB. Mechanistic peptidomics: factors that dictate the specificity on the formation of endogenous peptides in human milk. Molecular & Cellular Proteomics. 2014;13(12):3343–3351. doi: 10.1074/mcp.M113.036194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M, Stanton C, Fitzgerald GF, Ross RP. Putting microbes to work: dairy fermentation, cell factories and bioactive peptides. Part II: bioactive peptide functions. Biotechnology Journal. 2007;2(4):435–449. doi: 10.1002/biot.200700045. [DOI] [PubMed] [Google Scholar]
- Holton TA, Vijaykumar V, Dallas DC, Guerrero A, Borghese RC, Lebrilla CB, German JB, Barile D, Underwood MA, Shields DC, Khaldi N. Following the digestion of milk proteins from mother to baby. Journal of Proteome Research. 2014;13(12):5777–5783. doi: 10.1021/pr5006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesenkas H, Yerlikaya O, Ozer E. A functional milk beverage: kefir. Agro Food Industry Hi Tech. 2013;24(6):53–55. [Google Scholar]
- Khaldi N, Vijayakumar V, Dallas DC, Guerrero A, Wickramasinghe S, Smilowitz JT, Medrano JF, Lebrilla CB, Shields DC, German JB. Predicting the important enzyme players in human breast milk digestion. Journal of Agricultural and Food Chemistry. 2014;62(29):7225–7232. doi: 10.1021/jf405601e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok dJ, De Vos W. Genetics and biotechnology of lactic acid bacteria. Springer; 1994. The proteolytic system of lactic acid bacteria; pp. 169–210. [Google Scholar]
- Kunji ER, Mierau I, Hagting A, Poolman B, Konings WN. The proteotytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek. 1996;70(2–4):187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- Lee H, Friend BA, Shahani KM. Factors affecting the protein quality of yogurt and acidophilus milk. Journal of Dairy Science. 1988;71(12):3203–3213. [Google Scholar]
- Leite AMdO, Miguel MAL, Peixoto RS, Rosado AS, Silva JT, Paschoalin VMF. Microbiological, technological and therapeutic properties of kefir: a natural probiotic beverage. Brazilian Journal of Microbiology. 2013;44(2):341–349. doi: 10.1590/S1517-83822013000200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Nakagomi K, Tomizuka N, Suzuki H. Angiotensin I-converting enzyme inhibitor derived from an enzymatic hydrolysate of casein. II. Isolation and bradykinin-potentiating activity on the uterus and the ileum of rats. Agricultural and Biological Chemistry. 1985;49(5):1405–1409. [Google Scholar]
- Minervini F, Algaron F, Rizzello C, Fox P, Monnet V, Gobbetti M. Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Applied and Environmental Microbiology. 2003;69(9):5297–5305. doi: 10.1128/AEM.69.9.5297-5305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otles S, Cagindi O. Kefir: a probiotic dairy-composition, nutritional and therapeutic aspects. Pakistan Journal of Nutrition. 2003;2(2):54–59. [Google Scholar]
- Parker EA. PepEx. 2014. [Google Scholar]
- Prado B, Sombers S, Ismail B, Hayes K. Effect of heat treatment on the activity of inhibitors of plasmin and plasminogen activators in milk. International Dairy Journal. 2006;16(6):593–599. [Google Scholar]
- Pritchard GG, Coolbear T. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiology Reviews. 1993;12(1–3):179–206. doi: 10.1111/j.1574-6976.1993.tb00018.x. [DOI] [PubMed] [Google Scholar]
- Puri P, Mahapatra S, Bijlani R, Prasad H, Nath I. Feed efficiency and splenic lymphocyte proliferation response in yogurt-and milk-fed mice. International Journal of Food Sciences and Nutrition. 1994;45(4):231–235. doi: 10.3109/09637489609006952. [DOI] [PubMed] [Google Scholar]
- Quirós A, Hernández-Ledesma B, Ramos M, Amigo L, Recio I. Angiotensin-converting enzyme inhibitory activity of peptides derived from caprine kefir. Journal of Dairy Science. 2005;88(10):3480–3487. doi: 10.3168/jds.S0022-0302(05)73032-0. [DOI] [PubMed] [Google Scholar]
- Recio I, Visser S. Identification of two distinct antibacterial domains within the sequence of bovine a-s2-casein. Biochimica et Biophysica Acta (BBA)-General Subjects. 1999;1428(2):314–326. doi: 10.1016/s0304-4165(99)00079-3. [DOI] [PubMed] [Google Scholar]
- Regenberg B, Düring-Olsen L, Kielland-Brandt MC, Holmberg S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Current Genetics. 1999;36(6):317–328. doi: 10.1007/s002940050506. [DOI] [PubMed] [Google Scholar]
- Savijoki K, Ingmer H, Varmanen P. Proteolytic systems of lactic acid bacteria. Applied microbiology and biotechnology. 2006;71(4):394–406. doi: 10.1007/s00253-006-0427-1. [DOI] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in Skyline application to protein acetylation and phosphorylation. Molecular & Cellular Proteomics. 2012;11(5):202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DP, Peay KG. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PloS one. 2014;9(2):e90234. doi: 10.1371/journal.pone.0090234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M, Jolly N, Lambrechts M, Van Rensburg P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. Journal of Applied Microbiology. 2001;91(1):182–190. doi: 10.1046/j.1365-2672.2001.01379.x. [DOI] [PubMed] [Google Scholar]
- Vass A, Szakaly S, Schmidt P. Experimental study of the nutritional biological characters of fermented milks. Acta Medica Hungarica. 1983;41(2–3):157–161. [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Chen KN, Lo YM, Chiang ML, Chen HC, Liu JR, Chen MJ. Investigation of microorganisms involved in biosynthesis of the kefir grain. Food microbiology. 2012;32(2):274–285. doi: 10.1016/j.fm.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu X, Jiang H, Dong M. Analysis of the microflora in Tibetan kefir grains using denaturing gradient gel electrophoresis. Food microbiology. 2009;26(8):770–775. doi: 10.1016/j.fm.2009.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.