Abstract
Southern Africa continues to be the epicenter of the HIV/AIDS epidemic. This HIV-1 subtype C epidemic has a predominantly heterosexual mode of virus transmission and high (>15%) HIV prevalence among adults. The epidemiological dynamics of the HIV-1C epidemic in southern Africa are still poorly understood. Here, we aim at a better understanding of HIV transmission dynamics by analyzing HIV-1 subtype C sequences from Mochudi, a peri-urban village in Botswana.
HIV-1C env gene sequences (gp120 V1C5) were obtained through enhanced household-based HIV testing and counseling in Mochudi. More than 1,200 sequences were generated and phylogenetically distinct sub-epidemics within Mochudi identified. The Bayesian birth-death skyline plot was used to estimate the effective reproductive number, R, and the timing of virus transmission, to classify sub-epidemics as “acute” (those with recent viral transmissions) or “historic” (those without recent viral transmissions).
We identified two of the 15 sub-epidemics as “acute.” The median estimates of R among the clusters ranged from 0.72 to 1.77. The majority of HIV lineages, 11 out of 15 clusters with 5+ members, appear to have been introduced to Mochudi between 1996 and 2002. The median peak duration of viral transmissions was 7.1 years (range 2.9–9.7 years). The median life span of identified HIV sub-epidemics, i.e. the time between the inferred epidemic origin and its most recent sample, was 13.1 years (range 10.2–22.1 years). Most viral transmissions within the sub-epidemics occurred between 1997 and 2007. The time period during which infected people are infectious appears to have decreased since the introduction of the national ART program in Botswana.
Real-time HIV genotyping and breaking down local HIV epidemics into phylogenetically distinct sub-epidemics may help to reveal the structure and dynamics of HIV transmission networks in communities, and aid in the design of targeted interventions for members of the acute sub-epidemics that likely fuel local HIV/AIDS epidemics.
Keywords: HIV-1 subtype C, V1C5 sequences, HIV sub-epidemics, effective reproductive number R, Botswana
Introduction
Sub-Saharan Africa remains the area most severely affected by the HIV/AIDS epidemic, accounting for 70% of people living with HIV worldwide (UNAIDS, 2013b). Increased access to HIV treatment and advanced prevention of mother-to-child HIV transmission have helped reduce the number of HIV infections in southern African countries (UNAIDS, 2013b, 2014). At the same time, many southern African communities experience a devastating burden of HIV prevalence, at about 20% among general adult population (UNAIDS, 2013a, b, 2014). Better understanding the dynamics of HIV transmission networks and mechanisms of HIV transmission in these communities could assist in the design and evaluation of preventive HIV interventions.
Recent advances in molecular epidemiology and phylodynamics have made it possible to model the structure and dynamics of HIV epidemics (Bezemer et al., 2013; Bezemer et al., 2014; Bezemer et al., 2010; Frost and Volz, 2010; Kuhnert et al., 2014; Leigh Brown et al., 2011; Leventhal et al., 2014; Leventhal et al., 2012; Stadler and Bonhoeffer, 2013; Stadler et al., 2012; Stadler et al., 2013; Volz et al., 2013a; Volz et al., 2013b; Volz et al., 2012; Volz et al., 2009; Wertheim et al., 2014). Most of these studies were performed in HIV-1 subtype B settings in cohorts of men who have sex with men (MSM). It is likely that major principles and ideas developed in these studies are applicable to southern African communities, although the most prevalent viral subtype in southern African countries is HIV-1C, and the predominant mode of HIV transmission is heterosexual. Hence, these methods could be employed to better understand the epidemiological dynamics of HIV in southern Africa.
In this study we focus on circulating HIV lineages in a peri-urban Botswana community, the village of Mochudi. We utilize the well-established infrastructure and exceptional sample and data collection of the Mochudi Prevention Project. Using phylogenetic linkage to identify HIV lineages, we break down the local HIV epidemic into sub-epidemics and trace the spread of the phylogenetically distinct HIV lineages that caused these sub-epidemics over time.
Proper interpretation is needed to make epidemiological/biological conclusions based on phylogenetic clustering. Identification of HIV clusters and interpretation of HIV clustering results depend on specifics of sampling, genotyping, and phylogenetic inference. The relationship between phylogenetic clustering and transmission chains might be weak, or unreliable, if sampling is sparse and/or phylogenetic uncertainty is high. We previously showed that the extent of HIV clustering can be affected by sampling density (Novitsky et al., 2014). Importantly, a sampling density of approximately 70% was achieved in this study.
The approach of dividing the HIV epidemic into sub-epidemics has been used previously to study HIV transmissions within different risk groups (Barcherini et al., 1999; Cantoni et al., 1995; Feng et al., 2013; Graw et al., 2012; Kivela et al., 2010; Ng et al., 2013) (reviewed in (Tanser et al., 2014)). HIV lineages circulating in a given community or village can be identified and distinguished phylogenetically. A local HIV epidemic in a given community can be considered as a series of sub-epidemics caused by phylogenetically distinct HIV lineages that are likely to represent viral transmission chains. Mapping of HIV lineages/clusters followed by fitting recent HIV infections into these lineages can be used to trace HIV transmissions and associate viral transmissions with the spread of particular HIV variants. This approach ensures confidentiality, as HIV dynamics are studied entirely through virus variation, and not as directional HIV transmission between particular individuals participating in the prevention project.
Materials and Methods
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Health Research and Development Committee (HRDC) of the Republic of Botswana, and the Office of Human Research Administration (OHRA) of the Harvard School of Public Health. All adult study subjects provided written informed consent for participation in the study; all minor study subjects provided written informed assent, and each minor's guardian provided written informed consent, for their participation in the study.
Study subjects
The study subjects participating in the Mochudi Prevention Project (MPP) have been previously described (Novitsky et al., 2013). To estimate HIV-1 incidence and prevalence among 16–64-year-old residents, three rounds of home-based HIV testing and counseling (HTC) were conducted (at baseline and through two follow-up campaigns) in the northeastern sector (NES) of Mochudi during May 2010 – August 2013.
During the household visits, consented eligible residents were asked to donate a blood sample for a rapid HIV test, and quantification of HIV-1 RNA and viral genotyping (if HIV positive). HIV testing was performed in the household using Botswana HIV testing guidelines that include two rapid tests in parallel: Determine HIV-1/2 (Abbott Diagnostic Division, Belgium/Luxembourg) and Uni-Gold (Trinity Biotech, Wicklow, Ireland). Only concordant results in both tests were considered valid. HIV-infected individuals were referred to the Botswana national ART program (free-of-charge treatment of all adults with CD4 ≤350 cells/μL or WHO Stage III/IV). ART-naïve HIV-infected individuals (newly diagnosed, or linked to care) were invited to a clinic to determine their eligibility for initiation of ART; a clinic visit included collection of venous blood by phlebotomy for CD4 and HIV-1 RNA testing.
A total of 6,238 age-eligible individuals were tested during household-based HTC in Mochudi, and 1,240 of them were found HIV positive. HIV-1 prevalence was estimated at 19.9% (95% CI 18.9% to 20.9%). During the MPP, a total of 30 seroconverters were identified based on paired HIV-negative and HIV-positive tests.
HIV-1C env gp120 sequences
Nucleotide sequences spanning the HIV-1C env gp120 V1C5 region were generated by population-based (bulk) Sanger sequencing. All analyses in this study are based on a single sequence per subject, i.e., the number of viral sequences corresponds to the number of individuals. Details on nucleic acid extraction, amplification, sequencing and multiple sequence alignment have been presented elsewhere (Novitsky et al., 2013). The conserved and variable regions corresponding to functional domains within HIV-1 env gp120 were aligned separately by applying differential penalties for gap opening and gap extension. The aligned segments were concatenated into a final multiple sequence alignment.
Rationale for including sequences from Botswana only
Recently we analyzed clustering patterns between the 785 V1C5 sequences from Mochudi and 1,244 non-Botswana HIV-1C V1C5 sequences (Novitsky et al., 2013). We demonstrated that among 212 clustered Mochudi sequences, 191 (90.1%) were found in clusters with other Mochudi sequences, while 21 (9.9%) clustered with sequences from other parts of Botswana. Remarkably, none of the Mochudi sequences clustered with non-Botswana HIV-1C sequences, suggesting robustness of the observed clusters with Mochudi sequences, and providing a rationale for narrowing the analysis in the current study exclusively to HIV-1C V1C5 sequences with a Botswana sampling origin.
HIV-1 subtyping
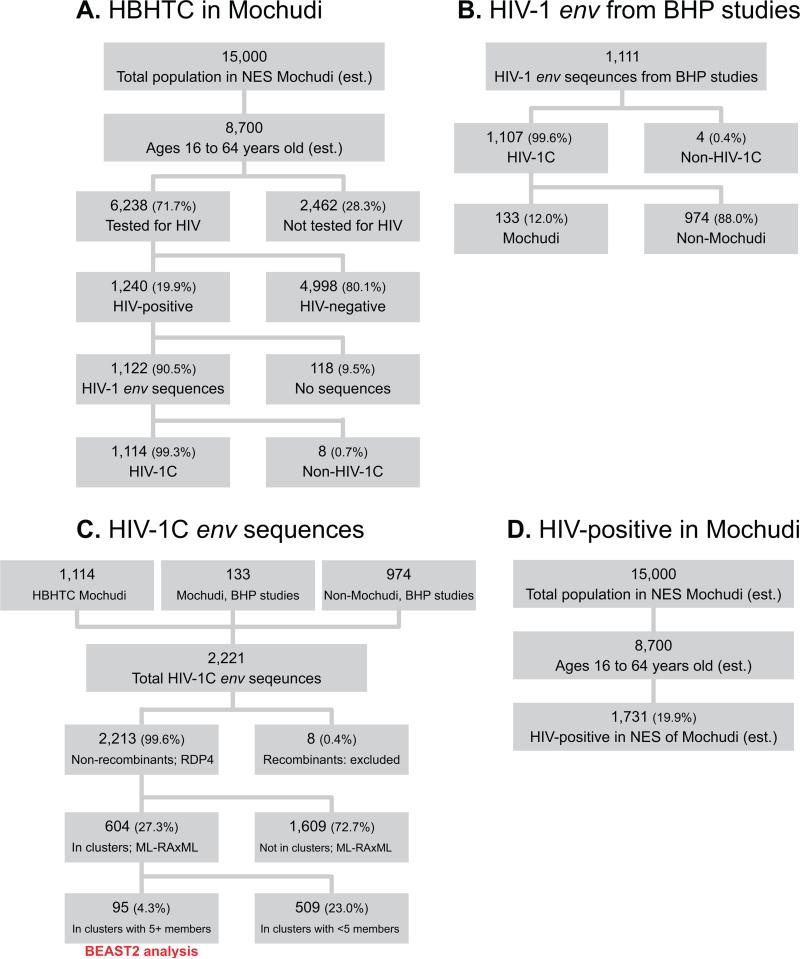
A total of 1,122 sequences were generated from 1,240 HIV-positive individuals from Mochudi (success rate of viral genotyping: 90.5%; 95% CI 88.7% – 92.0%; Fig. 1A). HIV-1 subtyping was performed using the REGA HIV-1 subtyping tool v3.0 (Pineda-Pena et al., 2013). The vast majority, 1,114 (99.3%; 95% CI 98.5% – 99.7%) of 1,122 generated HIV-1 env sequences, belong to HIV-1 subtype C. The 8 non-subtype C sequences included 4 A1, 1 A1/G, 1 D/CRF10_CD, 1 G/J, and 1 CRF11_cpx.
Figure 1.
Flowchart of home-based HTC (A), generation and preliminary analysis of HIV-1C env gp120 V1C5 sequences (B & C), and estimation of HIV-positive individuals in Mochudi (D).
HIV-1C V1C5 sequences included in the analyses
A total of 1,111 HIV-1 env V1C5 sequences with known geographic origin and time of sampling were utilized from multiple Botswana-Harvard AIDS Institute Partnership (BHP) studies performing viral genotyping. Most of these sequences, 1,107 (99.6%; 95% CI 99.0% – 99.9%; Fig. 1B), belong to HIV-1C, while 4 nonsubtype C sequences included 2 A1, 1 A1/G, and 1 CFR11_cpx. A subset of 133 (12.0%; 95% CI 10.2% – 14.1%) HIV-1C sequences originated from Mochudi (Fig. 1B).
The total set of 2,221 HIV-1C env sequences was comprised of 1,114 Mochudi sequences sampled during 2010–2013, 133 Mochudi sequences sampled during 2000–2008, and 974 non-Mochudi sequences sampled from other regions across Botswana during 1999–2014 (Fig. 1C).
Multiple sequence alignment
Multiple codon-based sequence alignment was generated by MUSCLE (Edgar, 2004) in Mega6 (Tamura et al., 2013) with gap opening penalty −3.2 and gap extension penalty −0.8. Variable loops V1, V2, V4, and V5 were independently realigned with reduced penalties, −0.8 for gap opening and −0.4 for gap extension, followed by concatenation in SeaView v.4 (Gouy et al., 2010) and minor manual adjustments in BioEdit (Hall, 1999).
Recombination analysis
The V1C5 sequences were analyzed for the presence of recombination signal using RDP4 (Martin et al., 2010), a software package for statistical identification of recombination events. The RDP4 package utilizes non-parametric recombination detection methods, such as RDP (Martin and Rybicki, 2000), GENECONV (Padidam et al., 1999), Bootscan/Recscan (Martin et al., 2005), MaxChi (Smith, 1992), Chimaera (Posada and Crandall, 2001), SiScan (Gibbs et al., 2000) and 3Seq (Boni et al., 2007). RDP4 does not require reference sequences, which makes analysis of viral sequences from epidemiologically unlinked patients more practical (Novitsky et al., 2011). A total of 8 sequences from Botswana (none from Mochudi) demonstrated evidence for recombination signal in RDP4 by at least 2 out of the 7 methods of analysis (data not shown). All recombination points were unique.
HIV-1C genotyping coverage
The total number of HIV-infected individuals in Mochudi was estimated at 1,731 based on HIV-1 prevalence of 19.9% among 16–64-year-old residents (Fig. 1D). HIV-1C env sequences were generated from blood samples collected in 1,114 tested residents in Mochudi during the 2010–2013 household-based HTC (Fig. 1A). In addition, 133 HIV-1C env sequences from Mochudi (Fig. 1B) were generated through other BHP studies, resulting in a total of 1,247 available HIV-1C env sequences of Mochudi origin. The overall HIV-1C genotyping coverage for Mochudi was estimated at 72.0% (95% CI 69.8% to 74.1%) as a proportion of generated HIV-1C V1C5 sequences (n=1,247; single sequence per person) to the estimated total number of HIV-infected individuals in Mochudi, n=1,731. The HIV-1C genotyping coverage at the lower end (0.70) was used as a prior of sampling rate s in the analysis of effective reproductive number R.
Definition of HIV cluster
We define HIV transmission clusters in terms of the mode of virus transmission, and specifics of sampling and genotyping. HIV transmission in Botswana and other southern African countries is predominantly heterosexual. In this study we define the HIV cluster as a viral lineage that gives rise to a monophyletic sub-tree of the overall phylogeny with strong statistical support in the context of high sampling density. High bootstrap support of splits is known to be an effective technique to test the relative stability of groups within a phylogenetic tree (Van de Peer, 2009). We use the bootstrapped maximum likelihood (ML) method (Felsenstein, 1985, 2004; Nei and Kumar, 2000) and internode certainty (Salichos and Rokas, 2013; Salichos et al., 2014) to determine the statistical support of clusters in Mochudi.
Short branches, genetic distances below a given threshold, monophyly, and estimated time to most recent common ancestor have been used to define HIV clusters (Brenner et al., 2008; Hue et al., 2004; Hue et al., 2005b; Hughes et al., 2009; Leigh Brown et al., 2011; Lewis et al., 2008). Bootstrap proportions can be used as a rough statistical estimate for a node in the phylogenetic tree (Andrieu et al., 1997; Buckley and Cunningham, 2002; Efron, 1979; Efron et al., 1996; Felsenstein and Kishino, 1993; Hillis and Bull, 1993; Lee, 2000; Sanderson, 1989; Swofford et al., 1996). We use bootstrap support to test the relative stability of groups within a phylogenetic tree (Van de Peer, 2009) in Mochudi where sampling density is relatively high (72%), which is key for tracing HIV spread. We assume that the tree inferred from densely sampled HIV sequences is more close to a true transmission tree (than a tree reconstructed from a low density sample), and the high bootstrap support in such a tree is more likely to be associated with true transmission chains. Thus, the identified phylogenetically distinct viral lineages in the context of high sampling density in a local community are likely represent HIV transmission chains, and if so, viral lineages could be used to trace virus spread in the community (e.g., Mochudi) over time.
For definition of HIV clusters, we follow the strategy of “bootstrap plus similarity” rather than “bootstrap vs. similarity.” This approach proved to be suitable in our recent study in Mochudi, in which HIV clusters identified by bootstrapped maximum likelihood had low intra-cluster distances (Novitsky et al., 2013). The distribution of intra-cluster distances in clusters with 3+ members was located on the left shoulder of the histogram of total pairwise distances in the sample set. The pairwise distances among dyads were located even further left.
Internode certainty measures the level of support for a given internal node by considering its frequency in a given set of trees jointly with the most prevalent conflicting bipartition in the same set of trees (Salichos and Rokas, 2013). Internode certainty values near zero indicate the presence of an almost equally supported bipartition that conflicts with the inferred internode, whereas values close to one indicate the absence of conflict (Salichos and Rokas, 2013; Salichos et al., 2014).
In this study, we define HIV clusters by a combination of bootstrapped ML (Felsenstein, 1985, 2004; Nei and Kumar, 2000) and internode certainty (Salichos and Rokas, 2013; Salichos et al., 2014). The bootstrap threshold was set to 0.80. While the internode certainty threshold was set at 0.70 (Salichos and Rokas, 2013; Salichos et al., 2014), clusters with bootstrap support of 0.80 and internode certainty between 0.50 and 0.70 were also considered.
Phylogenetic linkage analysis
Phylogenetic relatedness among HIV-1C env gp120 sequences was estimated by ML analysis (Nei and Kumar, 2000) using RAxML ver. 8 (Stamatakis, 2014). The best-fit model of nucleotide substitution, GTR+Γ4+Ι, the general time-reversible substitution model with a gamma distribution of among-sites rate variation (α-shape parameter at 0.56) and invariant sites (pinv at 0.05), was determined in MEGA6 (Tamura et al., 2013). The number of replicates was 1,000. The RAxML analysis was performed using the high-performance computing cluster Odyssey (http://rc.fas.harvard.edu/kb/high-performance-computing/architectural-description-of-the-odyssey-cluster/) at the Faculty of Arts and Sciences, Harvard University (https://rc.fas.harvard.edu/). The bootstrap support of splits was used as statistical support of monophyletic clades (subtrees, viral lineages, clusters). The bootstrap value of 0.80 was chosen as a threshold for identification of distinct viral lineages for the ML phylogeny. A relatively relaxed definition of clusters was used intentionally to avoid elimination of some viral sequences that show the capacity to cluster. This is reasonable because (i) the sample set included prevalent HIV infections with unknown time of transmission that likely diverged from the transmitted virus over time due to substantial HIV-1 intra-host evolution, (ii) the current analysis is based on the HIV-1 env gp120 V1C5 region, one of the most diversified regions across the HIV-1 genome, and (iii) the goal of ML screening was to select a subset of sequences with the capacity to form clusters in the subsequent Bayesian analysis.
HIV-1C clusters with 5+ members identified by ML screening were grouped for phylodynamic analysis. The Bayesian Markov Cain Monte Carlo (MCMC) phylogenetic inference implemented in the BEAST package v.2.1.3 (Bouckaert et al., 2014) was utilized to estimate time-scaled branch lengths and node heights. We make the simplifying assumption that the branching times in the tree reflect the timing of actual transmission events, the estimated branch lengths and node heights were interpreted as timing of HIV transmissions. The analysis employed a general time-reversible substitution model with a gamma distributed rate variation and proportion of invariant sites (GTR+Γ4+Ι), an uncorrelated log-normal relaxed molecular clock model (Drummond et al., 2006), and a Birth-Death Skyline Serial model (BDSKY) as a model for viral transmission (Stadler et al., 2012; Stadler et al., 2013). The following prior distribution of the BDSKY model parameters was used: LogNorm(0; 0.5) for effective reproductive number R; LogNorm(−0.5; 1) for the rate of becoming non-infectious δ; and Beta(35;15) for sampling rate s. The evolutionary rate for the analyzed V1C5 region of gp120 was set to 7.86E-03 (see Inter-host HIV-1C V1C5 evolutionary rate below and Supplementary Materials). The BEAST2 analyses were run until all relevant parameters converged, with 20% of the MCMC chains discarded as burn-in. Statistical confidence is represented by values for the 95% highest probability density (HPD). To generate the log file, five independent MCMC runs of 2×108 chain length were combined with LogCombiner (Drummond et al., 2012). Sampling dates were used to infer the tree height and internal node ages in the Maximum Clade Credibility (MCC) time-trees using BEAST2 (Bouckaert et al., 2014). Similarly to the log file, the time-trees from five independent runs of 2×108 chain length were combined with LogCombiner (Drummond et al., 2012) with 20% of the MCMC chains discarded as burn-in, and generation of MCMC time-trees in TreeAnnotator (Bouckaert et al., 2014; Drummond et al., 2012).
Inter-host HIV-1C V1C5 evolutionary rate
To estimate inter-host rate of nucleotide substitution per site within the V1C5 region of HIV-1 gp120, a subset of 58 viral sequences from Botswana sampled over 17 years, from 1996 to 2013, was utilized. The rate was estimated in the BEAST package v.2.1.3 (Bouckaert et al., 2014) using the general time-reversible substitution model with a gamma distributed rate variation (GTR+Γ4), an uncorrelated log-normal relaxed molecular clock model (Drummond et al., 2006), and a Birth-Death Skyline Serial model as a tree prior. A uniform prior was used for the origin parameter, with the upper value of 43 years, based on the assumption that HIV infection was introduced to Botswana after 1970 (2013 – 1970 = 43; see Supplementary materials). The mean inter-host evolutionary rate for the V1C5 region of HIV-1 gp120 was estimated at 7.86E-03 (median 7.84E-03) of nucleotide substitutions per site with 95% HPD from 7.19E-03 to 8.57E-03. This rate was used as a prior for inferring time-trees in this study.
Statistical analysis
All confidence intervals of estimated proportions are asymptotic 95% binomial confidence intervals (95% CI) computed with the prop.test function in R version 3.0.1 (R Core Team, 2013). P-values less than 0.05 were considered statistically significant and all hypothesis tests were two-sided. The Bonferroni correction was applied in recombination analysis due to the multiple methods used. Plots and histograms were produced in R. All figures were finalized in Adobe Illustrator CS6.
Results
HIV-1C gp120 V1C5 sequences in clusters
Maximum-likelihood was used to assess phylogenetic relationships among 2,213 non-recombinant HIV-1C env gp120 V1C5 sequences from Botswana, including 1,247 sequences from Mochudi (Fig. 1C; 1,114 + 133 = 1,247). The phylogeny was inferred using RAxML with 1,000 bootstrap replicates.
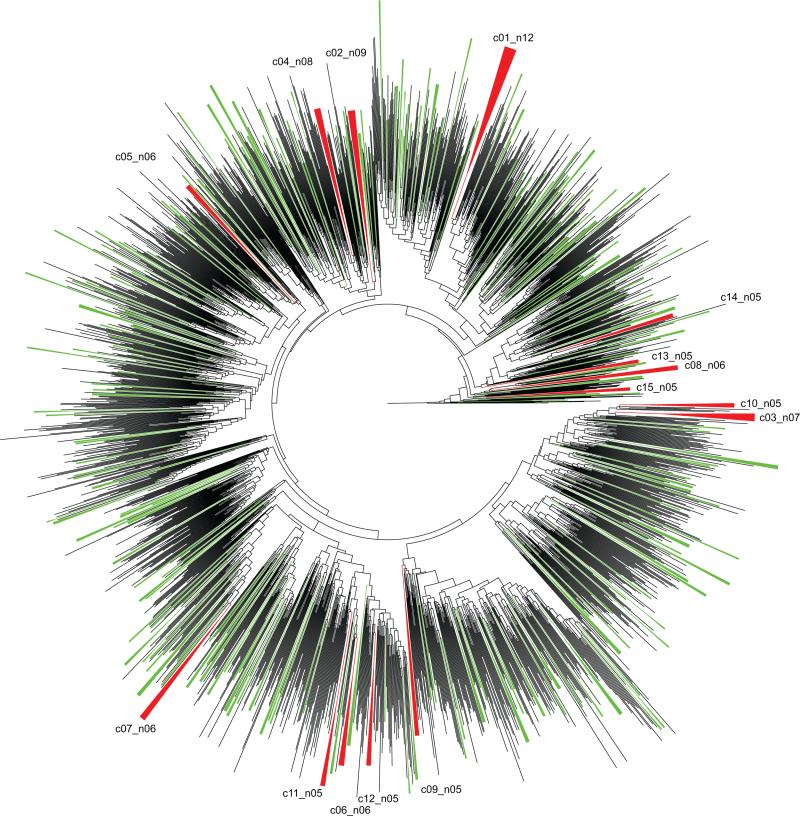
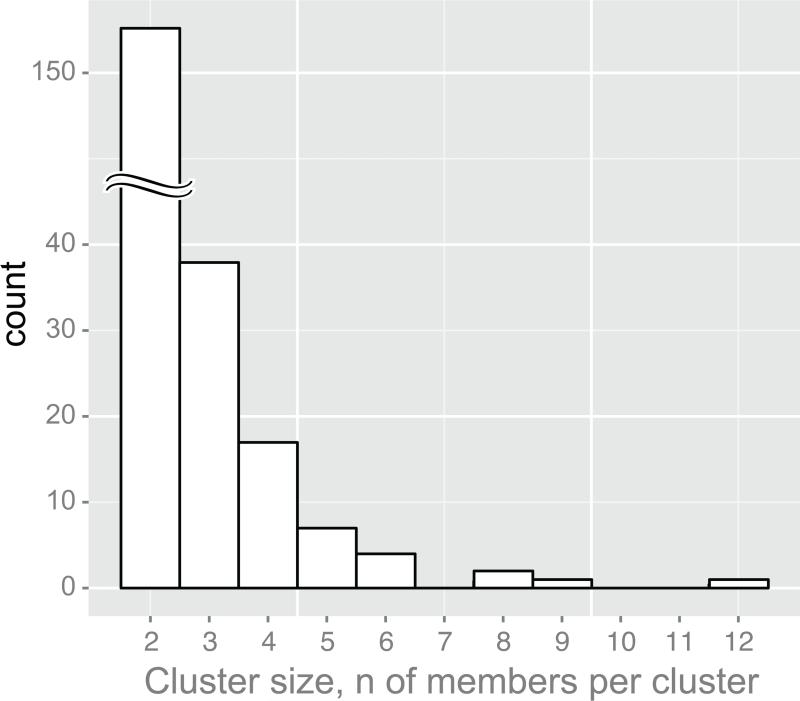
Viral lineages with bootstrap support of splits of ≥0.80 were selected (Fig. 2). A total of 604 V1C5 sequences were found in 233 clusters. The majority of HIV lineages, 163 of 233 (70%; 95% CI from 63.6% to 75.7%), were dyads. The cluster size distribution has a tail typical of a power law distribution (Clauset et al., 2009) (Fig. 3). A total of 15 viral lineages with 5+ members that included 95 HIV-1C V1C5 sequences from Botswana (82 from Mochudi and 13 from elsewhere in Botswana) were used for analysis in BEAST2 (Bouckaert et al., 2014). The selected HIV lineages included 7 clusters with 5 members, 4 clusters with 6 members, 1 cluster with 7 members, 1 cluster with 8 members, 1 cluster with 9 members, and 1 cluster with 12 members.
Figure 2.
Phylogenetic relationships of 2,213 HIV-1C V1C5 sequences from Botswana based on the bootstrapped Maximum Likelihood analysis and 1,000 bootstrap replicates. Clusters with bootstrap support of ≥0.80 are collapsed. Clusters with 5+ members are shown in red and enumerated. Clusters with 2 to 4 members are shown in green.
Figure 3.
Cluster size distribution. Axis x shows cluster size as the number of members per cluster. Axis y shows the number of identified clusters. Clusters with 5+ members (n=12) were analyzed in this study.
Trajectories of the effective reproductive number, R
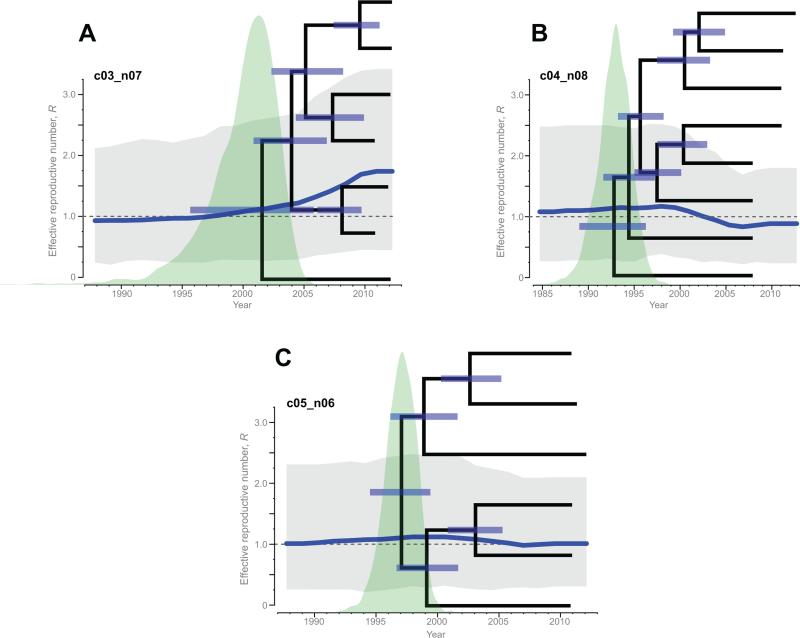
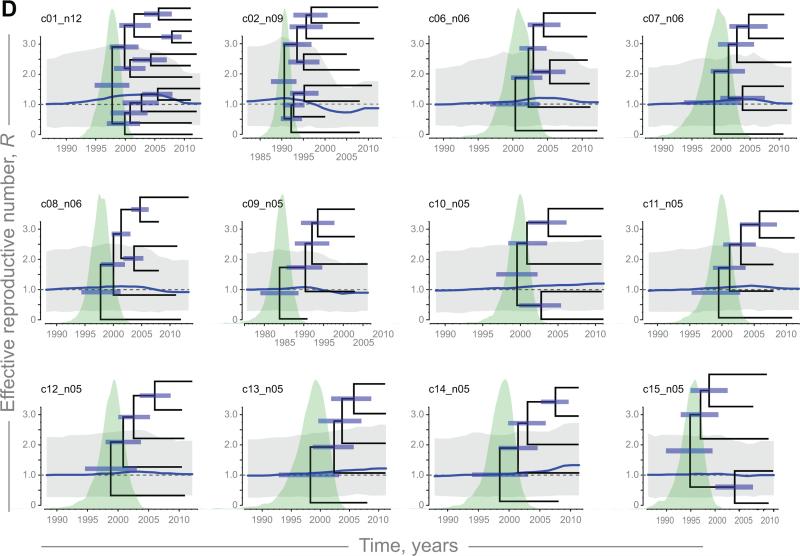
The R trajectories and corresponding 95% HPD were estimated for 15 HIV clusters identified in Mochudi with 5+ members (Fig. 4; blue curves and gray polygons). The MCC time trees with node bars indicate uncertainty in estimation of the convergence time per node and distribution of tMRCA are presented on the background of the effective reproductive number R trajectories for each of the 15 clusters. Slight increase (Fig. 4A) and decrease in R (Fig. 4B), and fluctuations of R around the threshold of 1 (Fig. 4C), exemplify differential behavior of the effective reproductive number for clusters c03_n07, c04_n08, and c05_n06, respectively. The trajectories of the remaining 12 clusters with 5+ members are presented in Figure 4D. The median estimates of R were within the range 0.72–1.77. The uncertainty illustrated by the 95% HPD intervals in grey (Fig. 4) varied across the identified HIV sub-epidemics in Mochudi.
Figure 4.
Trajectories of effective reproductive number R (blue curves) with 95% HPD (gray polygons), and MCC time-trees within analyzed HIV sub-epidemics with 5+ members. A: Increasing R in cluster c03_n07. B: Declining R in cluster c04_n08. C: Fluctuating R in cluster c05_n06. D (next page): Fluctuating R in other 12 clusters.
The estimated R trajectories allowed us to identify four HIV clusters (c03_n07, c10_n05, c13_n05, and c14_n05) with increasing R. These HIV lineages with the effective reproductive number R above the 1.0 threshold during the most recent time are interpreted as HIV sub-epidemics on the rise.
The R trajectories in four other HIV clusters (c02_n09, c04_n08, c08_n06, and c09_n05) had decreasing R below the 1.0 threshold at present; hence these HIV sub-epidemics appear to have peaked in the past and to be declining. The remaining seven HIV clusters (c01_n12, c05_n06, c06_n06, c07_n06, c11_n05, c12_n05, and c15_n05) had R values fluctuating near the threshold of 1.0.
The composition of analyzed clusters with 5+ members was not uniform. All 15 clusters with 5+ members included sequences from Mochudi, providing evidence of the spread for each analyzed viral lineage in the village of Mochudi. There were two types of clusters, the Mochudi-unique and mixed clusters (Table 1). Most of the HIV clusters with 5+ members, 11 of 15, were Mochudi-unique and included individuals from Mochudi only. In two mixed clusters (c04_n08 and c15_n05) individuals from Mochudi dominated, while in two other clusters (c02_n09 and c09_n05), Mochudi residents were a minority.
Table 1.
HIV-1C V1C5 clusters with 5+ members, bootstrap threshold ≥ 0.80
| Cluster ID | Members per cluster | Mochudi- unique, or mixed cluster |
ML bootstrap support |
Internode Certainty* |
The most recent HIV transmission |
Time to the most recent transmission |
Acute, or historic sub- epidemic** |
||
|---|---|---|---|---|---|---|---|---|---|
| Total | From Mochudi |
From outside Mochudi |
|||||||
| c01_n12 | 12 | 12 | 0 | unique | 0.92 | 0.71 | Nov 2007 Oct 2006 | 4.85.3 | historic |
| c02_n09 | 9 | 3 | 6 | mixed | 0.88 | 0.88 | Oct 1996 Jul 1994 | 15.517.8 | historic |
| c03_n07 | 7 | 7 | 0 | unique | 1 | 0.99 | Aug Feb 2009 | 2.73.1 | acute |
| c04_n08 | 8 | 6 | 2 | mixed | 0.8 | 0.84 | Feb 2002 Oct 2000 | 10.611.9 | historic |
| c05_n06 | 6 | 6 | 0 | unique | 0.99 | 0.98 | Jan 2003 Dec 2001 | 9.110.2 | historic |
| c06_n06 | 6 | 6 | 0 | unique | 1 | 1 | May 2005 Aug 2004 | 7.6.9 | historic |
| c07_n06 | 6 | 6 | 0 | unique | 0.86 | 0.53 | Oct 2004 Dec 2003 | 7.48.2 | historic |
| c08_n06 | 6 | 6 | 0 | unique | 0.9 | 0.89 | Jun 2004 Dec 2003 | 8.89.3 | historic |
| c09_n05 | 5 | 1 | 4 | mixed | 0.94 | 0.96 | Aug 1998 Jan 1997 | 12.614.2 | historic |
| c10_n05 | 5 | 5 | 0 | unique | 0.92 | 0.72 | Sep 20032002 | 78.4 | historic |
| c11_n05 | 5 | 5 | 0 | unique | 1 | 1 | Nov Feb 2005 | 6.18 | historic |
| c12_n05 | 5 | 5 | 0 | unique | 1 | 1 | Jan 2006 Apr 2005 | 6.18 | historic |
| c13_n05 | 5 | 5 | 0 | unique | 1 | 1 | Sep Jan 2005 | 5.6.2 | historic |
| c14_n05 | 5 | 5 | 0 | unique | 0.8 | 0.72 | Jun 2007 Nov 2006 | 3.94.5 | acute |
| c15_n05 | 5 | 4 | 1 | mixed | 0.81 | 0.62 | Feb 2004 Jan 2003 | 7.9.1 | historic |
most prevalent conflicting bipartitions
HIV sub-epidemic with (acute), or without (historic) HIV transmission over last 5 years
Acute and historic HIV sub-epidemics
We distinguish two types of HIV sub-epidemics based on the presence or absence of recent HIV transmissions, e.g., within the last 5 years, within targeted clusters. HIV lineages with recent HIV transmissions and R trajectories on the rise were considered acute HIV sub-epidemics. HIV lineages without evidence of recent HIV transmissions and declining or fluctuating R trajectories were considered historic HIV sub-epidemics. In this study, a five-year period (from the most recent sampling date in the cluster) was chosen as the recency period.
Using a combination of the 5-year recency threshold and the shape of R trajectories, we identified 2 acute and 13 historic HIV sub-epidemics with 5+ members in Mochudi (Table 1).
To test whether the R value at the root is smaller than the R value at present (expanding HIV sub-epidemic), we calculated the Bayes Factors (BF) for each cluster (Jeffreys, 1961) by comparing the R value in the earliest interval (at the root) to that of the last interval (at present) at each saved sample of the MCMC.
Two HIV sub-epidemics, c03_n09 and c14_n05, were identified as acute. In these sub-epidemics, the time between the most recent HIV transmission and the most recent sample is 3.1 and 4.5 years, respectively. Both sub-epidemics demonstrate an increase in R towards the present, with moderate evidence for cluster c03_n09 (BF=4.46) and weak evidence for cluster c14_n05 (BF=2.02).
With 5.3 years since the most recent viral transmission, cluster c01_n12 was close to the 5-year threshold. The R-trajectory in c01_n12 was elevated above the threshold of 1.0 in the early and mid-2000's, but declines toward 1.0 in the most recent time period. Based on the observed R-trajectory and the BF value of 1.18, c01_n12 was not considered an acute HIV sub-epidemic. All other clusters were above the 5-year recency threshold with BF values below 2, and were therefore considered to be historic HIV sub-epidemics (Table 1 and Figs. 4A–4D).
In this analysis, the 5-year recency was based on the assumption that the infectious period of an HIV-infected individual is approximately 5 years, although heterogeneity among individuals could vary broadly. Applying an alternative, more stringent recency period would result in fewer acute HIV sub-epidemics. For example, using a 3-year recency threshold would leave only one acute sub-epidemic, c03_n07, while tightening the recency threshold to 2 years would result in no acute HIV sub-epidemics identified among the analyzed HIV lineages in Mochudi.
Relaxing clustering definition
To examine how relaxing the clustering definition affects HIV transmission chains, we analyzed clusters with bootstrap support between 0.70 and 0.80. The number of HIV sequences in clusters increased from 604 (27.3%; 95% CI 25.5% to 29.2%) to 707 (31.9%; 95% CI 30.0% to 33.9%). The number of sequences in clusters with 5+ members increased from 94 (4.2%; 95% CI 3.5% to 5.2%) to 154 (7.0%; 95% CI 6.0% to 8.1%). Six additional clusters with 5+ members included one cluster with 22 members, one with 17, two with 8, one with 7, and one with 6 members. Our focus was on the first five of these clusters, as the smallest cluster with 6 members was essentially the same as previously described cluster c15_n05 with one additional sequence from Mochudi. The trajectories of the effective reproductive number R in HIV sub-epidemics with 5+ members that were identified by relaxing bootstrap support between 0.70 and 0.80 were similar to the R-trajectories described above for clusters identified by bootstrap of ≥ 0.80 (supplementary Figure S1).
All clusters with 5+ members that were identified by relaxed bootstrap threshold between 0.70 and 0.80 included up to 4 nested clusters identified with more stringent bootstrap support of ≥ 0.80. The size of the nested clusters was small, with 2–3 members per cluster. Five clusters with 5+ members were mixed by composition, although the proportion of non-Mochudi sequences was low (Table 2). Two of these clusters, c17_n22 and c21_n08 could be classified as acute HIV sub-epidemics based on their R-trajectories and estimated time of the most recent HIV transmission. However, both of them had low internode certainty, 0.38 and 0.48, respectively. There is no statistical evidence for an increase in R for the clusters identified by relaxing the bootstrap threshold below 0.80.
Table 2.
HIV-1C V1C5 clusters with 5+ members, relaxed bootstrap threshold between 0.70 & 0.80
| Cluster ID | Members per cluster | Mochudi- unique, or mixed cluster |
ML bootstrap support |
Internode Certainty* |
The most recent HIV transmission |
Time to the most recent transmission |
Acute, or historic sub- epidemic** |
||
|---|---|---|---|---|---|---|---|---|---|
| Total | From Mochudi |
From outside Mochudi |
|||||||
| c16_n17 | 17 | 14 | 3 | mixed | 0.76 | 0.84 | Jan 2005 | 7.0 | historic |
| c17_n22 | 22 | 18 | 4 | mixed | 0.70 | 0.38 | May 2010 | 1.9 | acute# |
| c19_n07 | 7 | 5 | 2 | mixed | 0.71 | 0.76 | Dec 2006 | 5.1 | historic |
| c20_n08 | 8 | 7 | 1 | mixed | 0.78 | 0.61 | Sep 2001 | 10.5 | historic |
| c21_n08 | 8 | 7 | 1 | mixed | 0.72 | 0.48 | Jun 2007 | 4.7 | acute# |
most prevalent conflicting bipartitions
HIV sub-epidemic with (acute), or without (historic) HIV transmission over last 5 years
Not supported by the BF values
Introduction of HIV-1C lineages to Mochudi
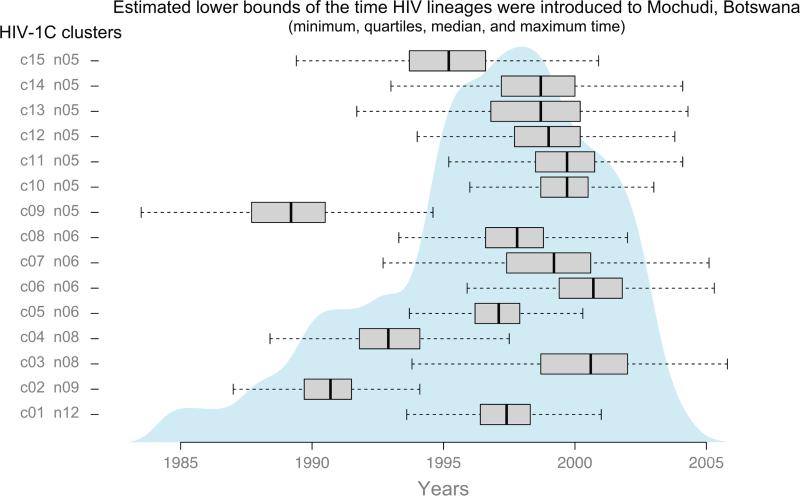
We used the time of the most recent common ancestor (tMRCA) for HIV sub-epidemics with 5+ members as lower bounds for the time HIV-1C lineages were introduced to Mochudi. The lower bounds for the timing of each introduction indicate the latest possible time, with the possibility of an earlier introduction. The tMRCA was estimated through the posterior distribution of the tree heights obtained for each HIV lineage. Figure 5 illustrates the summary statistics (median, quartiles, and range) for each sub-epidemic. The barplots show the estimated lower bounds of the time at which each sub-epidemic was introduced to Mochudi. The shaded density plot is a smoothed summary over all sub-epidemics, obtained by dividing time into small units (0.1 years) and assigning to each time point the number of sub-epidemics for which the 95% HPD of its time of introduction contains this time point. This summary suggests that among clusters with 5+ members, the majority of HIV-1C lineages were introduced to Mochudi .at, or before 1996 and 2002.
Figure 5.
Estimated lower bounds of the introduction time for 15 HIV-1C lineages with 5+ members in Mochudi. Boxplots show medians and quartiles of the estimated time. Dashed lines indicate ranges of the estimated time per HIV lineage. Gray area on the background summarizes estimated lower bounds of time for 15 HIV-1C lineages.
Timing of viral transmissions within HIV-1C sub-epidemics in Mochudi
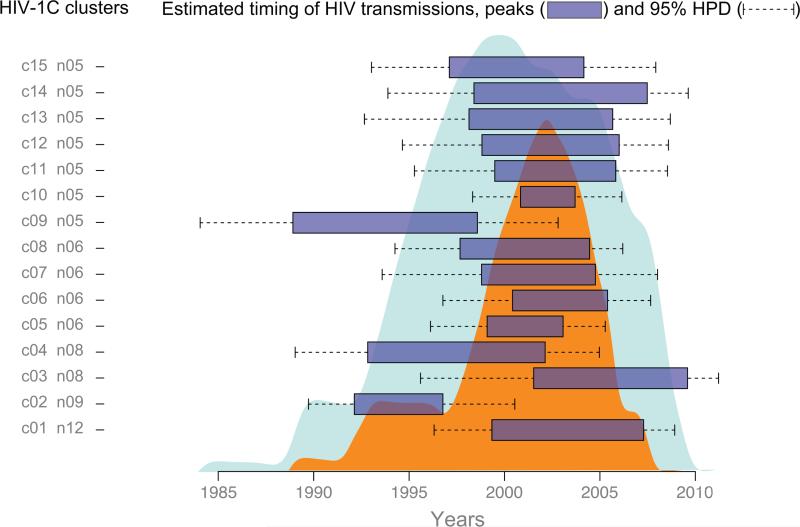
The internal nodes in the MCC time tree reflect the estimated time of coalescence of any two lineages. Assuming that a substantial fraction of HIV transmissions occurs during early stage of infection (Wawer et al., 2005) due to high levels of HIV-1 RNA (Quinn et al., 2000), we use this as a proxy for the time of viral transmission. The projection of internal nodes on the time scale gives an idea of the potential time of transmission events. The uncertainty of the coalescent times is reflected in the 95% HPD intervals. Overall, this analysis provides estimates for the time of HIV transmissions within each HIV sub-epidemic in Mochudi.
We utilized the MCC time-trees to estimate the time of viral transmission within 15 HIV-1C sub-epidemics in Mochudi. Specifically, we focused on the time interval between the oldest and the most recent viral transmission within each cluster with 5+ members. For each sub-epidemic, this time interval was estimated by projecting internal nodes associated with terminal branches to the time line in the MCC time-tree (Fig. 6). The time interval was interpreted as peak viral transmission for a particular HIV lineage (shown as horizontal boxes). The corresponding 95% HPD values of flanking nodes were interpreted as confidence intervals (shown as dashed lines).
Figure 6.
Estimated time of viral transmissions within 15 HIV-1C lineages with 5+ members in Mochudi. Boxplots show peaks, while dashed lines indicate 95% HPD for each sub-epidemic. Smaller (orange) area on the background summarizes peaks of HIV transmission. Larger (blue) area on the background summarizes 95% HPD's.
The cumulative effect of HIV transmissions was estimated as a sum of viral transmissions within each sub-epidemic per year. The smaller (orange) area shows cumulative peaks of HIV transmissions, while the broader (light blue) area indicates 95% HPD, accounting for uncertainty in estimates of the time of HIV transmissions. The analysis suggests that the majority of HIV transmissions in Mochudi (within the 15 sub-epidemics analyzed) peaked over about a decade from about 1997 to 2007. The 95% HPD intervals spread across a broader time interval from about 1993 to 2009.
Discussion
Insight into the dynamics of HIV transmission networks is critical for better understanding of HIV incidence in communities, and for the design and evaluation of HIV prevention strategies. Phylogenetic mapping and linkage of circulating viruses could help in developing strategies aimed to reduce onward HIV transmissions in communities. The structure and dynamics of HIV transmission networks in communities could be inferred through HIV cluster analysis. A robust and cost-effective methodology for HIV cluster analysis is particularly important for scale-up of HIV prevention strategies.
In this study we used phylogenetic linkage as a tool for breaking down the HIV epidemic in a single peri-urban village in Botswana into a series of phylogenetically distinct HIV sub-epidemics. Fitting of newly diagnosed cases of HIV infection into phylogenetically mapped clusters could help to identify “acute” sub-epidemics. The ultimate goal of breaking down a local epidemic into HIV sub-epidemics is to trace virus spread and to extinguish HIV transmission chains, one by one, by targeted interventions. If this approach works at the community level, it could be expanded to the global HIV epidemic.
The important clinical and public health relevance of the HIV sub-epidemics approach lies in its ability to track the main driver of HIV transmissions in communities. To identify circulating HIV lineages in real time, spread of the virus in communities should be traced proactively. Knowledge of HIV spread is time-sensitive. Targeted interventions, such as enhanced HTC, linkage to care, and initiation of ART, could be applied to “acute” HIV sub-epidemics, i.e., sub-epidemics with recent viral transmissions. Targeted treatment-as-prevention (TasP) for the members of acute sub-epidemics could be more efficient and cost-effective than uniformly applied TasP, particularly if the scale-up of TasP takes time.
In this study we showed differential transmission dynamics in HIV sub-epidemics caused by different HIV lineages. We identified acute HIV sub-epidemics with evidence of recent HIV transmissions and distinguished them from historic sub-epidemics without recent HIV transmissions. Importantly, in the context of high sampling density, our results suggest that only a small fraction of circulating HIV lineages are acute (with recent HIV transmissions). It is likely that only the acute sub-epidemics fuel HIV spread in communities at any given time.
In contrast to acute sub-epidemics, HIV sub-epidemics without recent infections represent historic HIV transmissions that happened in the past. As this type of sub-epidemic does not contribute to the current spread of HIV in communities, it might require less attention and fewer resources and dedicated interventions. The ultimate goal of public health interventions could be to transform acute HIV sub-epidemics into historical ones.
For each HIV sub-epidemic with 5+ members, we estimated the effective reproductive number R by linking evolutionary analysis with the birth-death model (Stadler et al., 2012; Stadler et al., 2013).
Information on the sampling origin of clustered viral sequences is critical for better understanding of HIV transmission dynamics, and transmission mixing, in particular. Most of the analyzed clusters with 5+ members, 11 of 15, included HIV-1C sequences from Mochudi only, suggesting local spread of these viral lineages in Mochudi. However, four clusters included sequences from other parts of Botswana outside of Mochudi indicating broader transmission range of some viral lineages.
We provided evidence that the life span of each HIV sub-epidemic with a predominantly heterosexual mode of virus transmission is limited. Within the sub-epidemic, the number of HIV transmissions rises, peaks, and declines over time. This notion provides a reasonable hope that each acute HIV sub-epidemic could be ended, and transformed into a historical sub-epidemic. The median life span of historical sub-epidemics – time of HIV transmissions within the sub-epidemic within the boundaries of 95% HPD – was 13.3 years. The median peak of HIV transmissions was 6.8 years, ranging from 2.9 to 9.7 years.
Taken together, the study provides evidence for the presence of multiple phylogenetically distinct HIV-1C lineages circulating in the community and the limited life span of particular HIV-1C lineages. The temporal nature of HIV sub-epidemics may be related to the patterns of partnerships forming and to the specifics of heterosexual transmission of HIV in southern African communities. Our results suggest that in contrast to the stable progressive growth of HIV transmission clusters over time described previously in the predominantly MSM and IDU epidemics (Brenner et al., 2013; Brenner et al., 2011; Leigh Brown et al., 2011; Poon et al., 2014), HIV sub-epidemics in the predominantly heterosexual transmission setting, such as southern African communities, have limited life spans and temporal dominance at any given time.
One of the study limitations is the focus on HIV clusters with 5+ members, which represent a relatively small subset of HIV-1C circulating in Mochudi, Botswana. This limitation is related to the methodology of inferring epidemiological parameters from virus sequence data. Due to the small size of the clusters we combined all clusters in a single analysis and let them share one of the epidemiological parameters, the rate to become non-infectious, as well as the evolutionary rate, which was fixed to 7.86E-03. Nevertheless, we recommend cautious interpretation of results originating from clusters with very few members, and suggest that additional validation of the methodological approach used in this study might be warranted. We were not able to take into account the difference between sampling densities within and outside Mochudi due to the small size of the clusters, and particularly the small number of non-Mochudi sequences, which did not allow us separate estimation.
Sampling events were assumed to remove infected individuals from the infectious pool. To test if this has any impact on the results, we repeated the analysis using the sampled ancestor version of the birth-death skyline plot (Gavryushkina et al., 2014). However, none of the clusters supported the existence of sampled ancestors within our samples (data not shown).
Trajectories of the effective reproductive number R in this study had relatively broad 95% HPD. We calculated the BF values for each cluster, testing whether the R value at the root is smaller than the R value at present. When there is no or only weak evidence for an increase in R through time, it is possible that a decreasing R-trajectory might have provided a similarly good fit to the data as an increasing R-trajectory.
The definition of an HIV cluster may be another limitation of this study. The high (72%) sampling density utilized in Mochudi is combined with the bootstrapped ML inference of HIV phylogeny in this study. While pairwise distances were taken into account, the model-based phylogeny (the bootstrap threshold in ML analysis) was the primary criterion for identification of HIV clusters. The rationale was based on HIV transmission mode, specifics of sampling, and targeted region of the HIV-1 genome. The predominant mode of virus transmission in the HIV epidemic in Mochudi, as in most southern African countries, is heterosexual. The Mochudi specimens represented a broad range of HIV infection stages including numerous chronic infections. The diversity within the targeted V1C5 region of HIV-1C env gp120 is one of the highest across the viral genome. Due to high intra-host diversity within the targeted V1C5 region, using pairwise distance threshold as the primary criterion for cluster definition could lead to elimination of the vast majority of viral sequences from analysis. However, the criteria for definition of an HIV transmission cluster (e.g., bootstrap support, tree certainty, or pairwise distance threshold) in a predominantly heterosexual epidemic remain uncertain. The methodology of phylogenetic inference and thresholds used for cluster identification in this study match well with previous studies (Bezemer et al., 2013; Bezemer et al., 2014; Bezemer et al., 2010; Hue et al., 2004, 2005a; Hue et al., 2005b; Hughes et al., 2009; Kosakovsky Pond et al., 2008; Leigh Brown et al., 2011; Lewis et al., 2008; Volz et al., 2013a; Volz et al., 2013b; Volz et al., 2012; Volz et al., 2009). Further studies are needed to identify optimal, or “state of the art,” approaches for biologically meaningful HIV cluster analysis.
This study used relatively short viral sequences, ~1,200 bp, spanning the V1C5 region of HIV-1C env gp120. It is possible that bootstrapped ML inference of the short-range sequence set selected HIV lineages that represent only small sub-chains of much larger transmission chains in the population. Recently we demonstrated that viral sequence length plays an important role in HIV cluster analysis (Novitsky et al., 2015). It is likely that using long-range sequences could refine clustering and reveal more extensive clustering.
The relaxed definition of clusters with lower bootstrap threshold (≥0.70) resulted in a larger number of identified clusters including clusters with 5+ members. This is consistent with our recent studies on sampling density (Novitsky et al., 2014) and importance of virus sequence length (Novitsky et al., 2015) in HIV cluster analysis. Two additional acute HIV sub-epidemics were found among clusters with 5+ members and bootstrap support between 0.70 and 0.80, although both of these clusters had low internode certainty. Classification of these clusters as acute was driven primarily by recent HIV transmissions in the MCC time-trees, and should be interpreted cautiously due to low internode certainty (0.38 and 0.48), low bootstrap support (0.70 and 0.72), and a lack of support by the BF values.
Finally, we are assuming that the transmission tree coincides with the phylogeny, which is a fair assumption when superinfection is relatively low and the within-host coalescence time is short compared to the viral transmission time.
In summary, we demonstrated that proactive viral genotyping and breaking down a local HIV epidemic into a series of phylogenetically distinct sub-epidemics might be a useful approach for identification of acute HIV sub-epidemics in communities. This approach might help to reveal the structure and dynamics of HIV transmission networks in communities, and could aid in the design of targeted interventions for members of acute sub-epidemics.
Supplementary Material
A local southern African HIV epidemic was broken into phylogenetically distinct sub-epidemics.
Effective reproductive number trajectories estimated for HIV sub-epidemics with 5+ members.
“Acute” sub-epidemics were distinguished from “historic” sub-epidemics.
Real-time HIV genotyping and sub-epidemic analysis could aid in design of targeted interventions.
Acknowledgements
We are very grateful to all participants of the Mochudi Prevention Project in Botswana and community leaders in Mochudi, Botswana. We thank the study field team and the leader of the field team, Sarah Masole, for dedication and outstanding work in outreach. We thank the Botswana Ministry of Health for their collaboration. We thank Jesse Rizutko for superior IRB-related support. Finally, we thank Lendsey Melton for excellent editorial assistance. The Mochudi Prevention Project in Botswana was supported and funded by NIH grant R01 AI083036, An HIV Prevention Program for Mochudi, Botswana. DK was supported by an ETH Zurich postdoctoral fellowship. LO was supported in part by NIH grant D43 TW000004.
We thank reviewers for their valuable input.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
VN, DK and ME conceived and designed the study. SM, LO and EW implemented the study and collected specimens and data. SM performed laboratory tests. EW designed and implemented electronic data collection and storage. VN and DK performed the statistical analysis and wrote the manuscript.
References
- Andrieu G, Caraux G, Gascuel O. Confidence intervals of evolutionary distances between sequences and comparison with usual approaches including the bootstrap method. Mol Biol Evol. 1997;14:875–882. doi: 10.1093/oxfordjournals.molbev.a025829. [DOI] [PubMed] [Google Scholar]
- AVERT History of HIV & AIDS in Africa. 2014 http://www.avert.org/history-hiv-aids-africa.htm.
- Barcherini S, Cantoni M, Grossi P, Verdecchia A. Reconstruction of human immunodeficiency virus (HIV) sub-epidemics in Italian regions. Int J Epidemiol. 1999;28:122–129. doi: 10.1093/ije/28.1.122. [DOI] [PubMed] [Google Scholar]
- Bezemer D, Faria NR, Hassan AS, Hamers RL, Mutua G, Anzala O, Mandaliya KN, Cane PA, Berkley JA, Rinke de Wit TF, Wallis CL, Graham SM, Price MA, Coutinho R, Sanders EJ. HIV-1 transmission networks amongst men having sex with men and heterosexuals in Kenya. AIDS Res Hum Retroviruses. 2013 doi: 10.1089/aid.2013.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezemer D, Ratmann O, van Sighem A, Dutilh BE, Faria N, van den Hengel R, Gras L, Reiss P, de Wolf F, Fraser C, ATHENA observational cohort . Ongoing HIV-1 Subtype B Transmission Networks in the Netherlands. CROI 2014; Boston, MA.: 2014. [Google Scholar]
- Bezemer D, van Sighem A, Lukashov VV, van der Hoek L, Back N, Schuurman R, Boucher CA, Claas EC, Boerlijst MC, Coutinho RA, de Wolf F, cohort A.o. Transmission networks of HIV-1 among men having sex with men in the Netherlands. AIDS. 2010;24:271–282. doi: 10.1097/QAD.0b013e328333ddee. [DOI] [PubMed] [Google Scholar]
- Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botswana Ministry of Health BOTSWANA NATIONAL POLICY ON HIV/AIDS. 1993 http://hivhealthclearinghouse.unesco.org/sites/default/files/resources/iiep_botswana_national_hivaids_policy_93.pdf.
- Bouckaert R, Heled J, Kühnert D, Vaughan TG, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST2: A software platform for Bayesian evolutionary analysis. PLOS Computational Biogogy. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B, Wainberg MA, Roger M. Phylogenetic inferences on HIV-1 transmission: implications for the design of prevention and treatment interventions. AIDS. 2013;27:1045–1057. doi: 10.1097/QAD.0b013e32835cffd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner BG, Roger M, Moisi DD, Oliveira M, Hardy I, Turgel R, Charest H, Routy JP, Wainberg MA. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS. 2008;22:2509–2515. doi: 10.1097/QAD.0b013e3283121c90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner BG, Roger M, Stephens D, Moisi D, Hardy I, Weinberg J, Turgel R, Charest H, Koopman J, Wainberg MA. Transmission Clustering Drives the Onward Spread of the HIV Epidemic Among Men Who Have Sex With Men in Quebec. J Infect Dis. 2011;204:1115–1119. doi: 10.1093/infdis/jir468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley TR, Cunningham CW. The effects of nucleotide substitution model assumptions on estimates of nonparametric bootstrap support. Mol Biol Evol. 2002;19:394–405. doi: 10.1093/oxfordjournals.molbev.a004094. [DOI] [PubMed] [Google Scholar]
- Cantoni M, Cozzi Lepri A, Grossi P, Pezzotti P, Rezza G, Verdecchia A. Use of AIDS surveillance data to describe subepidemic dynamics. Int J Epidemiol. 1995;24:804–812. doi: 10.1093/ije/24.4.804. [DOI] [PubMed] [Google Scholar]
- Clauset A, Shalizi CR, Newman MEJ. Power-law distributions in empirical data. SIAM Review. 2009;51:661–703. [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Bootstrap Methods: Another Look at the Jackknife. Annals of Statistics. 1979;7:1–26. [Google Scholar]
- Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci U S A. 1996;93:7085–7090. doi: 10.1073/pnas.93.14.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using a bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies. Sinauer Associates, Inc.; 2004. [Google Scholar]
- Felsenstein J, Kishino H. Is there something wrong with the bootstrap on phylogenies? A reply to Hillis and Bull. Syst Biol. 1993;42:193–200. [Google Scholar]
- Feng Y, He X, Hsi JH, Li F, Li X, Wang Q, Ruan Y, Xing H, Lam TT, Pybus OG, Takebe Y, Shao Y. The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS. 2013;27:1793–1802. doi: 10.1097/QAD.0b013e328360db2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SD, Volz EM. Viral phylodynamics and the search for an ‘effective number of infections’. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2010;365:1879–1890. doi: 10.1098/rstb.2010.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavryushkina A, Welch D, Stadler T, Drummond AJ. Bayesian inference of sampled ancestor trees for epidemiology and fossil calibration. PLoS Comput Biol. 2014;10:e1003919. doi: 10.1371/journal.pcbi.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Graw F, Leitner T, Ribeiro RM. Agent-based and phylogenetic analyses reveal how HIV-1 moves between risk groups: injecting drug users sustain the heterosexual epidemic in Latvia. Epidemics. 2012;4:104–116. doi: 10.1016/j.epidem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Hue S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- Hue S, Clewley JP, Cane PA, Pillay D. Investigation of HIV-1 transmission events by phylogenetic methods: requirement for scientific rigour. AIDS. 2005a;19:449–450. doi: 10.1097/01.aids.0000161778.15568.a1. [DOI] [PubMed] [Google Scholar]
- Hue S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci U S A. 2005b;102:4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog. 2009;5:e1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys H. Theory of Probability. 3rd ed. Oxford University Press; Oxford, U.K.: 1961. [Google Scholar]
- Kivela PS, Krol A, Salminen MO, Ristola MA. Determinants of late HIV diagnosis among different transmission groups in Finland from 1985 to 2005. HIV medicine. 2010;11:360–367. doi: 10.1111/j.1468-1293.2009.00783.x. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Poon AF, Leigh Brown AJ, Frost SD. A maximum likelihood method for detecting directional evolution in protein sequences and its application to influenza A virus. Mol Biol Evol. 2008;25:1809–1824. doi: 10.1093/molbev/msn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert D, Stadler T, Vaughan TG, Drummond AJ. Simultaneous reconstruction of evolutionary history and epidemiological dynamics from viral sequences with the birth-death SIR model. J R Soc Interface. 2014;11:20131106. doi: 10.1098/rsif.2013.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS. Tree robustness and clade significance. Syst Biol. 2000;49:829–836. doi: 10.1080/106351500750049879. [DOI] [PubMed] [Google Scholar]
- Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT, Collaboration UHDR. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis. 2011;204:1463–1469. doi: 10.1093/infdis/jir550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal GE, Gunthard HF, Bonhoeffer S, Stadler T. Using an epidemiological model for phylogenetic inference reveals density dependence in HIV transmission. Mol Biol Evol. 2014;31:6–17. doi: 10.1093/molbev/mst172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal GE, Kouyos R, Stadler T, Wyl V, Yerly S, Boni J, Cellerai C, Klimkait T, Gunthard HF, Bonhoeffer S. Inferring epidemic contact structure from phylogenetic trees. PLoS Comput Biol. 2012;8:e1002413. doi: 10.1371/journal.pcbi.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. 2005;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York, NY.: 2000. [Google Scholar]
- Ng KT, Ong LY, Lim SH, Takebe Y, Kamarulzaman A, Tee KK. Evolutionary history of HIV-1 subtype B and CRF01_AE transmission clusters among men who have sex with men (MSM) in Kuala Lumpur, Malaysia. PLoS One. 2013;8:e67286. doi: 10.1371/journal.pone.0067286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Bussmann H, Logan A, Moyo S, van Widenfelt E, Okui L, Mmalane M, Baca J, Buck L, Phillips E, Tim D, McLane MF, Lei Q, Wang R, Makhema J, Lockman S, DeGruttola V, Essex M. Phylogenetic Relatedness of Circulating HIV-1C Variants in Mochudi, Botswana. PLoS One. 2013;8:e80589. doi: 10.1371/journal.pone.0080589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Moyo S, Lei Q, DeGruttola V, Essex M. Impact of Sampling Density on the Extent of HIV Clustering. AIDS Res Hum Retroviruses. 2014;30:1226–1235. doi: 10.1089/aid.2014.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Moyo S, Lei Q, DeGruttola V, Essex M. Importance of Viral Sequence Length and Number of Variable and Informative Sites in Analysis of HIV Clustering. AIDS Res Hum Retroviruses. 2015 doi: 10.1089/aid.2014.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, Ndung'u T, Klein I, Chang SY, Peter T, Thior I, Foley BT, Gaolekwe S, Rybak N, Gaseitsiwe S, Vannberg F, Marlink R, Lee TH, Essex M. HIV-1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Margolin L, Baca J, Rossenkhan R, Moyo S, van Widenfelt E, Essex M. Transmission of Single and Multiple Viral Variants in Primary HIV-1 Subtype C Infection. PLoS One. 2011;6:e16714. doi: 10.1371/journal.pone.0016714. PMCID: PMC3048432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Woldegabriel E, Wester C, McDonald E, Rossenkhan R, Ketunuti M, Makhema J, Seage GR, 3rd, Essex M. Identification of primary HIV-1C infection in Botswana. AIDS Care. 2008;20:806–811. doi: 10.1080/09540120701694055. NIHMSID # 79283 PMCID: PMC2605733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky VA, Montano MA, McLane MF, Renjifo B, Vannberg F, Foley BT, Ndung'u TP, Rahman M, Makhema M, J., Marlink R, Essex M. Molecular cloning and phylogenetic analysis of HIV-1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 1999;73:4427–4432. doi: 10.1128/jvi.73.5.4427-4432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- Pineda-Pena AC, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, Gomez-Lopez A, Camacho RJ, de Oliveira T, Vandamme AM. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol. 2013;19:337–348. doi: 10.1016/j.meegid.2013.04.032. [DOI] [PubMed] [Google Scholar]
- Poon AF, Joy JB, Woods CK, Shurgold S, Colley G, Brumme CJ, Hogg RS, Montaner JS, Harrigan PR. The impact of clinical, demographic and risk factors on rates of HIV transmission. A population-based phylogenetic analysis in British Columbia, Canada. J Infect Dis. 2014 doi: 10.1093/infdis/jiu560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan MO, Lutalo T, Gray RH. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. http://www.R-project.org/ [Google Scholar]
- Salichos L, Rokas A. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature. 2013;497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- Salichos L, Stamatakis A, Rokas A. Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol Biol Evol. 2014;31:1261–1271. doi: 10.1093/molbev/msu061. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. Confidence limits on phylogenies: the bootstrap revisited. Cladistics. 1989;5:113–129. doi: 10.1111/j.1096-0031.1989.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Smith MJ. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Stadler T, Bonhoeffer S. Uncovering epidemiological dynamics in heterogeneous host populations using phylogenetic methods. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2013;368:20120198. doi: 10.1098/rstb.2012.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T, Kouyos R, von Wyl V, Yerly S, Boni J, Burgisser P, Klimkait T, Joos B, Rieder P, Xie D, Gunthard HF, Drummond AJ, Bonhoeffer S. Estimating the basic reproductive number from viral sequence data. Mol Biol Evol. 2012;29:347–357. doi: 10.1093/molbev/msr217. [DOI] [PubMed] [Google Scholar]
- Stadler T, Kuhnert D, Bonhoeffer S, Drummond AJ. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc Natl Acad Sci U S A. 2013;110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DM, Motitz C, Mable BK, editors. Molecular systematics. 2nd edition. Sinauer; Sunderland, Mass.: 1996. pp. 407–514. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanser F, de Oliveira T, Maheu-Giroux M, Barnighausen T. Concentrated HIV subepidemics in generalized epidemic settings. Curr Opin HIV AIDS. 2014;9:115–125. doi: 10.1097/COH.0000000000000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS AIDS by the numbers. 2013a file:///Users/vladimirnovitsky/Documents/PubMed__2014/PubMed%20part%20L-Z/UNAIDS/2013_JC2571_AIDS_by_the_numbers_en.pdf.
- UNAIDS Global Report. UNAIDS Report on the global AIDS epidemic 2013. 2013b [Google Scholar]
- UNAIDS The Gap Report. 2014 file:///Users/vladimirnovitsky/Documents/PubMed__2014/PubMed%20part%20L-Z/UNAIDS/2014/UNAIDS_Gap_report_en.pdf.
- Van de Peer Y. Phylogenetic inference based on distance methods. In: Lemey P, Salemi M, Vandamme AM, editors. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2nd edition Cambridge University Press; 2009. [Google Scholar]
- Volz EM, Ionides E, Romero-Severson EO, Brandt MG, Mokotoff E, Koopman JS. HIV-1 Transmission during Early Infection in Men Who Have Sex with Men: A Phylodynamic Analysis. PLoS Med. 2013a;10:e1001568. doi: 10.1371/journal.pmed.1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz EM, Koelle K, Bedford T. Viral phylodynamics. PLoS Comput Biol. 2013b;9:e1002947. doi: 10.1371/journal.pcbi.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz EM, Koopman JS, Ward MJ, Brown AL, Frost SD. Simple Epidemiological Dynamics Explain Phylogenetic Clustering of HIV from Patients with Recent Infection. PLoS Comput Biol. 2012;8:e1002552. doi: 10.1371/journal.pcbi.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz EM, Kosakovsky Pond SL, Ward MJ, Leigh Brown AJ, Frost SD. Phylodynamics of infectious disease epidemics. Genetics. 2009;183:1421–1430. doi: 10.1534/genetics.109.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, Kiwanuka N, Kigozi G, Kiddugavu M, Lutalo T, Nalugoda F, Wabwire-Mangen F, Meehan MP, Quinn TC. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DM, Kosakovsky Pond SL. The global transmission network of HIV-1. J Infect Dis. 2014;209:304–313. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.