Abstract
The peptidyl nucleoside blasticidin S (BS) isolated from Streptomyces griseochromogenes was the first non-mercurial fungicide used on a large scale to prevent rice blast. In the biosynthesis of BS, leucylblasticidin S (LBS) was suggested as the penultimate metabolite with 20-fold less inhibitory activity than the final product BS. Incomplete conversion of LBS to BS at a variable efficiency ranging from 10% to 90% was observed either in the native strain S. griseochromogenes or a heterologous producer Streptomyces lividans WJ2. In this study, we determined that maturation of BS from LBS is not a spontaneous process but is governed by a standalone peptidase PepN, which hydrolyzes LBS in a pH-sensitive way with most appropriate of pH 7~8 but is inactive when the pH is below 5 or above 10. PepN1 and PepN2, two neighboring PepN homologs from Streptomyces lividans were purified in E. coli but displayed ca.100-fold difference in LBS hydrolytic activity. Overexpression of pepN1 in WJ2 enhanced BS yield by 100% and lowered the ratio of LBS to BS from 2:1 to 2:3. This work presents the expansion of the biological role for PepN in antibiotic maturation and the first report of hydrolysis of beta amide linkage by this conserved enzyme.
Blasticidin S (BS) is a nucleoside analog consisting of a cytosine linked to a dideoxyhexose and bonded to a modified arginine. BS was isolated from the broth of Streptomyces griseochromogenes in an effort to identify natural non-mercurial fungicides1. BS has been used on a large scale to control rice blast by foliar application against the virulent fungus (Pyricularia oryzae) which was the cause of the disease in Asia2. BS was found to strongly inhibit the production of aflatoxin by Aspergillus flavus without impairing fungal growth3.
Blasticidin S exerts its inhibitory activity by interfering with ribosomal protein synthesis in host cells. Unlike other peptidyltransferase inhibitors, such as clinically used chloramphenicol, linezolid, the lincosamides lincomycin and clindamycin, which bind to the A site of the large ribosomal subunit4,5,6, BS occupies the P site, where it induces conformational changes of the P-site tRNA7.
As BS displayed non-selective toxicity to prokaryotic and eukaryotic cells, it is of great interest to understand the resistance mechanism, particularly the mechanism for self-resistance. There have been two groups of BS resistance genes reported in non-BS producers. The first group featured by BSD or BSR encodes blasticidin S deaminase8,9,10,11,12 that converts BS to non-toxic deaminohydroxyblasticidin S. This group of resistance genes along with BS are widely used in the selection of the transformed cells, particularly in eukaryotic transgenic studies13,14,15. The other group is blasticidin S acetyltransferase that uses acetyl-coenzyme A to acetylate the amino group at C-13 of blasticidin S to give N-acetylblasticidin S16,17,18. In some fungi, two groups of resistance genes are present to convert BS to N-acetyldeaminohydroxyblasticidin S3. Impermeability of the cell membrane constitutes another barrier to delivery of BS to its intracellular targets. But some nutrient uptake systems could be hijacked for BS delivery, such as an ABC transporter named NppA1A2BCD19 and the leucine-rich repeat-containing 8(LRRC8) proteins20 in mammalian cells.
In studies of blasticidin S, demethylblasticidin S (DBS)21 and leucylblasticidin S (LBS)22 were isolated as the intermediates in the broth of S. griseochromogenes when the pH of the culture medium was kept below 4. However, LBS was later isolated from Streptomyces sp. SCC 1785 and S. griseochromogenes under the normal laboratory conditions23,24. Addition of leucine to C-13 of the beta-amino group of BS was expected as a new self-resistance mechanism because the antibiotic activity of LBS against indicator strain Bacillus circulans was decreased by 20 folds compared to BS23. LBS was suggested as the direct precursor for BS biosynthesis as the washed cells22 or cell lysate23 of S. griseochromogenes can convert LBS into BS (Fig. 1A). It is of particular interest to know where this conversion occurs to avoid self-destruction of the host by the generated BS, as well as identifying the protein responsible for this hydrolysis in vivo.
Figure 1. Transformation of LBS to BS is significantly affected by fermentation conditions.
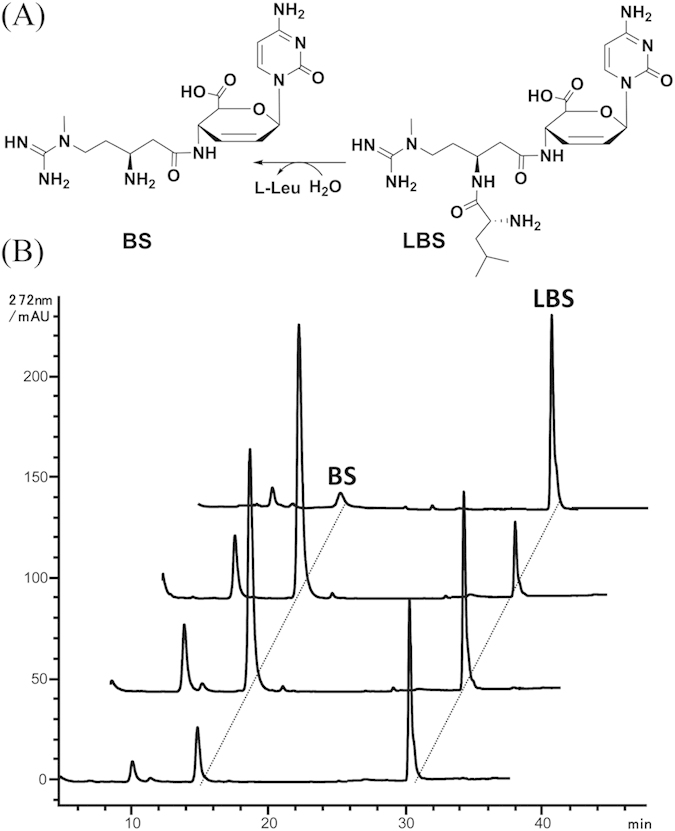
(A) Schematic representation of transformation of LBS to BS; (B) HPLC analysis of metabolites produced by S. lividans WJ2 which was engrafted with the expanded BS biosynthetic gene cluster in different fermentations. BS is abbreviated for the blasticidin S, LBS is for leucylblasticidin S. The ratio between LBS to BS varied from 2:9 to 9:1 in different fermentations.
In this study, we determined that PepN from E. coli and Streptomyces lividans can catalyze the hydrolysis of LBS into BS. The activity of PepN is pH-dependent wherein it shows the highest hydrolytic activity at pH 7–8 and loses its activity when pH is below 5 or above 10. The variation in the ratio between LBS to BS in the fermentation broth of S. lividans is attributed to the changes in activity of PepN that is affected by the medium pH in different fermentations.
Results
LBS and BS productivity is significantly affected by fermentation conditions
The blasticidin S native producer Streptomyces griseochromogenes has a very strong barrier system against introduction of foreign DNA. Heterologous expression of the BS biosynthetic gene cluster in Streptomyces lividans initially only yielded LBS, but not BS25. As the published BS gene cluster contains nine putatively unrelated genes upstream of blsS but stops at blsN, immediately downstream of essential gene blsM, we speculated that there might exist a peptidase gene downstream of blsM and thus cloned 8780-bp sequences (Accession number is JX244070) after blsN through chromosomal walking. We engrafted the expanded BS biosynthetic gene cluster onto S. lividans to generate WJ2, which is able to produce BS11. We then conjectured that the 8780-bp sequence might encode the putative LBS peptidase gene.
blsN, bls-1 and bls-2 were then independently knocked out in S. lividans WJ2, and analysis of the metabolites and biological activity assay of three mutants revealed no difference from strain WJ2 (Supplementary Figure S1A,B), implying they are not peptidase genes or there exist multiple peptidase gene(s) elsewhere. Unexpectedly, we observed that all these mutants as well as WJ2 can produce LBS in addition to BS. The ratio between LBS to BS varied from 2:9 to 9:1 in different fermentations (Fig. 1B). It therefore raised two possibilities with respect to the maturation of BS from LBS: i) a LBS hydrolase gene locates outside of BS gene cluster and its activity is significantly affected by the fermentation condition; ii) LBS hydrolysis is a spontaneous process that is greatly affected by fermentation conditions.
Many non-BS producing cells can hydrolyze LBS into BS
The washed cells of S. griseochromogenes, the BS native producer, were reported to hydrolyze LBS into BS22. We repeated this experiment using the cells of S. lividans WJ2 (harboring the BS gene cluster) and its parent strain S. lividans HXY16 (without the BS gene cluster) as the negative control. Surprisingly, the cells of both strains are capable of hydrolyzing LBS to BS in similar effective way. We further found that cells from E. coli DH10B and Saccharomyces sake are also able to accomplish this process. The cell free extracts of these strains were all found to possess LBS hydrolytic activity as well (Fig. 2). However, the boiled CFE of E. coli DH10B cannot perform this process, demonstrating that LBS hydrolysis is not a spontaneous process and that there is a conserved hydrolase gene in charge of the maturation of LBS into active BS.
Figure 2. HPLC analysis of the reaction product by incubating LBS with the cell free extracts (CFE) of Streptomyces griseochromogenes, Saccharomyces sake, E. coli DH10B.
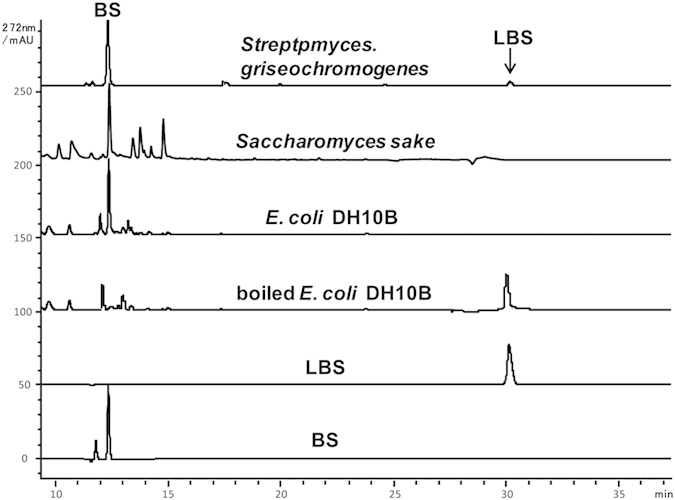
All of them were found to convert LBS into BS. Boiled CFE of DH10B was used as negative control.
Identification of PepN in E. coli and Streptomyces lividans as the peptidase involved in LBS hydrolysis
As shown in Fig. 2, E. coli, the most studied bacterium, displayed relatively high efficiency in LBS hydrolysis and was thus chosen as the start strain to clone LBS hydrolase gene. The CFE of E. coli were precipitated with different concentration of ammonium sulfate wherein the fraction precipitated by 40% (V/V) ammonium sulfate displayed high catalytic activity, and further subjected to sequential separation on HiTrap Q HP (GE Healthcare, 5 ml) and on MonoQ HR 5/5 column (GE Healthcare). The eluate fractions from No.15 till No.19 showed LBS hydrolysis activity. These fractions and the inactive fraction No. 14 were analyzed by gel electrophoresis. There are some comparatively thicker bands in the most active No.16 fraction, such as bands of approximate 95 kD and 66 kD (indicated by arrows in Fig. 3). The fractions of No.14 and No.16 were analyzed by LC-MS/MS for their protein compositions (Supplementary Figure S2). 296 specific proteins were identified in the fraction of No.16 wherein the five most abundant peptidases were chosen as the candidates for the LBS hydrolase (Table 1).
Figure 3. SDS-PAGE analysis of the protein fractions eluted from MonoQ HR 5/5 column. “−” represents this fraction has no LBS hydrolysis activity.
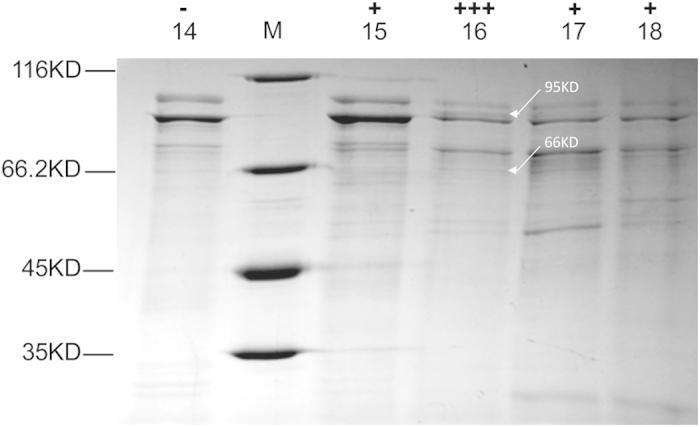
“ + ” represents positive in LBS hydrolysis. There are some comparatively thicker bands in the most active No.16 fraction, such as bands of approximate 95 kD and 66 kD indicated by arrows.
Table 1. Five most abundant peptidases identified in fraction 16.
| NO | Protein | Score | Mass | emPAI |
|---|---|---|---|---|
| 9 | Aminopeptidase N (PepN) | 2311 | 99313 | 1.56 |
| 139 | Predicted peptidase | 244 | 45288 | 0.33 |
| 147 | Predicted hydrolase | 227 | 23200 | 0.71 |
| 176 | Predicted hydrolase | 172 | 34153 | 0.74 |
| 201 | Predicted peptidase required for the maturation of the antibiotic peptide MccB17 | 138 | 48625 | 0.39 |
NO. represents the number of proteins in the order of decreasing score in fraction 16.
emPAI represents protein abundance.
The five peptidase genes were then disrupted in E. coli BW25113, an E. coli derivative facilitating homologous recombination between linear DNA fragment and chromosome. The CFE of the five mutants were assayed for LBS hydrolytic activity. Only the pepN mutant lost all LBS hydrolytic activity while the other four mutants retained the same efficacy in LBS hydrolysis as the wild type (Fig. 4). This result demonstrates that PepN is responsible for LBS hydrolysis in E. coli.
Figure 4. HPLC analysis of the reaction products for incubation of LBS with the CFEs of mutants of peptidase genes.
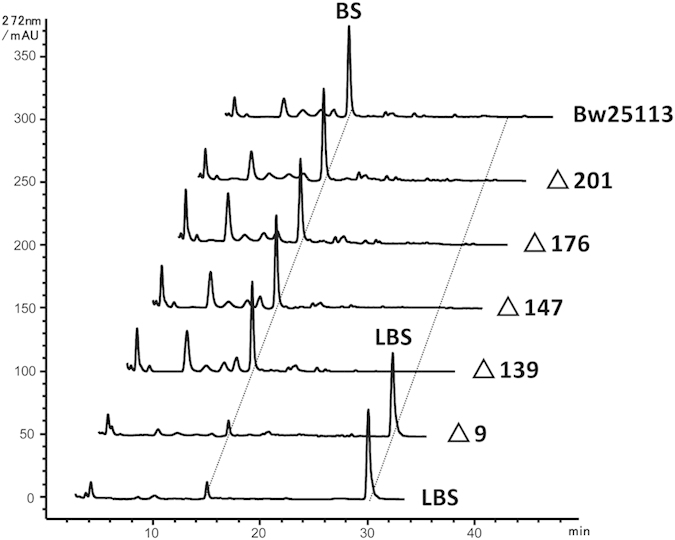
Only the pepN (∆9) mutant lost all LBS hydrolytic activity while the other four mutants retained the same efficacy in LBS hydrolysis as the wild type.
In vitro activities of PepNs from S. lividans WJ2 and E. coli are affected by pH variation
PepN homologs are widespread in prokaryotes and eukaryotes and may function as the LBS hydrolase in various strains. Two closely located PepN homologs from Streptomyces lividans were identified through blast search; PepN1 is 27% identical (114/427) and 42% positive (180/427), and PepN2 is 26% identical (91/346) and 42% positive (146/346) to PepN, respectively. We thus expressed and purified PepN, PepN1 and PepN2 in E. coli BL21 (DE3) to assay their LBS hydrolysis activity (Left panel, Supplementary Figure.S3). PepN and PepN1 exhibited similar LBS hydrolyzing efficiency which is at least 100-fold higher than that for PepN2 (Right panel, Supplementary Figure S3). The transcription level of pepN1 is comparable to that of blsD as well as constitutively expressed hrdB, but is at least 3 folds higher than that of pepN2 in S. lividans WJ2 (Supplementary Figure S4), therefore PepN1 is the major LBS hydrolase in S. lividans WJ2 during fermentation.
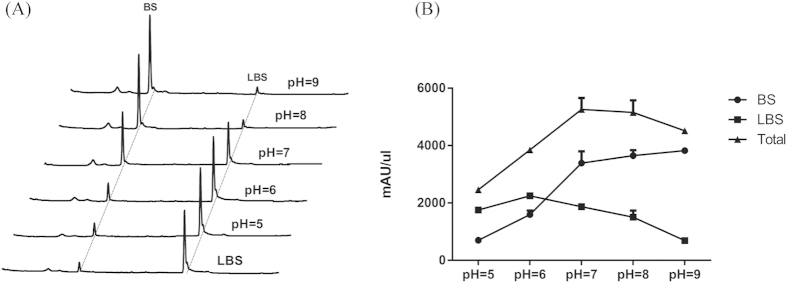
As in different fermentations, the proportion of LBS in the final metabolites varies, implying that PepN activity is affected by the changing medium conditions, particularly the pH value as it was affected very much by cell contamination or sterilizations process. PepN and PepN1 were then measured for their activity in hydrolysis of LBS at pH ranging from 5 to 9. PepN and PepN1 behave similarly at the same pH, and the pattern change of activity are also similar, namely with most appropriate pH for in vitro reaction at 7.5 and least activity above 10 or below 5 (Fig. 5A). These observation are consistent with that pH variation in the fermentation broth affected the LBS hydrolase activity (Fig. 5B) and thus determined the proportion of LBS to BS in the final metabolites.
Figure 5. pH is the key factor in regulating PepN activity in vitro and in vivo.
(A) HPLC analysis of reaction products for purified PepN1 with excessive LBS at pH ranging from 5 to 9 for 5 minutes, the most appropriate pH for in vitro reaction at 7.5 and least active above 10 or below 6, PepN activity is significantly affected by pH. (B) The production of BS and LBS and their total amount by S. lividans WJ2 when the pH of the fermentation medium was pre-adjusted to 5, 6, 7, 8 and 9, the pH is adjusted after medium sterilization.
Overexpression of PepN1 in S. lividans WJ2 to improve the yield of BS
As LBS is 20-fold less active than the final product BS23, in vivo conversion of LBS to BS by overexpression of pepN1 in S. lividans WJ2 is desired. pepN1 under the control of constitutive expression promoter PermE* were constructed and introduced into WJ2 to generate mutant strains S. lividans YGY6. The ratio of BS to LBS yield in the YGY6 is 3:2 as compared to that of 1:2 in WJ2::pIB139, the absolute yield of BS increased 100% upon the overexpression of PepN1(Fig. 6).These results demonstrated that overexpression of pepN1 is an efficient way to enhance BS production.
Figure 6. The yield of BS and LBS in S. lividans YGY7 (WJ2::pIB139), S. lividans YGY6 (WJ2::pIB139 + pepN1).
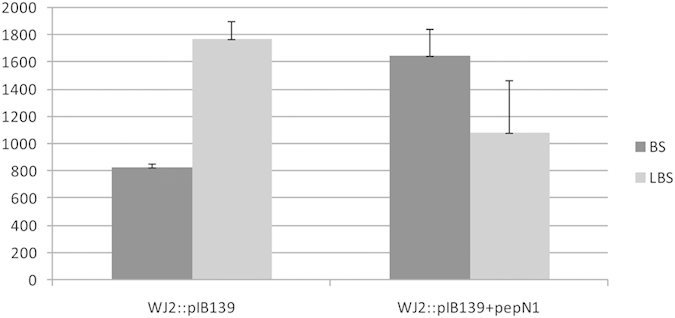
The ratio of BS to LBS yield in the YGY6 is 3:2 as compared to that of 1:2 in YGY7, the yield of BS has nearly doubled.
Discussion
Removal of the leucyl group from Leucylblasticidin S was proposed as the final step in BS biosynthesis23. Heterologous expression of blasticidin S biosynthetic gene cluster in Streptomyces lividans occasionally produces the mixture of LBS and BS. It remains a puzzle whether the LBS hydrolysis is a spontaneous process or the LBS hydrolase gene is included in the published BS biosynthetic gene cluster or locates elsewhere on the chromosome. In the process to clone the possible LBS hydrolase gene, we observed that CFE of some none-BS producers can efficiently covert LBS to BS whereas the boiled CFE of these strains cannot perform this process, demonstrating that LBS is not a spontaneous process and LBS hydrolase gene is outside of BS gene cluster. Hence, the published BS biosynthetic gene cluster actually encodes all essential proteins for LBS rather than BS. BS is now widely used commercially as a selection reagent in transgenic studies due to its higher inhibition activity than LBS, thus the study on the maturation mechanism for LBS into BS is of great application value.
We here report the identification of PepN as the LBS hydrolase by tracking the LBS hydrolytic activity in the CFE of E. coli and confirmed that PepN is the sole peptidase of E. coli to cleave the leucine from the beta-amino group of arginine. PepN in E. coli is a major aminopeptidase that is involved in downstream protein processing during cytosolic protein degradation and has a preference to cleave basic and small amino acids in an ATP-independent manner26. It is well conserved in all life kingdoms. Now that PepN1 and PepN2 were present and expressed in Streptomyces lividans, it is odd that WJ2 sometimes produces LBS at a ratio to BS as high as 9:1, but in some cases as low as 2:9, indicating that the fermentations conditions either affect the expression level of PepNs or the catalytic efficiency of PepNs in LBS hydrolysis. LBS was previously isolated when the fermentation broth was kept below pH 4 but later studies revealed it is unnecessary. This study again suggested that pH might plays an important role in regulation of the activity of PepNs. Consistent with this assessment is that the yield of BS is highest when the fermentation broth was kept at pH 8 as compared to lowest yield at pH 5. This finding provides a good way to promote the conversion of LBS into BS in vivo by simply adjusting the fermentation broth to pH 8. Given the above data, the possibility that pH plays a role in regulation of expression level of PepN in vivo could not be excluded.
An important concern on LBS hydrolysis is the location of where PepN acts on the substrate LBS. In the proposed pathway for BS biosynthesis, leucine is first loaded to the beta-amino group of L-arginine to form demethylleucylblasticidin S (LDBS), followed by methylation of the guanidino group to generate LBS. LDBS and LBS are both easily converted to DBS and BS by whole cells or the CFE of BS producer23. Thus, PepNs is unlikely to be co-localized with the enzyme forming LDBS, otherwise there could be a futile cycle of adding and removing leucine in the final steps of BS biosynthesis. Because analysis using HMMTOP (http://www.enzim.hu/hmmtop/index.php) revealed that PepNs has a short transmembrane region spanning from residues 384 to 401 and because, washed cells are capable of converting LBS and LDBS into BS and DBS, respectively, PepNs may be membrane-associated with its catalytic domain exposed to the cell exterior. The proposal is that LBS is formed within the cytoplasm either for self-protection or to facilitate export out of the cell where it is then hydrolyzed by PepNs that is anchored on the cell surface.
As PepN in E. coli is the major aminopeptidase that is responsible for the majority activity of aminopeptidase and is implicated in cytosolic protein degradation. This study showed that PepN is also capable of cleaving the amido bond between the carboxyl group of leucine and the beta-amino group of arginine that is bonded to the 4’-amino group of hexose ring of blasticidin S. It might also play a role in processing other secondary metabolites harboring moiety of amino acid or peptides.
Methods
Bacterial strains, plasmids and culture conditions
Strains, plasmids, and primers used in this study are listed in Supplementary Table S1 in the supplemental material. To purify the LBS hydrolase in the cell lysate, Streptomyces strains mycelia was cultured in 25 ml TSBY (tryptic soy broth [TSB] supplemented with 10.3% sucrose and 1% yeast extract, w/v) medium at 30 °C in 250-ml baffled flasks and at 220 rpm, and harvested after 36 hours incubation. For sporulation, Streptomyces strains were grown at 30 °C on SFM (soybean flour 2%, mannitol 2%, agar 1%, pH 7.2–7.4, w/v) agar plate for 5–7 days. SFM agar was also used for the conjugation between Escherichia coli and Streptomyces lividans WJ2. E. coli strains were grown at 37 °C in Luria Broth medium and on Luria Broth agar.
Fermentation and HPLC analysis of blasticidin S and its derivatives
For blasticidin S and its intermediate metabolites analysis, the fermentation of strain S. lividans WJ2 and its derivatives was performed as previously described11. Cell broths were centrifuged at 10,000 × g for 10 min and the supernatants were applied to Supelclean LC-SCX solid-phase extraction (SPE) columns to remove unwanted non-BS derivatives. HPLC was operated on an Agilent series 1260 with a Thermo Golden C18 column (2.1 × 150 mm, 3 μm). The analytes were eluted at a flow rate of 0.2 ml/min with a gradient of 0% to 11% 20 mM ammonium acetate (pH 5.5) (buffer A) in the first 40 minutes and 95% acetonitrile (buffer B) in the following 5 minutes and were detected with UV absorption 272 nm.
Preparation of cell-free extracts
Mycelia of Streptomyces lividans WJ2 or cells of E. coli and Saccharomyces sake were pelleted and washed twice in ten volumes of lysis buffer (50 mM K3PO4, pH 8.0, 1 mM dithiothreitol). The cells were then suspended in two volumes of lysis buffer in an ice bath and lysed by sonication. The cell lysate was centrifuged at 10,000 × g for 10 min at 4 °C, and the supernatant was either used for further purification or for in vitro enzymatic assays. The cell-free extract (CFE) was boiled in water for 10 min to inactivate the enzyme and then used as the negative control in the enzymatic assays.
Ammonium sulfate precipitation and anion exchange column separation
CFE of E. coli was fractionated using 20%, 30%, 40%, and 50% ammonium sulfate (final concentration) respectively. The precipitates were collected and dissolved in dialysis buffer (50 mM NaCl, 50 mM Tris-Cl, pH 8.0), generating 20%, 30%, 40%, 50% (v/v) gradient protein components, respectively. These dissolved proteins were dialyzed against dialysis buffer overnight in a 4 °C cold room.
The most active protein fraction was further separated by a MonoQ HR 5/5 column (GE Healthcare) on the AKTA FPLC (GE Healthcare) system using a linear gradient of 50–500 mM NaCl. The elution was collected and the hydrolysis activity towards LBS was determined. LBS was prepared as reported in Zhang et al.23. The most active fraction and the one without LBS hydrolytic activity were compared on a 12% SDS-PAGE gel and sent for LC-MS/MS analysis (Shanghai BoYuan Biotechnology Co., LTD, China).
Assay of the LBS hydrolytic activity of the CFE and protein eluates
1 μM LBS was incubated with 10 μl CFE or protein elutes in a total volume of 50 ul at 30 °C overnight (8–12 h), whereas in the control experiments boiled CFE was used. The reactions were quenched by adding 50 ul chloroform. The supernatant was analyzed by HPLC analysis as described above.
PCR target disruption of the candidate genes encoding LBS hydrolase in E. coli BW25113 chromosome
PCR-targeted gene inactivation was achieved using REDIRECTR Technology27. The pairs of primers Tar9-F/R, Tar139-F/R, Tar147-F/R, Tar176-F/R, Tar201-F/R with 5′-terminus paring with the putative hydrolase genes and 3′-terminus complementary to resistance cassette aac(3)IV (Supplementary Table S1) were used to generate linear fragments for targeted gene disruption. pIJ773 harboring aac(3)IV coding for apramycin resistance and FRT site-specific recombination sites was used as the template. 1.38 kb linear DNA fragments were generated and transformed separately by electroporation into E. coli BW25113. Mutants were selected by apramycin of 50 μg/ml final concentration and PCR verified using primers C-tar9F/R, C-tar139F/R, C-tar147F/R, C-tar176F/R, C-tar201F/R (Supplementary Table S1, Supplementary Figure S5).
Heterologous expression and purification of PepN from E. coli and S. lividans WJ2
pepN genes from E. coli and S. lividans 1326 were amplified with primers PepN-F/R, PepN1-F/R and PepN2-F/R, respectively. They were sequenced and ligated into NdeI/XhoI-digested pET44b to generate pYGY1, pYGY2 and pYGY3 with C-terminal 6xHis tags. The recombinant proteins were expressed in E. coli BL21 (DE3) in LB medium supplied with 50 μg/ml ampicillin and 0.2 mM IPTG at 16 °C for 21 h. The recombinant proteins were purified to greater than 95% in purity determined by 12% SDS-PAGE using Ni-Sepharose™ 6 Fast Flow (GE). Protein concentrations were determined by Bradford Protein Assay Kit (Bio-Rad). LBS hydrolysis assays by purified PepN is the same as that described above for CFE assays.
Overexpression of gene pepN1 in S. lividans WJ2
For overexpression of pepN1 in S. lividans WJ2, pepN1 was PCR amplified from S. lividans WJ2 with primer pair pepN1-139-F/R (Supplementary Table S1), and inserted into expression vector pIB139 between NdeI and XbaI under the control of constitutive promoter PermE*. The recombinant plasmid pYGY4 and pIB139 were transformed into E. coli ET12567/pUZ8002 and transferred into S. lividans WJ2 by conjugation. The resultant mutants S. lividans::pYGY4 and S. lividans::pIB139 were cultured and compared in the LBS conversion rates with the wild type strain.
Additional Information
How to cite this article: Yu, G. et al. The standalone aminopeptidase PepN catalyzes the maturation of blasticidin S from leucylblasticidin S. Sci. Rep. 5, 17641; doi: 10.1038/srep17641 (2015).
Supplementary Material
Acknowledgments
This work received support from the National Natural Science Foundation of China (31470195), 973 program from the Ministry of Science and Technology (2012CB721004).
Footnotes
Author Contributions Y.G, L.L, L.X. and L.G. performed experiments. J.M, H.X. and D.Z. analyzed the data. H.X. and M.Z. wrote the manuscript. All authors reviewed, revised, commented on and approved the final version of the manuscript.
References
- Takeuchi S., Hirayama K., Ueda K., Sakai H. & Yonehara H. Blasticidin S, a new antibiotic. The Journal of antibiotics 11, 1–5 (1958). [PubMed] [Google Scholar]
- Misato T., Ishii I., Asakawa M., Okimoto Y. & Fukunaga K. Antibiotics as protectant fungicides against rice blast. II. The therapeutic action of blasticidin S. Ann Phytopathol Soc Jpn 24, 5 (1959). [Google Scholar]
- Yoshinari T. et al. New metabolic pathway for converting blasticidin S in Aspergillus flavus and inhibitory activity of aflatoxin production by blasticidin S metabolites. Journal of agricultural and food chemistry 61, 7925–7931 (2013). [DOI] [PubMed] [Google Scholar]
- Wilson D. N. On the specificity of antibiotics targeting the large ribosomal subunit. Annals of the New York Academy of Sciences 1241, 1–16, 10.1111/j.1749-6632.2011.06192.x (2011). [DOI] [PubMed] [Google Scholar]
- Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annual review of biochemistry 74, 649–679, 10.1146/annurev.biochem.74.082803.133130 (2005). [DOI] [PubMed] [Google Scholar]
- Blaha G. M., Polikanov Y. S. & Steitz T. A. Elements of ribosomal drug resistance and specificity. Current opinion in structural biology 22, 750–758, 10.1016/j.sbi.2012.07.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svidritskiy E., Ling C., Ermolenko D. N. & Korostelev A. A. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proceedings of the National Academy of Sciences of the United States of America 110, 12283–12288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi I., Shibata H., Seto H. & Misato T. Isolation and purification of blasticidin S deaminase from Aspergillus terreus. The Journal of antibiotics 28, 7–14 (1975). [DOI] [PubMed] [Google Scholar]
- Nawa K. et al. Inactivation of blasticidin S by Bacillus cereus. V. Purification and characterization of blasticidin S-deaminase mediated by a plasmid from blasticidin S resistant Bacillus cereus K55-S1. Biological & pharmaceutical bulletin 18, 350–354 (1995). [DOI] [PubMed] [Google Scholar]
- Nawa K., Tanaka T., Kamakura T., Yamaguchi I. & Endo T. Inactivation of blasticidin S by Bacillus cereus. VI. Structure and comparison of the bsr gene from a blasticidin S-resistant Bacillus cereus. Biological & pharmaceutical bulletin 21, 893–898 (1998). [DOI] [PubMed] [Google Scholar]
- Li L., Wu J., Deng Z., Zabriskie T. M. & He X. Streptomyces lividans blasticidin S deaminase and its application in engineering a blasticidin S-producing strain for ease of genetic manipulation. Applied and environmental microbiology 79, 2349–2357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Sekido S., Isogai Y. & Yamaguchi I. Expression, purification, and characterization of blasticidin S deaminase (BSD) from Aspergillus terreus: the role of catalytic zinc in enzyme structure. Journal of biochemistry 127, 955–963 (2000). [DOI] [PubMed] [Google Scholar]
- Kamakura T., Yoneyama K. & Yamaguchi I. Expression of the blasticidin S deaminase gene (bsr) in tobacco: fungicide tolerance and a new selective marker for transgenic plants. Mol Gen Genet 223, 332–334 (1990). [DOI] [PubMed] [Google Scholar]
- Freitas A. C., Bento F. M., Ramesh N., Osborne W. R. & Han S. W. Modified blasticidin S resistance gene (bsrm) as a selectable marker for construction of retroviral vectors. Journal of biotechnology 95, 57–62 (2002). [DOI] [PubMed] [Google Scholar]
- Brooks D. R., McCulloch R., Coombs G. H. & Mottram J. C. Stable transformation of trypanosomatids through targeted chromosomal integration of the selectable marker gene encoding blasticidin S deaminase. FEMS microbiology letters 186, 287–291 (2000). [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Takeda A., Paik S. Y., Nimi O. & Nomi R. Acetylation of blasticidin S by its producing actinomycetes. The Journal of antibiotics 39, 827–832 (1986). [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Takeda A., Paik S. Y. & Nimi O. Isolation and properties of a blasticidin S acetylating enzyme from a producer organism. The Journal of antibiotics 42, 135–137 (1989). [DOI] [PubMed] [Google Scholar]
- Perez-Gonzalez J. A., Ruiz D., Esteban J. A. & Jimenez A. Cloning and characterization of the gene encoding a blasticidin S acetyltransferase from Streptoverticillum sp. Gene 86, 129–134 (1990). [DOI] [PubMed] [Google Scholar]
- Pletzer D. et al. The Pseudomonas aeruginosa PA14 ABC transporter NppA1A2BCD is required for uptake of peptidyl nucleoside antibiotics. Journal of bacteriology 197, 2217–2228, 10.1128/JB.00234-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. C., Freinkman E., Sabatini D. M. & Ploegh H. L. The protein synthesis inhibitor blasticidin S enters mammalian cells via leucine-rich repeat-containing protein 8D. The Journal of biological chemistry 289, 17124–17131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto H. & Yonehara H. Studies on the biosynthesis of blasticidin S. VII. Isolation of demethylblasticidin S. The Journal of antibiotics 30, 1022–1024 (1977). [DOI] [PubMed] [Google Scholar]
- Seto H., Yamaguchi I., Ōtake N. & Yonehara H. Studies on the Biosynthesis of Blasticidin S II. Agricultural and Biological Chemistry 32, 7 (1968). [Google Scholar]
- Zhang Q., Cone M. C., Gould S. J. & Zabriskie T. M. Reevaluation of the final steps in the biosynthesis of blasticidin S by Streptomyces griseochromogenes and identification of a novel self-resistance mechanism. Tetrahedron 52, 9 (2000). [Google Scholar]
- Cooper R., Conover M. & Patel M. Sch 36605, structure of a novel nucleoside. The Journal of antibiotics 41, 123–125 (1988). [DOI] [PubMed] [Google Scholar]
- Cone M. C., Yin X., Grochowski L. L., Parker M. R. & Zabriskie T. M. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. Chembiochem 4, 821–828 (2003). [DOI] [PubMed] [Google Scholar]
- Chandu D. & Nandi D. PepN is the major aminopeptidase in Escherichia coli: insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology 149, 3437–3447 (2003). [DOI] [PubMed] [Google Scholar]
- Gust B., Challis G. L., Fowler K., Kieser T. & Chater K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proceedings of the National Academy of Sciences of the United States of America 100, 1541–1546, 10.1073/pnas.0337542100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.