Abstract
Dynamic interactions of nuclear lamins with chromatin through lamin-associated domains (LADs) contribute to spatial arrangement of the genome. Here, we provide evidence for prepatterning of differentiation-driven formation of lamin A/C LADs by domains of histone H2B modified on serine 112 by the nutrient sensor O-linked N-acetylglucosamine (H2BS112GlcNAc), which we term GADs. We demonstrate a two-step process of lamin A/C LAD formation during in vitro adipogenesis, involving spreading of lamin A/C–chromatin interactions in the transition from progenitor cell proliferation to cell-cycle arrest, and genome-scale redistribution of these interactions through a process of LAD exchange within hours of adipogenic induction. Lamin A/C LADs are found both in active and repressive chromatin contexts that can be influenced by cell differentiation status. De novo formation of adipogenic lamin A/C LADs occurs nonrandomly on GADs, which consist of megabase-size intergenic and repressive chromatin domains. Accordingly, whereas predifferentiation lamin A/C LADs are gene-rich, post-differentiation LADs harbor repressive features reminiscent of lamin B1 LADs. Release of lamin A/C from genes directly involved in glycolysis concurs with their transcriptional up-regulation after adipogenic induction, and with downstream elevations in H2BS112GlcNAc levels and O-GlcNAc cycling. Our results unveil an epigenetic prepatterning of adipogenic LADs by GADs, suggesting a coupling of developmentally regulated lamin A/C-genome interactions to a metabolically sensitive chromatin modification.
Genome organization in eukaryotic interphase nuclei involves dynamic associations of chromatin with the nuclear lamina (Gay and Foiani 2015), a meshwork of intermediate filaments called A- and B-type lamins (Burke and Stewart 2013). Nuclear lamins interact with chromatin primarily through so-called lamin-associated domains (LADs) (Guelen et al. 2008). LADs have initially been identified by DamID (Guelen et al. 2008) and later by chromatin immunoprecipitation-sequencing (ChIP-seq) (Sadaie et al. 2013; Shah et al. 2013), with robust overlap (Lund et al. 2014) despite differences between the two methods. LADs cover ∼0.1–10 Mb and are overall gene-poor and transcriptionally inactive (Guelen et al. 2008; Meuleman et al. 2013; Lund et al. 2014). Accordingly, LADs are enriched in histone modifications marking silent chromatin, including H3 lysine 9 di- or trimethylation (H3K9me2/3) (Guelen et al. 2008; Kind et al. 2013; Sadaie et al. 2013; Shah et al. 2013), in line with the heterochromatic environment at the nuclear periphery (Reddy et al. 2008; Mattout et al. 2011; Towbin et al. 2012; Solovei et al. 2013). Targeting and anchoring of loci at the nuclear periphery in Caenorhabditis elegans or mouse cells requires H3K9me2/3, H3K27me3, lamin A/C (in mammals), short DNA sequences, and protein factors (Towbin et al. 2012; Zullo et al. 2012; Harr et al. 2015), suggesting that the formation of LADs is tightly regulated.
In contrast to B-type lamins (lamins B1 and B2), which are restricted to the nuclear periphery, A-type lamins (lamins A and C, or lamin A/C, splice variants of the LMNA gene) also exist as a nucleoplasmic pool (Dechat et al. 2000, 2010; Kolb et al. 2011). This intranuclear pool of lamin A/C is dependent on the nucleoplasmic protein lamina-associated polypeptide LAP2, isoform alpha (LAP2; encoded by the TMPO gene), which directly binds lamin A/C (Dechat et al. 2000; Naetar and Foisner 2009). Nucleoplasmic lamin A/C and LAP2 affect retinoblastoma protein function (Johnson et al. 2004) and promote cell-cycle arrest in tissue progenitor cells (Naetar and Foisner 2009; Gotic and Foisner 2010; Gotic et al. 2010). Interestingly, LAP2 overexpression in mouse preadipocytes elicits adipogenic differentiation (Dorner et al. 2006), suggesting that maintenance of an intranuclear lamin A/C pool is important for adipogenesis. Nucleoplasmic lamin A/C appears therefore to be critical for differentiation of tissue progenitor cells and may play important roles regulating chromatin and gene expression in the nuclear interior (Dechat et al. 2010; Gesson et al. 2014).
Whereas most LADs seem to be conserved between cell types, others are cell-type–specific (Peric-Hupkes et al. 2010; Meuleman et al. 2013; Harr et al. 2015). Cell-type–specific facultative fLADs (also called variable vLADs) differ from constitutive cLADs by their higher gene density and lower A/T and LINE content (Meuleman et al. 2013). LAD variability has been attributed to shifts in LAD borders (Meuleman et al. 2013; Harr et al. 2015), which are flanked by regions of H3K27me3 (Guelen et al. 2008) that appear to be important for association with the nuclear lamina (Harr et al. 2015). A- and B-type lamins may also temporally interact with genes and regulatory regions during embryonic and somatic stem cell differentiation (Peric-Hupkes et al. 2010; Lund et al. 2013). Recent work shows that in adipose tissue stem cells (ASCs), proadipogenic gene promoters are released from lamin A/C after differentiation into adipocytes, whereas many nonadipogenic, lineage-specific promoters retain lamin association (Lund et al. 2013). A similar uncoupling of a myogenic promoter from C. elegans lamin is associated to muscle-specific gene activation (Mattout et al. 2011). These results suggest that subsets of nuclear lamin–genome interactions are under developmental regulation.
Cell metabolism is coupled to post-translational modifications of histones and chromatin remodeling proteins affecting chromatin structure and gene expression (Keating and El-Osta 2015). Metabolic intermediates often act as cofactors or substrates of histone modifying enzymes. The hexosamine biosynthetic pathway (HBP) is responsive to intracellular levels of amino acids, fatty acids, and carbohydrates (Hanover et al. 2012) and constitutes an important link between glucose metabolism and chromatin. Approximately 3%–5% of glucose taken up by the cell is directed to the HBP (Marshall et al. 1991) and converted to UDP-N-acetylglucosamine (UDP-GlcNAc), the donor of O-linked GlcNAc for O-GlcNAcylation of proteins. GlcNAcylation is catalyzed by the O-GlcNAc transferase OGT, while O-GlcNAcase (OGA) hydrolyzes O-GlcNAc (Gambetta and Müller 2015). All core histones can be GlcNAcylated (Zhang et al. 2011; Fong et al. 2012; Vella et al. 2013), indicating that chromatin organization is influenced by the OGT/OGA balance. GlcNAcylation of H2B on serine 112 (S112) has been reported to promote H2BK120 monoubiquitination (H2BK120ub1), suggesting a link to gene activity (Fujiki et al. 2011). Notwithstanding, localization of H2BS112 on the nucleosome surface raises the possibility that H2BS112GlcNAc promotes additional chromatin-associated processes.
Here, we used an in vitro adipogenic differentiation system from human adipocyte progenitors (Shah et al. 2014) to address the relationship between H2B GlcNAcylation and association of lamin A/C with chromatin at key time points of adipogenesis. Our results suggest a coupling of developmentally regulated lamin A/C–genome interactions to H2BS112GlcNAc, a metabolically sensitive chromatin modification.
Results
Adipogenic differentiation reorganizes genome-wide lamin A/C–chromatin interactions
We first investigated the dynamics of A-type lamin interactions with the genome by ChIP-seq analysis of lamin A/C in ASCs at five stages of in vitro adipogenic differentiation. We examined (1) proliferating undifferentiated ASCs (2 d before adipogenic induction; D−2); (2) cell cycle-arrested undifferentiated ASCs on D0 (immediately before adipogenic induction); and (3) cells in the undergoing differentiations on D1, D3, and D9 after adipogenic induction (Supplemental Fig. 1A).
Using Enriched Domain Detector (EDD), an algorithm we developed for analysis of broad genomic domains of protein enrichment (Lund et al. 2014), we identify throughout differentiation approximately 200–500 lamin A/C LADs ranging from ∼100 kb to ∼10 Mb (Fig. 1A–C). We find that on D0, cells display a threefold increase in LAD number and coverage relative to D−2 cells (Fig. 1B,D) despite constant intracellular lamin A/C levels (Supplemental Fig. 1B). On D0, ASCs have notably reached cell-cycle arrest as a result of replating to confluency and growth factor removal 48 h earlier. Most lamin A/C–chromatin interactions in proliferating ASCs are maintained on D0 (Fig. 1D, D−2 gray bar), so we conclude that cell-cycle arrest, which precedes adipogenic induction, is accompanied by a net gain in LADs. This is consistent with the reorganization of chromatin that occurs after mitosis exit (Naumova et al. 2013).
Figure 1.
Lamin A/C LADs are reorganized by induction of adipogenic differentiation. (A) Lamin A/C ChIP-seq enrichment profiles showing ChIP/input ratios (scale −0.4 to +0.4 centered on 0), LADs mapped with EDD, and RNA-seq gene expression profiles (scale 0–1000 FPKM). (B) Number of LADs identified at each differentiation stage. (C) Median LAD lengths at all differentiation time points confounded (All) and at each time point. (D) Genome coverage by LADs (color bars) and conservation of LAD coverage between two consecutive time points (gray bars). (E) Coefficient of overlap of small, medium, and large LADs (Jaccard indices) with large (L), medium (M), and small (S) LADs between time points (x-axis). (F) Gene density within LADs and in the whole genome. (G) Median expression level of genes within LADs and of RefSeq genes: (*) P < 10−5, t-test with Bonferroni correction. (H) Proportion of lamin A/C LAD coverage at each time point, which overlaps with lamin B1 LADs mapped by DamID in fibroblasts (Guelen et al. 2008).
Although EDD calls lamin A/C LADs conservatively, we also detect non-LAD regions of interspersed positive and negative bins of enrichment (see Fig. 1A); these can be interpreted as enriched and depleted lamin A/C–chromatin interactions that are distinct from truly depleted regions. It is at present unclear whether these “wobbly” areas represent noise caused by stochastic lamin–chromatin interactions or whether they have biological significance for the formation of LADs.
Genome-wide reorganization of lamin A/C–chromatin associations is not restricted to the preadipogenic phase. We find that within day 1 of stimulation with a proadipogenic cocktail (D0-D1 transition), lamin A/C–chromatin interactions are strikingly redistributed, with poor LAD overlap between undifferentiated and differentiated stages (D−2/D0 versus D1/D3/D9) (Fig. 1A,D, gray bars). This LAD redistribution is not elicited by mitotic disassembly and reassembly of the nuclear envelope, which could reset lamin–genome contacts (Kind et al. 2013), because ASCs do not divide once induced to differentiate (Supplemental Fig. 1C). Altogether, these changes reveal a genome-wide reorganization of lamin A/C–chromatin associations elicited by adipogenic induction.
The wide range of LAD sizes detected (Fig. 1C) suggests differences in behavior and genomic properties between LADs of various sizes during differentiation. To address this, we categorized LADs into small (<0.32 Mb), medium (0.32–1.13 Mb), and large (>1.1 Mb) domains based on their quantile size distribution (Supplemental Fig. 1D) and evaluated the extent to which these domains overlap between differentiation time points by computing Jaccard similarity coefficients. We find that small and medium LADs poorly overlap between differentiation time points (Fig. 1E), suggesting that they may represent unstable or stochastic lamin–chromatin interactions. The overlap between large LADs is minimal between proliferation (D−2) and cell cycle-arrested (D0) stages in undifferentiated cells, because most D0 LADs are not detected on D−2 (Fig. 1D). However, once large LADs form on D1, they are maintained thereafter (Fig. 1A,E).
We conclude that adipogenic commitment of human primary ASCs elicits high-order genome rearrangements through a large-scale reorganization of lamin A/C LADs. This involves a spreading of LADs after cell-cycle arrest and a genome-wide remodeling of LADs upon adipogenic induction.
Gene content of LADs depends on LAD size and differentiation status
In line with earlier data on lamin B1 LADs (Guelen et al. 2008) and lamin A/C LADs (Lund et al. 2014), we find that lamin A/C LADs are overall gene poor; however, this is only after induction of differentiation (D1-D9). Indeed in undifferentiated cells, LADs are strikingly gene rich, averaging 9–13 genes/Mb (Fig. 1F; Supplemental Fig. 1E). RNA-seq shows that in undifferentiated ASCs, genes within LADs are expressed at the same level as all expressed RefSeq genes as a whole (Fig. 1G). In contrast, on D1 and D3, LADs are gene poor (fewer than two genes/Mb) and genes within LADs are more weakly expressed than RefSeq genes (Fig. 1G). Thus, ASC LADs display genomic properties distinct from LADs of lineage-committed cells. Supporting this view, we further find that LADs in differentiated ASCs overlap with lamin B1 LADs (Guelen et al. 2008) to a greater extent than predifferentiation LADs (Fig. 1H). These results are consistent with a view that lamin A/C LADs can occur in different chromatin contexts in undifferentiated and differentiated cells, and lamin A/C association with genes does not always coincide with a transcriptionally repressed state.
Domains of GlcNAcylated histone H2B constitute stable chromatin landmarks during adipogenic differentiation
Modifications in chromatin architecture during adipogenesis have been shown to be linked to cellular metabolic state (Siersbaek et al. 2012). This suggests that resetting of lamin A/C-genome associations upon adipogenic induction may be linked to metabolically linked chromatin modifications. To investigate this possibility, we mapped by ChIP-seq the profile of H2BS112GlcNAc (which we refer to as “H2BGlcNAc” here), as a putative nutrient sensor on chromatin (Hart 2014), using an antibody raised against an H2B peptide GlcNAcylated on S112.
We first evaluated the specificity of the anti-H2BS112GlcNAc antibody in several ways: (1) We show differential immunodetection sensitivity of immobilized H2BS112GlcNAc and control unmodified H2B peptides (Fig. 2A; Supplemental Fig. 2A); (2) anti-H2BS112GlcNAc immunoreactivity is abrogated after peptide incubation with recombinant OGA (Fig. 2A); (3) immunofluorescence analysis shows differential detection of H2BS112GlcNAc in a competition assay using an H2BS112GlcNAc or a control unmodified H2B competitor peptide (Fig. 2B); (4) incubation of an ASC lysate with recombinant OGA abrogates H2BS112GlcNAc, but not pan-H2B, immunoreactivity (Fig. 2C); (5) overexposure of anti-H2BS112GlcNAc Western blots from cell and nuclear lysates reveal no other detectable histones (Supplemental Fig. 2B), indicating that the antibody does not recognize unmodified or GlcNAcylated histones other than H2B; (6) expression of an EGFP-H2BS112A mutant, which cannot be GlcNAcylated on S112, leads to a weaker detection (but not abolishment) of EGFP-H2BS112A than wild-type EFGP-H2B using anti-H2BS112GlcNAc antibodies, whereas control immunoreactivity with anti-H2B or anti-GFP antibodies is not affected (Fig. 2D); thus, shifts in H2BGlcNAc detected by ChIP (see below) do not necessarily represent absolute changes in GlcNAcylated H2B levels; (7) moreover, ChIP-seq of H2BS112GlcNAc, pan-H2B, and of a control IgG reveals distinct enrichment profiles for pan- and modified H2B (Supplemental Fig. 2C; see also below); although we note some sites of coincidence between H2B and H2BS112GlcNAc (Supplemental Fig. 2B), we find that genome-wide, only 0.8% of H2BS112GlcNAc-enriched domains display pan-H2B enrichment, representing only 6.5 Mb of genome, which is negligible (Supplemental Fig. 2D); this indicates H2BS112GlcNAc domains are not merely due to H2B enrichment, in fact, 91% of the total area enriched in H2BS112GlcNAc relative to input chromatin is also enriched in H2BS112GlcNAc relative to pan-H2B (Supplemental Fig. 2E); and (8) this differential enrichment in H2B versus H2BS112GlcNAc was corroborated by independent ChIP-PCR validation of 20 loci, of which 16 were found to be enriched in H2BGlcNAc and four not enriched, based on ChIP-seq data (Supplemental Fig. 2F; Supplemental Table 1). These results collectively indicate that the H2BS112GlcNAc antibody recognizes a GlcNAc epitope on H2BS112 despite some cross reactivity with unmodified H2B.
Figure 2.

Assessment of specificity of the anti-H2BS112GlcNAc antibody. (A) Immunoreactivity of the anti-H2BS112GlcNAc antibody on H2BS112GlcNAc and corresponding unmodified H2B peptides (KHAVS112EGTK) immobilized on nitrocellulose. Peptides were preincubated without or with 2 ng/µL recombinant OGA prior to immunodetection. (B) Immunofluorescence peptide competition assay using H2BS112GlcNAc and unmodified H2B peptides; immunodetection using anti-H2BS112GlcNAc. Scale bar, 10 μm. (C) Immunoblot of ASC extracts preincubated with 0 or 2 ng/µL recombinant OGA for 30 min. (D) Expression of an EGFP-H2BS112A mutant, but not a control wild-type EGFP-H2B, in HeLa cells reduces H2BS112GlcNAc immunoreactivity (left) without affecting detection of total H2B (middle) or EGFP (right).
We identify throughout adipogenesis 600 to nearly 900 H2BGlcNAc-enriched domains, ranging from 60 kb to ∼10 Mb, which we call GADs (Fig. 3A,B; Supplemental Fig. 3A,B). GADs primarily localize to intergenic regions and sharply demarcate clusters of transcribed genes (Fig. 3A). Accordingly, GADs are gene poor (Fig. 3C), and genes within GADs are repressed (P < 10−15 relative to all genes and genes outside GADs; Wilcoxon test) (Fig. 3D; Supplemental Fig. 3C). H2BS112GlcNAc has been proposed to be connected to H2BK120ub1 (Fujiki et al. 2011), a modification linked to transcription; however, ChIP-qPCR of H2BK120ub1 on 20 regions high or low in H2BGlcNAc in ASCs shows no obvious relationship between the two marks in this cell type (Supplemental Fig. 3D). H2BGlcNAc enrichment in intergenic regions does not result from increased density of H2B-bearing nucleosomes (see also above) because (1) it was corroborated when we determined enrichment of H2BGlcNAc over H2B in these regions (P = 2.2 × 10−16; Wilcoxon test) but not in genes (Supplemental Fig. 3E); and (2) genes within regions enriched in H2BGlcNAc over H2B are not expressed (Supplemental Fig. 3F). Furthermore, overlap analysis reveals that large GADs (>1.13 Mb in size) (Fig. 3B) are overall maintained during adipogenic differentiation (Fig. 3E). Since features of GADs and of lamin B1 LADs are similar, we compared the overlap between GADs in ASCs and lamin B1 LADs in fibroblasts (Guelen et al. 2008). Data show that ∼70% of GAD coverage is shared with lamin B1 LADs (Fig. 3F). ChIP-qPCR analysis confirms that in ASCs, lamin B1 occupies regions enriched in H2BGlcNAc but not in nonenriched regions (Fig. 3G).
Figure 3.
H2B GlcNAcylated domains (GADs) are maintained during adipogenic differentiation. (A) H2BGlcNAc ChIP-seq profiles (ChIP/input ratios, scale −0.4/+0.4), GADs identified using EDD, and RNA-seq profiles (scale 0–1000 FPKM). (B) Median GAD length and partitioning based on quantile length distribution. (C) Gene density in GADs and in the whole genome. (D) Median expression level of genes within GADs, outside GADs, and in the whole genome (All). (E) Jaccard index of overlap of small, medium, and large GADs with large (L), medium (M), and small (S) GADs between consecutive differentiation time points (x-axis). (F) Percentages of GAD coverage shared with lamin B1 LADs identified in fibroblasts (Guelen et al. 2008). (G) ChIP-qPCR analysis of lamin B1 in ASCs on D0 and D1 of differentiation, at 20 loci shown by ChIP-seq to be H2BGlcNAcylated (amplicons 1–16) or not (amplicons 17–20) (see also Supplemental Fig. 2F).
Adipogenic induction transiently elevates H2BGlcNAc levels and up-regulates glycolytic genes
Overall GAD conservation through differentiation does not preclude that H2BGlcNAcylation is dynamic. We find that adipogenic induction is manifested by a transient increase in GAD number and coverage between D0 and D1 (Supplemental Fig. 3A,B). Increased H2BGlcNAc enrichment is also shown by ChIP-qPCR (Fig. 4A) and mirrors transient elevations in total cellular H2BGlcNAc and O-GlcNAc levels (Fig. 4B; Supplemental Fig. 3G). ASC stimulation with each component of the adipogenic cocktail separately, which by itself does not elicit adipogenesis, failed to elevate O-GlcNAc or H2BGlcNAc (data not shown). Thus, the transient elevation in H2BGlcNAc results from the adipogenic stimulus, although a stress response cannot formally be ruled out. Concomitant with O-GlcNAc increase, levels of OGT and OGA proteins are also transiently up-regulated on D1 (Fig. 4B), providing evidence for enhanced O-GlcNAc cycling (Hart et al. 2007) in the early stages of adipogenesis.
Figure 4.
Adipogenic induction results in up-regulation of metabolic genes, lamin A/C dissociation from these genes, and in a transient increase in H2BGlcNAc. (A) ChIP-qPCR analysis of H2BGlcNAc on enriched and nonenriched loci on D0, D1, and D3 of differentiation. (B) Western blot analysis of H2BGlcNAc, total O-GlcNAc (RL2 antibody), OGT, OGA, and H2B throughout differentiation. Graph shows mean H2BGlcNAc levels at each time point relative to D−2 (levels standardized to H2B; mean ± SD of four experiments). (C) Gene ontology enrichment classification for the 2716 genes up-regulated on D1 (see Supplemental Fig. 4A,B). Note the high proportions of metabolic genes. (D,E) Localization of D0-D1 up-regulated genes in GADs (D) and lamin A/C LADs (E), and maintenance of these genes within GADs or LADs between consecutive differentiation time points (gray bars). (F) H2BGlcNAc and lamin A/C ChIP-seq profiles on PDK4 and indicated glycolytic HIF1A-target genes. Arrows point to losses of lamin A/C from promoters, coinciding with transcriptional up-regulation of the genes. RNA-seq profiles are shown (scale 0–1000 FPKM).
Providing biological significance to the elevation in H2BGlcNAc in the transition from preadipogenic to adipogenic stages, gene ontology (GO) analyses show that the 2716 genes up-regulated at the D0-D1 transition (Supplemental Fig. 4A) are for the most part involved in metabolic functions pertaining to cellular metabolic process including oxidation-reduction processes, lipid synthesis and glucose metabolism (Fig. 4C; Supplemental Fig. 4B; see Supplemental Table 2 for gene lists; Shah et al. 2014). The vast majority of these genes reside outside GADs on D0 (Fig. 4D; only 46 genes are in GADs at this stage), consistent with their expressed status on D0 (Supplemental Fig. 4C), and remain outside GADs throughout differentiation (Fig. 4D). Moreover, <10% of the 2716 up-regulated genes are in LADs on D−2, yet 752 are found in LADs on D0 (Fig. 4E), in line with the broad increase in LAD coverage after cell-cycle arrest (Fig. 1D). Remarkably however, these genes lose lamin A/C association on D1 (Fig. 4E), which coincides with their transcriptional up-regulation (Fig. 4F; Supplemental Fig. 4A).
Interestingly, adipogenic stimulation up-regulates hypoxia inducible factor 1, alpha subunit (HIF1A), which positively regulates the glucose transporter SLC2A1 and genes of the glycolytic pathway (Supplemental Fig. 4D,E). HIF1A also up-regulates the pyruvate dehydrogenase PDK4 gene, which inhibits mitochondrial pyruvate oxidation (Supplemental Fig. 4D,E). Consistent with our previous findings, the PDK4 gene is de-GlcNAcylated concomitant to its activation on D1 (Fig. 4F); however, none of the glycolytic genes are marked by H2BGlcNAc on D0, as predicted from their expressed state (Fig. 4F; data not shown). Additionally, up-regulation of PDK4 and of glycolytic genes on D1 consistently also correlates with loss of lamin A/C interaction from promoter regions (Fig. 4F, arrows), consistent with the exchange of LADs taking place at the D0/D1 transition. A 2-Mb window around these genes unveils the loss of lamin A/C association on D1 (TPI1, ENO1, LDHA, PGAM1, PFKL), in a non-GAD context, and maintenance of these genes outside LADs and GADs throughout differentiation (Supplemental Fig. 5).
These results argue that the passage from quiescence to adipogenic commitment up-regulates the glycolytic pathway, a metabolic transition characterizing ASCs committed to adipogenesis in vivo (Shyh-Chang et al. 2013). Since the HBP is coupled to glycolysis, our data suggest a link between cellular metabolic state, lamin A/C–chromatin interactions, and changes in gene expression at an early stage of adipogenesis in a manner consistent with changes in H2BGlcNAc levels on D1.
De novo LADs formed on D1 of adipogenesis are nonrandomly established on GADs
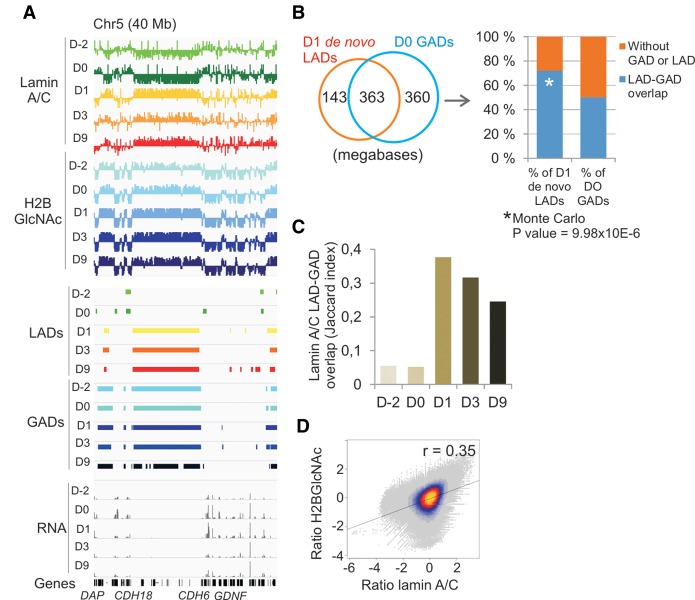
Several features of lamin A/C and H2BGlcNAc enrichment patterns during differentiation suggest that LADs and GADs may be functionally related. First, LADs formed on D1 strongly coincide with domains enriched in H2BGlcNAc (Fig. 5A). We find that 71% of de novo LAD coverage on D1 overlaps with domains that are H2BGlcNAcylated already on D0 (Fig. 5B). Monte Carlo simulations show that LAD formation on preexisting GADs is not random, but is rather highly significant (P = 9.98 × 10−5) (Fig. 5B). This suggests that establishment of de novo lamin A/C–chromatin interactions on D1 of adipogenesis is prepatterned by domains of H2BGlcNAc. Formation of lamin A/C LADs on GADs on D1 is further demonstrated by the increase in the Jaccard index of overlap between LADs and GADs at this time point (Fig. 5C). In addition, genome-wide lamin A/C levels positively correlate with H2BGlcNAc levels when taking into account enrichment ratios rather than LADs and GADs (Fig. 5D). We conclude that lamin A/C LAD formation after adipogenic induction occurs nonrandomly on GADs, consistent with a view of epigenetic prepatterning of de novo adipogenic lamin A/C LADs by GADs.
Figure 5.
Lamin A/C LADs form de novo on chromatin domains pre-enriched in H2BGlcNAc within 1 day of adipogenic induction. (A) Browser views of LADs and GADs during adipogenic differentiation. (B) Venn diagram analysis of overlap of de novo LADs formed on D1 with preexisting D0 GADs maintained on D1 (in megabases): Data show that 71% of de novo LAD coverage overlaps with preexisting GADs, reflecting a nonrandom association (permutation t-test P value = 9.98 × 10−6), and this de novo association concerns 50% of GADs. (C) Jaccard index of overlap of lamin A/C LADs with GADs during differentiation. (D) Genome-wide H2BGlcNAc levels positively correlate with lamin A/C levels (D1).
Chromatin states underlining removal and formation of LADs during differentiation
To provide insight into the interplay between LADs and GADs during differentiation, we identified chromatin states underlining GADs and the de novo formation of LADs in particular. We used ChromHMM (Ernst and Kellis 2012), a Hidden Markov Modeling algorithm, to model chromatin states from recurrent combinations of six histone modifications (H3K4me1, H3K4me2, H3K4me3, H3K27me3, H3K27ac, H3K36me3), insulator protein CCCTC-binding factor (CTCF), lamin A/C, and H2BGlcNAc. Histone H3 modifications and CTCF were previously mapped by ChIP-seq in human ASCs on D−2, D0, D3, and D9 of adipogenic differentiation in a separate study (Mikkelsen et al. 2010).
Using ChromHMM, we identify a 15-chromatin state model, i.e., 15 different combinations of chromatin marks characterizing the chromatin landscape of in vitro adipogenesis (Fig. 6A). From its composition in modified histones, CTCF, and lamin A/C, each chromatin state could be “annotated” into a given genomic feature (Fig. 6A). We identify four chromatin states (states 2, 3, 4, 15) containing lamin A/C (Fig. 6A): lamin A/C together with H3K36me3 (annotated as “lamin A/C on transcribed gene bodies”; state 2); lamin A/C alone (state 3); lamin A/C with H3K4me1/2/3 and/or H3K27ac (“lamin A/C on promoters and enhancers”; state 4); and lamin A/C with H2BGlcNAc and no other mark (state 15). This analysis reveals prominent chromatin features of lamin A/C LADs and indicates that a pool of lamin A/C associates with genes and gene regulatory elements, some of which are transcriptionally active.
Figure 6.
Modeling of chromatin states reveals the dynamics of LADs and GADs during adipogenic differentiation. (A) ChromHMM emission parameters: heat map of the relative abundance of chromatin states (numbered 1–15) in each indicated chromatin mark. The four lamin A/C-containing states are labeled green (states 2, 3, 4, 15). (B) Heat map of the relative abundance of the 15 states on predefined genomic regions on D0 and D3 of differentiation. Distribution of the four lamin A/C-containing states is shown in green areas in D0. On D3, red and yellow areas depict significant reductions (red) or increases (yellow) in enrichment levels of the lamin A/C-containing states. D−2, D0, D3, and D9 time point data and statistics are shown in Supplemental Figure 6A,C. (C) A two-step model of formation of lamin A/C LADs during adipogenic differentiation. In proliferating adipocyte progenitors, LAD coverage is relatively limited and does not necessarily involve GADs. After cell-cycle arrest, a necessary step for adipogenic differentiation, LAD coverage is extended independently of GADs. In undifferentiated cells, lamin A/C LADs contain both repressed chromatin, presumably harboring H3K9me2/3 (Guelen et al. 2008; Kind et al. 2013; Harr et al. 2015) and active chromatin domains. Adipogenic differentiation elicits an exchange of lamin A/C LADs; this involves the formation of LADs predominantly on H2BGlcNAc domains, consistent with an epigenetic prepatterning of de novo adipogenic lamin A/C LADs by GADs. The recently shown involvement of H3K27me3-enriched regions in LAD borders on the maintenance of LADs in mouse cells (Guelen et al. 2008; Kind et al. 2013; Harr et al. 2015) raises the possibility that de novo lamin A/C LADs formed during adipogenesis also entail a contribution from trimethylated H3K27 in LAD borders. The overall repressed state of these de novo LADs, together with their strong overlap with lamin B1 LADs, suggests that they become enriched in heterochromatin marked by di- or trimethylated H3K9.
Next, we identified changes in chromatin composition (chromatin states) within LADs as cells differentiate. We determined on D−2, D0, D3, and D9 of differentiation the relative abundance of each chromatin state in genomic elements predefined in ChromHMM (CpG islands, exons, genes, transcription start sites [TSS], transcription end sites [TES], and lamin B1 LADs) (Fig. 6B; Supplemental Fig. 6A). The data show that chromatin state enrichment patterns across the various genomic elements are similar in undifferentiated cells (D−2/D0); this includes patterns of the four lamin A/C-containing states (Supplemental Fig. 6A, green areas). On D3, however, we note a strong reduction of all lamin A/C-containing states on CpG islands, genes, exons, TSSs and TESs (Fig. 6B, red areas; Supplemental Fig. 6A–C). In contrast, there is an increase in enrichment of lamin A/C-containing states 2, 3, and 4 on genomic domains predefined in ChromHMM as “lamin B1 LADs” (Fig. 6B; yellow area). Supporting these results, we have earlier shown a significant increase in the fraction of lamin A/C LADs overlapping with lamin B1 LADs (see Fig. 1H). This importantly shows dissociation of lamin A/C from genic regions, including active promoter and enhancer elements, after induction of differentiation. De novo formation of lamin A/C LADs in contrast implicates gene-poor and repressed heterochromatin domains.
Discussion
We provide evidence for a prepatterning of lamin A/C LADs by domains of GlcNAcylated H2B (GADs) during adipogenic differentiation of human primary adipocyte progenitors. We propose a model of a two-step process of lamin A/C LAD formation during adipogenesis (Fig. 6C). Our model entails spreading of LADs lamin A/C–chromatin interactions after the transition from progenitor cell proliferation to cell-cycle arrest and redistribution of these interactions through an exchange of LADs within hours of adipogenic induction. In undifferentiated cells, lamin A/C LADs are enriched in active genes and regulatory elements and do not necessarily involve GADs. However, post-differentiation LADs contain repressive chromatin features and strongly coincide with GADs. As lamin A/C is found both at the nuclear periphery and in the nuclear interior (Dechat et al. 2010; Gesson et al. 2014), the heterochromatic versus euchromatic nature of lamin A/C–chromatin interactions evidenced in this study is likely influenced by the dual localization of A-type lamins: Peripheral LADs are enriched in heterochromatin (Guelen et al. 2008; Reddy et al. 2008), while nucleoplasmic LADs would be more euchromatic. Extending the dogma that LADs are mainly gene poor and transcriptionally inactive, our results indicate that lamin A/C LADs can be found both in active and repressive chromatin contexts, which can be influenced by cell differentiation status.
Remodeling of LADs after adipogenic induction raises the question of how this reorganization is orchestrated. Chromosome movements in response to changes in gene expression (Bickmore and van Steensel 2013) regulate radial positioning of loci and may contribute to forming de novo LADs (Harr et al. 2015). Alternatively, pools of lamin A/C may translocate to the nuclear periphery (Gesson et al. 2014), a process regulated by LAP2 (Naetar et al. 2008), and this could potentially generate new LADs. Interestingly, LAP2 also modulates the balance between tissue progenitor cell proliferation and differentiation (Dorner et al. 2006; Naetar and Foisner 2009; Gotic et al. 2010). This raises the possibility that LAP2-lamin A/C interactions, and the equilibrium between nucleoplasmic and peripheral pools of lamin A/C, participate in the regulation of cell fate including adipogenic differentiation (Dorner et al. 2006).
H2BS112GlcNAc emerges as a new epigenetic mark underlying lamin A/C associations with chromatin. However, since 50% of GADs do not harbor lamin A/C after differentiation, GADs are not the only component specifying lamin A/C LADs. Several factors including DNA sequences in LAD borders, transcription factors, histone modifiers, and repressive histone modifications regulate sequence positioning to the nuclear periphery in various cellular contexts (Towbin et al. 2012; Zullo et al. 2012; Kind et al. 2013; Harr et al. 2015). LADs have been shown to be enriched in H3K9me2/3 (Guelen et al. 2008; Kind et al. 2013; Harr et al. 2015) and targeting of sequences to the nuclear periphery in C. elegans requires H3K9 histone methyltransferase activity (Towbin et al. 2012). There is no evidence that the H3K9 methyltransferase G9a (encoded by EHMT2) is preferentially active at the nuclear periphery in human cells; however, G9a plays a role in promoting chromatin–lamina contacts (Kind et al. 2013). The overall repressed state of GADs identified in ASCs and their overlap with lamin B1 LADs is both compatible with their enrichment in heterochromatin marked by H3K9me2/3. However, this may also speculatively reflect a cell-specific state since we find that lamin A/C LADs in undifferentiated ASCs contain active genes (Fig. 6C). It would be interesting to assess the role of H3K9me di- or trimethylation on the formation of adipogenic LADs on GADs.
Whether the Polycomb facultative heterochromatin mark H3K27me3 is also involved in adipogenic de novo lamin A/C LAD formation remains unknown but is a possibility, in light of its involvement (together with H3K9me2/3) in the formation of LADs in mouse cells (Harr et al. 2015). Interestingly, short DNA sequences (Zullo et al. 2012) located in LAD borders are important for chromatin–lamina interactions (Harr et al. 2015). LAD borders, but not LAD interiors, have been found to be enriched in H3K27me3 in human and mouse cells (Guelen et al. 2008; Simon et al. 2013; Harr et al. 2015), and abrogation of H3K27me3 by down-regulation of the H3K27 methyltransferase EZH2 impairs LAD maintenance (Harr et al. 2015). Thus, in addition to H3K9me2/3 within LADs, H3K27me3 in LAD borders could be a key player in de novo lamin A/C LAD formation during differentiation (Fig. 6C). The role of protein O-GlcNAcylation in Polycomb-mediated gene repression in Drosophila (Gambetta and Müller 2014), although this remains to be shown in mammals, and the discovery that O-GlcNAc is enriched at Polycomb response elements (Gambetta et al. 2009; Sinclair et al. 2009), suggest a view of an interplay between different histone modifications and the formation or maintenance of nuclear lamin–genome associations. Several levels of regulation involving combinations of multiple factors and H2BGlcNAc patterns may therefore determine lamin A/C–chromatin interactions elicited by differentiation in a cell- and tissue-specific manner.
Through several means of association of OGT with chromatin, GlcNAcylation is connected to multiple functions. In immortalized cell lines, OGT and H2BS112GlcNAc have been linked to transcriptional activity via targeting of OGT to chromatin by ten-eleven translocation enzymes (Chen et al. 2013; Deplus et al. 2013; Vella et al. 2013). However, none of the TET genes are expressed in ASCs or adipocytes (Shah et al. 2014), and we find no clear correlation between H2BGlcNAc and H2BK120ub1 (which is linked to transcriptional activity) in ASCs. Instead, at least in ASCs, GADs correspond to intergenic regions and silent chromatin. In line with our findings, OGT is implicated in gene silencing (Gambetta et al. 2009; Sinclair et al. 2009; Darwanto et al. 2010; Gambetta and Müller 2014), for instance by modulating stability and function of EZH2 (Chu et al. 2014), and through its recruitment to chromatin by transcriptional corepressors (Yang et al. 2002; Vella et al. 2013). Thus, distinct mechanisms may link OGT to chromatin and OGT activity toward histones in different cellular contexts experiencing different metabolic states.
Inasmuch as in vivo quiescent tissue progenitor cells under hypoxic conditions in organs produce ATP through glycolysis, rapidly dividing cells in culture also use glycolysis more than oxidative phosphorylation (Shyh-Chang et al. 2013). Accordingly, glycolytic enzymes are expressed in undifferentiated proliferating ASCs in vitro. Consistent with this finding, the transcription factor MEIS1 is expressed in ASCs, enabling up-regulation of its target HIF1A (Simsek et al. 2010) on D1 of adipogenic induction, and activation of HIF1A targets including glycolytic enzymes (Simsek et al. 2010; Shyh-Chang et al. 2013). Our results argue that adipogenic stimulation up-regulates the glycolytic pathway, consistent with the in vivo adipogenic commitment of adipocyte precursors. Enhanced glycolysis up-regulates the hexosamine biosynthetic pathway, in turn increasing O-GlcNAc levels and O-GlcNAc cycling. It is tempting to speculate that adipogenic differentiation and fat accumulation are influenced by the rate of ATP synthesis from glycolysis, and this may involve O-GlcNAcylation of H2B and formation of adipogenic LADs. Indeed, deregulation of O-GlcNAc cycling by down-regulation or mutation of OGA leads to glucose intolerance and insulin resistance (Lehman et al. 2005) and obesity in mice (Keembiyehetty et al. 2015).
These syndromes are also linked to mutation in the LMNA gene causing partial lipodystrophies (Guenantin et al. 2014), coupling A-type lamins to cellular metabolic state. Specific lipodystrophic LMNA mutation hot spots disrupt lamin A/C interaction with DNA in vitro (Stierlé et al. 2003) and could affect LADs. The common metabolic disorders caused by deregulation of protein O-GlcNAcylation and lamin A-linked lipodystrophies raise the possibility of a relationship between A-type lamins and chromatin modifications modulated by histone GlcNAcylation. This relationship may speculatively link spatial genome organization to cellular metabolic state and may have implications in the etiology of laminopathies.
Methods
Antibodies
Antibodies used were against H2BS112GlcNAc (ab130951, Abcam), H2B (ab1790, Abcam), H2BK120Ub1 (5546, Cell Signaling), O-GlcNAc (RL2, Abcam), lamin A/C (sc7292, Santa Cruz), lamin B1 (sc6216, Santa Cruz), α-tubulin (T5168, Sigma-Aldrich), EGFP (11814460001, Roche), and γ-tubulin (T5326, Sigma-Aldrich). Control IgGs used for immunoprecipitation and ChIP-seq were from Millipore (12-370).
ASCs and adipogenic differentiation
Human normal adipose tissue stromal cells (ASCs) (Boquest et al. 2005) were cultured under proliferative conditions in DMEM/F12 (17.5 mM glucose) with 10% fetal calf serum, 10 ng/mL epidermal growth factor, and 20 ng/mL basic fibroblast growth factor (Sigma-Aldrich). Cells at passage 6–10 were used for differentiation in 2–5 independent experiments. Two days before adipogenic induction, cells were harvested and used for analysis (“D−2”) or replated confluent in DMEM/F12/10% serum without growth factors. Adipogenesis was induced on D0 by adding 0.5 µM 1-methyl-3 isobutylxanthine, 1 µM dexamethasone, 200 µM indomethacin, and 10 µg/mL insulin, and cells were cultured for up to 9 d.
Vectors
Human H2B cDNA was amplified by PCR from pH2B-diHcRed (a gift from Jan Ellenberg, EMBL, Heidelberg) using a sense primer containing a HindIII site at the 5′ end (5′-CGCAAGCTTTACCAGAGCCAGCGAAGTCTGCTCCC-3′) and an antisense primer with a SacII site at the 5′ end (5′-AGTCCGCGGTCACTTAGCGCTGGTGTACTTGGTGAC-3′). Products were digested with HindIII and SacII and ligated into the HindIII and SacII cloning sites of pEGFP-C1 (Clontech). Plasmid encoding EGFP-H2B(S112A) was made from pEGFP-H2B by mutagenesis using the following primers: 5′-GCCAAGCACGCCGTGGCCGAGGGTACTAAGG-3′ and 5′-CCTTAGTACCCTCGGCCACGGCGTGCTTGGC-3′.
OGA treatment of cell extract
ASCs were lysed in 20 mM Hepes/HCl, pH 8.2, 50 mM NaCl, 5 mM MgCl2, and a protease inhibitor mix for 25 min and sonication with a probe sonicator (Braun Biotech; 60% power, 0.5 sec intervals for 30 sec). Lysates were incubated for 30 min at room temperature with 2 ng/µL recombinant OGA (6779GH020, R&D Systems) and dissolved in 2× Laemmli buffer for SDS-PAGE and immunoblotting.
Dot blot assay and immunoblotting
Synthetic H2B (residues 108–117) and H2BS112GlcNAc (residues 108–116 with S112GlcNAc) peptides (Abcam) were spotted on a nitrocellulose membrane, air-dried, and blocked with Odyssey blocking buffer (Licor). IRDye-800-coupled secondary antibodies and Odyssey imaging were used for visualization. For immunoblotting, proteins were separated by SDS-PAGE, blotted onto Immobilon-FL membranes (Millipore), and blocked with Odyssey blocking buffer. Proteins were visualized using IRDye-800- or IRDye-680-coupled antibodies (Licor) and by Odyssey imaging.
Immunofluorescence and microscopy
Cells on coverslips were fixed with 3% paraformaldehyde for 15 min followed by 2 min in ice-cold methanol. Coverslips were blocked with 2% BSA, 0.1% Tween 20 in PBS (PBST-BSA) for 90 min, and incubated with primary antibodies overnight at 4°C. Coverslips were incubated with Cy2, Cy3 (Jackson ImmunoResearch), or Alexa 488, Alexa 594 (Life Technologies) coupled secondary antibodies for 30 min before washing with PBST-BSA and mounting with DAPI Fluoromount G (Southern Biotech). Images were acquired on an Olympus IX71 microscope fitted with the DeltaVision system or through a SuperApochromat 60X/1.35 objective on an Olympus FluoView 1000 confocal microscope. Images were processed with ImageJ v1.42q.
Chromatin immunoprecipitation
Cells (107/ChIP) were cross-linked with 1% formaldehyde for 10 min, lysed for 10 min in ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 7.5, proteinase inhibitors, 1 mM PMSF), and sonicated 4 × 10 min in a Bioruptor (Diagenode) to generate 200–500 bp DNA fragments. After sedimentation at 10,000g for 10 min, the supernatant was collected and diluted 10 times in RIPA buffer (140 mM NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM PMSF, protease inhibitors) (input chromatin). Chromatin was incubated overnight at 4°C with antibodies to lamin A/C (5 µg/106 cells; Santa Cruz sc7292), lamin B1 (Abcam ab16048; 2.5 µg/106 cells), H2BS112GlcNAc (Abcam ab130951; 2.5 µg/106 cells), H2BK120ub1 (Cell Signaling 5546; 2.5 µg/106 cells), H2B (Abcam ab1790; 2.5 µg/106 cells), or with control IgGs (2.5 µg/106 cells) coupled to magnetic Dynabeads Protein A/G (Invitrogen). ChIP samples were washed four times in ice-cold RIPA buffer and incubated with 0.5 µg/mL RNase A for 20 min at 37°C. Crosslinks were reversed, and DNA was eluted for 6 h at 68°C in 50 mM NaCl, 20 mM Tris-HCl pH 7.5, 5 mM EDTA, 1% SDS, and 50 ng/µL Proteinase K. DNA was purified and dissolved in H2O. Sequencing libraries were prepared as per Illumina protocol and sequenced on an Illumina HiSeq 2500. For H2BS112GlcNAc ChIP, buffers were supplemented with 10 µM of the OGA inhibitor PugNAc (Sigma-Aldrich). ChIP DNA was also used as template for quantitative (q)PCR using primers to genic and intergenic regions (Supplemental Table 1). PCR was done using SYBR Green (BioRad) for 3 min at 95°C and 40 cycles of 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C.
RNA sequencing and data analysis
RNA-seq data for D−2, D0, D3, and D9 of differentiation were published earlier (Shah et al. 2014) and downloaded from NCBI GEO GSE60237. RNA-seq data for D1 were generated as part of the same data set. Each RNA-seq data set was generated from duplicate differentiation experiments and sequence reads combined. RNA-seq reads were processed using Tuxedo (Trapnell et al. 2010). TopHat (Trapnell et al. 2012) was used to align reads with no mismatch against the hg19 reference genome with default settings, applying the Bowtie 2 (Langmead and Salzberg 2012) preset “very sensitive.” Cufflinks and cuffdiff were run using default settings and bias correction. Gene ontology analyses were done using GOSlim (Ashburner et al. 2000) and Gorilla (Eden et al. 2009) packages.
ChIP-seq data processing and identification of LADs and GADs
All scripts were written in Perl (Stajich et al. 2002) or R (R Core Team 2015). H2BGlcNAc and lamin A/C ChIP-seq reads were mapped to hg19 using Bowtie v1.0.0 (Langmead et al. 2009) with default settings and option “-best” enabled. Note that using hg19 instead of the more recent GRCh38 annotation did not affect our conclusions because GRCh38 updated annotations for mitochondrial gene, centromeric, and other repeat regions, to which sequencing reads are not mapped. Mapped reads were used to call peaks using Enhanced Domain Detector (EDD) (Lund et al. 2014). Browser files were generated by getting a ratio of ChIP/input for each of 1-kb (H2BGlcNAc) or 10-kb (lamin A/C) bins with input normalized to the ratio of total ChIP reads/total input reads. Lengths of lamin A/C and H2BGlcNAc peaks (domains) were categorized small (below lower quantile), large (above upper quantile), and medium (inter-quantile range) (Supplemental Script 1). Gene density was determined by computing numbers of genes per megabase in LADs and GADs (Supplemental Script 2).
Analysis of chromatin states
ChIP-seq data sets of histone marks and CTCF were from a previous study (Mikkelsen et al. 2010). Reads were remapped using Bowtie and peaks called using MACS v1.4.2 (Zhang et al. 2008). ChromHMM (Ernst and Kellis 2012) was used to identify chromatin states using the called peaks. Options were selected to learn a 15-state model using the Baum-Welch training algorithm (Miklós and Meyer 2005).
Statistical analysis
Gene intersects were generated by using “intersectBed” from BEDTools v2.21.0 (Quinlan and Hall 2010) on gene lists from RNA-seq data and ChIP-seq peaks. Genome coverage, Jaccard indices, and overlap between GADs and LADs were calculated using “genomeCoverageBed,” “coverageBed,” and “BEDTools jaccard” (Quinlan et al. 2010) (Supplemental Scripts 3, 4). Significance of the overlap between GADs and LADs was calculated by performing 10,034 Monte Carlo simulations of randomly shuffling LADs along the genome using “shuffleBed” and generating a P-value based on the number of times the permutations were equally or more extremely colocalized (Supplemental Script 5). Significance for difference in expression between genes within peaks, outside peaks, and RefSeq genes was determined by a Wilcoxon rank-sum test and Bonferroni corrected. Significance for difference in enrichment of predefined genomic regions in chromatin states was tested by generating a distribution of the difference in ratios of bases of a given state for the region, divided by the total bases of the chromatin state between the time points shuffled in a case-control manner, and a two-tailed t-test with the original value as the testing value.
Data viewing
Browser views of gene tracks, ChIP-seq data, and chromatin states are shown using the Integrated Genomics Viewer (Robinson et al. 2011). Genes are from Illumina iGenomes gene annotation with UCSC source for hg19. All plots were generated using the ggplot2 or LSD libraries in R.
Data access
The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE63346.
Acknowledgments
We thank Dr. Gunnar Kvalheim (Oslo University Hospital) for adipose stromal cells and Dr. Jan Ellenberg (EMBL, Heidelberg) for pH2B-diHcRed. This work was supported by the University of Oslo (A.S., T.R.), The Research Council of Norway, the Norwegian Cancer Society (E.D.), South East Health Norway (A.R.O.), and the Norwegian Center for Stem Cell Research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.193748.115.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152: 1270–1284. [DOI] [PubMed] [Google Scholar]
- Boquest AC, Shahdadfar A, Frønsdal K, Sigurjonsson O, Tunheim SH, Collas P, Brinchmann JE. 2005. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell 16: 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Stewart CL. 2013. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol 14: 13–24. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. 2013. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, Kang ML, Wong CH, Juan LJ. 2014. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci 111: 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwanto A, Curtis MP, Schrag M, Kirsch W, Liu P, Xu G, Neidigh JW, Zhang K. 2010. A modified “cross-talk” between histone H2B Lys-120 ubiquitination and H3 Lys-79 methylation. J Biol Chem 285: 21868–21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. 2000. Lamina-associated polypeptide 2α binds intranuclear A-type lamins. J Cell Sci 113: 3473–3484. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gesson K, Foisner R. 2010. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb Symp Quant Biol 75: 533–543. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. 2013. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 32: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner D, Vlcek S, Foeger N, Gajewski A, Makolm C, Gotzmann J, Hutchison CJ, Foisner R. 2006. Lamina-associated polypeptide 2α regulates cell cycle progression and differentiation via the retinoblastoma–E2F pathway. J Cell Biol 173: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, Sifers RN. 2012. β-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem 287: 12195–12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, et al. 2011. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Müller J. 2014. O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev Cell 31: 629–639. [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Müller J. 2015. A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma. 10.1007/s00412-015-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Müller J. 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325: 93–96. [DOI] [PubMed] [Google Scholar]
- Gay S, Foiani M. 2015. Nuclear envelope and chromatin, lock and key of genome integrity. Int Rev Cell Mol Biol 317: 267–330. [DOI] [PubMed] [Google Scholar]
- Gesson K, Vidak S, Foisner R. 2014. Lamina-associated polypeptide (LAP)2α and nucleoplasmic lamins in adult stem cell regulation and disease. Semin Cell Dev Biol 29: 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotic I, Foisner R. 2010. Multiple novel functions of lamina associated polypeptide 2α in striated muscle. Nucleus 1: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotic I, Schmidt WM, Biadasiewicz K, Leschnik M, Spilka R, Braun J, Stewart CL, Foisner R. 2010. Loss of LAP2α delays satellite cell differentiation and affects postnatal fiber-type determination. Stem Cells 28: 480–488. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Guenantin AC, Briand N, Bidault G, Afonso P, Bereziat V, Vatier C, Lascols O, Caron-Debarle M, Capeau J, Vigouroux C. 2014. Nuclear envelope-related lipodystrophies. Semin Cell Dev Biol 29: 148–157. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. 2012. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol 13: 312–321. [DOI] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. 2015. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 208: 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW. 2014. Three decades of research on O-GlcNAcylation—a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front Endocrinol 5: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. 2007. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Nitta RT, Frock RL, Mounkes L, Barbie DA, Stewart CL, Harlow E, Kennedy BK. 2004. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc Natl Acad Sci 101: 9677–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating ST, El-Osta A. 2015. Epigenetics and metabolism. Circ Res 116: 715–736. [DOI] [PubMed] [Google Scholar]
- Keembiyehetty C, Love DC, Harwood KR, Gavrilova O, Comly ME, Hanover JA. 2015. Conditional knock-out reveals a requirement for O-linked N-Acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J Biol Chem 290: 7097–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Kolb T, Maass K, Hergt M, Aebi U, Herrmann H. 2011. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus 2: 425–433. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman DM, Fu DJ, Freeman AB, Hunt KJ, Leach RJ, Johnson-Pais T, Hamlington J, Dyer TD, Arya R, Abboud H, et al. 2005. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc–selective N-acetyl-β-D glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 54: 1214–1221. [DOI] [PubMed] [Google Scholar]
- Lund E, Oldenburg A, Delbarre E, Freberg C, Duband-Goulet I, Eskeland R, Buendia B, Collas P. 2013. Lamin A/C-promoter interactions specify chromatin state–dependent transcription outcomes. Genome Res 23: 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Oldenburg AR, Collas P. 2014. Enriched domain detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res 42: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. 1991. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem 266: 4706–4712. [PubMed] [Google Scholar]
- Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. 2011. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol 21: 1603–1614. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. 2013. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143: 156–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklós I, Meyer IM. 2005. A linear memory algorithm for Baum-Welch training. BMC Bioinformatics 6: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naetar N, Foisner R. 2009. Lamin complexes in the nuclear interior control progenitor cell proliferation and tissue homeostasis. Cell Cycle 8: 1488–1493. [DOI] [PubMed] [Google Scholar]
- Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, Gotic I, Fuchs P, Cohen TV, Bittner R, et al. 2008. Loss of nucleoplasmic LAP2α-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol 10: 1341–1348. [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the mitotic chromosome. Science 14: 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https:/www.R-project.org/. [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young AR, Narita M, Pérez-Mancera PA, Bennett DC, Chong H, et al. 2013. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 27: 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Oldenburg A, Collas P. 2014. A hyper-dynamic nature of bivalent promoter states underlies coordinated developmental gene expression modules. BMC Genomics 15: 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. 2013. Stem cell metabolism in tissue development and aging. Development 140: 2535–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, Mandrup S. 2012. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab 23: 56–64. [DOI] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504: 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA. 2010. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7: 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc Natl Acad Sci 106: 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierlé V, Couprie J, Östlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin JC, Duband-Goulet I. 2003. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry 42: 4819–4828. [DOI] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. 2013. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell 49: 645–656. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang F, Kudlow JE. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110: 69–80. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Roche K, Nasheuer HP, Lowndes NF. 2011. Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem 286: 37483–37495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. 2012. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149: 1474–1487. [DOI] [PubMed] [Google Scholar]