Abstract
The hepatitis B virus (HBV) core protein is a dynamic and versatile protein that directs many viral processes. During capsid assembly, core protein allosteric changes ensure efficient formation of a stable capsid that assembles while packaging viral RNA–polymerase complex. Reverse transcription of the RNA genome as well as transport of the capsid to multiple cellular compartments are directed by dynamic phosphorylation and structural changes of core protein. Subsequently, interactions of the capsid with the surface proteins and/or host proteins trigger envelopment and release of the viral capsids or the transport to the nucleus. Held together by many weak protein–protein interactions, the viral capsid is an extraordinary metastable machine that is stable enough to persist in the cellular and extracellular environment but dissociates to allow release of the viral genome at the right time during infection.
Hepatitis B virus (HBV) capsids are stable both inside and outside the cell but dissociate during infection. Their assembly and egress relies heavily on the HBV core protein and its interactions with other host and viral proteins.
Hepatitis B virus (HBV) belongs to the family of Hepadnaviridae and is an enveloped virus that contains a partially double-stranded DNA (dsDNA) genome that is surrounded by an icosahedral protein capsid. HBV virions enter the cell via the recently identified receptor sodium taurocholate cotransporting polypeptide (NTCP), presumably losing their envelope during endocytosis (Yan et al. 2012). Within the cytosol capsids are transported to the nucleus during which uncoating and release of viral genome is initiated. In the nucleus, the partially double-stranded genome is then repaired into a covalently closed circular DNA (cccDNA) genome. The nuclear cccDNA serves as a template for the pregenomic RNA (pgRNA) and subgenomic RNAs transcripts, encoding all viral proteins (Beck and Nassal 2007).
One copy of pgRNA together with the viral polymerase polypeptide (P) is packaged concomitant with capsid assembly. After formation of the core particle, P initiates reverse transcription of the RNA genome leading to the synthesis of minus-strand DNA and digestion of pgRNA by the ribonuclease H (RNaseH) domain of P. Minus-strand DNA then serves as a template for the synthesis of plus–strand DNA, resulting in the formation of a relaxed circular, partially dsDNA genome. Mature capsids are either enveloped and secreted from the cell or transported to the nucleus, amplifying the pool of cccDNA. Secretion of virus particles is mediated by interactions of the capsid with the large hepatitis B surface antigen (L-HBsAg) at the endoplasmic reticulum (ER). Independent of infectious virus particles (Dane particles), noninfectious subviral particles (SVPs) containing small, medium, and large forms of HBsAg are also released from the infected cell.
This review describes mechanisms of core assembly and virion release. We also discuss findings from in vitro studies of core protein that have greatly enhanced our understanding of the molecular mechanisms of capsid assembly.
CAPSID ASSEMBLY IN VIVO
Regulation of capsid formation and packaging of the viral genome is not yet fully understood. Within the cytosol of infected cells, HBV capsids contain 120 or 90 copies of the core protein, yielding particles with T = 4 and T = 3 icosahedral symmetry with diameters of ∼34 and ∼30 nm, respectively. The T = 4 particles represent the predominant form in vivo (Stannard and Hodgkiss 1979). The core protein is 183 or 185 amino acids (aa) long depending on the virus isolate, and contains an assembly domain (aa 1–149) and an arginine-rich carboxy-terminal domain (CTD), essential for interactions with RNA and DNA (aa 150–183/5) (Birnbaum and Nassal 1990; Nassal 1992).
Specific packaging of the viral RNA depends on the formation of a ribonucleoprotein (RNP) complex that requires interaction of P with the packaging signal ɛ near the 5′ end of pgRNA (Bartenschlager and Schaller 1992; Pollack and Ganem 1994). ɛ is a 137-nucleotide-long sequence motif that forms a stem loop structure that is conserved among all known hepadnaviruses (Junker-Niepmann et al. 1990; Pollack and Ganem 1993; Flodell et al. 2002). Sequence motifs within ɛ as well as the distance between the 5′ cap of pgRNA and ɛ are essential determinants for RNA packaging (Pollack and Ganem 1993; Fallows and Goff 1995; Jeong et al. 2000; Hu and Boyer 2006). A recent cryo-EM study of RNA-filled cores, performed without imposing symmetry on the complex, showed two concentric layers of core protein surrounding RNA. Inside the RNA layer, multiple densities were observed, likely corresponding to the P protein bound to pgRNA and host factors packaged into the virion (Wang et al. 2014). Interestingly, because RNA and non-RNA densities were well ordered in the reconstruction, it suggests that organization of protein and RNA in HBV is uniform from particle to particle.
Multiple host factors have also been implicated in capsid assembly and RNA packaging in vivo. Among them is the molecular chaperone Hsp90, which binds to P. Inhibition of Hsp90 can decrease capsid formation and packaging of viral RNA in cell cultures (Hu and Seeger 1996; Hu et al. 2004; Shim et al. 2011). Other chaperone proteins, such as Hsc70 and Hsp40, have been found to be associated with viral capsids by mass spectrometry (Wang et al. 2009). Finally, nucleophosmin (B23) has been implicated in interacting with HBV core proteins (Okuwaki 2008; Lee et al. 2009; Jeong et al. 2014).
Encapsidated host factors that can inhibit HBV replication include the DDX3 DEAD-box helicase and the human cytidine deaminase APOBEC3G (A3G) (Turelli et al. 2004; Baumert et al. 2007; Nguyen et al. 2007; Nguyen and Hu 2008; Wang et al. 2009). Both proteins appear to block early stages of DNA synthesis that occur subsequent to RNA packaging (Turelli et al. 2004; Nguyen et al. 2007; Wang et al. 2009). Recently, it has been suggested that the HBV core protein recruits APOBEC3A and APOBEC3B to cccDNA within the nucleus (Lucifora et al. 2014).
Core Protein Determinants for Encapsidation
Detailed genetic analyses of the core protein revealed that efficient packaging of pgRNA does not require a complete CTD. For example, core proteins truncated at residue 172, lacking 11 residues, show normal RNA packaging and reverse transcription activities (Nassal 1992; Chua et al. 2009). However, core proteins terminating at residues 163 and 164 are defective for RNA packaging (Nassal 1992; Beames and Lanford 1993; Kock et al. 2004). Interestingly, the 19-residue deletion predominantly incorporated a spliced RNA that was reverse transcribed into a 2-kb dsDNA (Kock et al. 2004; Le Pogam et al. 2005). Empty particles have also been found in the cytosol of HBV-producing cells, indicating that core proteins can assemble without RNA packaging (Gerin et al. 1975; Kaplan et al. 1976; Alberti et al. 1978; Ning et al. 2011). It is still unclear why core protein assembly in vivo is highly specific for viral RNA, whereas random RNAs are readily packaged in Escherichia coli and in vitro. It can be hypothesized that specificity for pgRNA in vivo is achieved by yet-to-be-determined regulatory factors (Chen et al. 2011).
A complicated system of dynamic CTD phosphorylation regulates RNA packaging reverse transcription, and intracellular trafficking. Residues S155, S162, and S170 are essential phosphorylation sites for pgRNA packaging (Liao and Ou 1995; Gazina et al. 2000). Alanine mutations of these residues, except an S155 point mutation, showed reduced pgRNA packaging (Kann and Gerlich 1994; Lan et al. 1999; Gazina et al. 2000; Kock et al. 2004; Le Pogam et al. 2005; Lewellyn and Loeb 2011b). The kinase(s) mediating CTD phosphorylation is (are) the subject of conflicting reports. It is possible that more than one kinase plays a role. Protein kinase C (PKC), protein kinase A (PKA), and cyclin-dependent kinase 1 and 2 (CDK1 and CDK2) have been identified based on inhibitor assays (Kann and Gerlich 1994; Ludgate et al. 2012). Ludgate and coworkers provided immunological evidence for CDK2, but could not exclude others (Ludgate et al. 2012). Other kinases such as GAPDH kinase, a 46-kDa serine kinase, serine protein kinase 1 and 2 (SRPK1 and SRPK 2) have been implicated (Duclos-Vallee et al. 1998; Kau and Ting 1998; Daub et al. 2002; Zheng et al. 2005). Although not discussed in the literature, dephosphorylation of the CTD associated with reverse transcription implies the presence of an encapsidated phosphatase (Yu and Summers 1994b; Perlman et al. 2005).
ENVELOPMENT AND RELEASE
Maturation Signal
Particles at all stages of reverse transcription are found within infected cells. Depending on the level of maturity, they can contain predominantly RNA, ssDNA, or dsDNA. The dsDNA particles contain mature, partially double-stranded, relaxed circular DNA (rcDNA) and, at much lower levels, double-stranded linear DNA (dslDNA) (Staprans et al. 1991). Empty and mature particles are directed for envelopment (Summers and Mason 1982; Gerelsaikhan et al. 1996; Perlman and Hu 2003; Ning et al. 2011; Cui et al. 2013). The signal for capsid envelopment has not yet been well defined. However, an increase in internal pressure caused by the relative rigidity of dsDNA compared with ssRNA could lead to capsid instability that triggers envelopment (Tzlil et al. 2003; Hagan 2013). Enveloped empty particles are found in the serum of infected patients, cell cultures, and infected chimpanzees, suggesting that empty and mature particles share a common maturation signal (Gerin et al. 1975; Kaplan et al. 1976; Alberti et al. 1978; Ning et al. 2011; Cui et al. 2013). In empty capsids, the lack of genome could result in instability possibly attributable to electrostatic repulsion by CTDs. Indeed, empty and mature capsids, but not immature particles, are sensitive to proteinase K (Cui et al. 2013).
Emphasizing the importance of capsid structure during envelopment, there are core protein mutations and truncations that block envelopment, in some cases despite complete synthesis of dsDNA (Koschel et al. 2000; Le Pogam et al. 2000; Ponsel and Bruss 2003). Highlighting the importance of core dynamics, a buried F97L core mutant results in secretion of immature virions (Yuan et al. 1999).
Not all mature capsid particles are enveloped and released from the cell. A portion of capsids are transported back to the nucleus to replenish the pool of cccDNA as shown for duck hepatitis B virus (DHBV) (Tuttleman et al. 1986). Transport to the nucleus depends on genome maturation, exposure of a nuclear localization sequence (NLS) on the CTD, as well as phosphorylation of the CTD (Tuttleman et al. 1986; Eckhardt et al. 1991; Kann et al. 1999; Rabe et al. 2003). Indeed, in capsids derived from cell cultures, CTDs were more readily exposed in mature and empty particles than immature ones (Rabe et al. 2003; Wang et al. 2012; Yu et al. 2013). Kann and coworkers have suggested that phosphorylation of the CTD can act to facilitate CTD exposure to nuclear transport proteins, importin α and importin β (Rabe et al. 2003, 2009; Wittkop et al. 2010).
Envelope Proteins
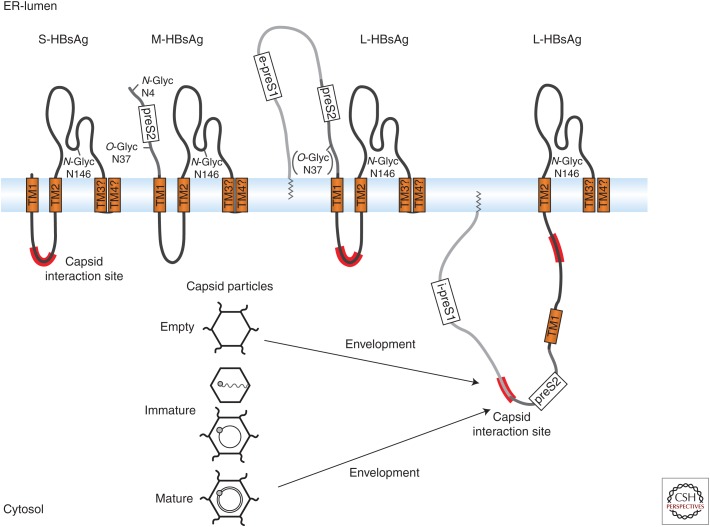
Three envelope, or surface, proteins (HBsAg) of increasing size called small (S-HBsAg, S), medium (M-HBsAg, M), and large (L-HBsAg, L) are expressed from one open reading frame during HBV infection (Fig. 1). M is dispensable for the production of infectious virions (Schlicht et al. 1987; Bruss and Ganem 1991; Fernholz et al. 1993). All three proteins are cotranslationally integrated into the ER membrane and form disulfide-linked dimers and heterodimers (Mangold and Streeck 1993; Mangold et al. 1995; Wounderlich and Bruss 1996). Membrane integration of the amino-terminal domain of the S domain is directed by the first and second transmembrane (TM1 and TM2) segments (Fig. 1) (Eble et al. 1987, 1990; Short et al. 2009). M has the same topology as S with the 55-amino-acid preS2 domain located in the ER lumen. The large envelope protein, L has myristic acid at its amino terminus and is attached to membranes. Moreover, it has dual topology important for assembly and infectivity of virions. During expression, the preS1 and preS2 domains of L remain in the cytosol (i-preS1). In about 50% of expressed L proteins, the preS1-preS2 domains are posttranslationally translocated into the ER lumen, anchored into the membrane by TM1 (e-preS1) (Fig. 1). The e-preS form is exposed on the surface of virions, where it is believed to facilitate subsequent attachment and entry of the viral particle (Gripon et al. 1995; Le Seyec et al. 1999; Lepere-Douard et al. 2009). A short conserved region of the cytosolic i-preS1 form between residues 96–116 and part of a cytosolic loop from the S domain are important for the interaction with capsids and subsequent virion envelopment (Fig. 1) (Bruss 1997; Poisson et al. 1997; Le Seyec et al. 1998; Loffler-Mary et al. 2000).
Figure 1.
Morphology of the small hepatitis B surface antigen S-HBsAg (S), the medium M-HBsAg (M), and the large L-HBsAg (L) (adapted from Schädler and Hildt 2009). The large envelope protein has three domains called PreS1, PreS2, and S, whereas M-HBsAg contains PreS2 and S, and S-HBsAg comprises the S domain. Envelope proteins are translocated through the endoplasmic reticulum (ER), anchoring the transmembrane (TM)1/2 and TM3/4 domains into the membrane. L-HBsAg displays dual topology. In i-preS1, preS1 remains in the cytosol, whereas in e-preS1, preS1 is translocated through the membrane into the ER-lumen. N-glycosylation (N-glyc) of the S domain can occur at residue Asn146 (Peterson et al. 1982). M-HBsAg harbors another glycosylation site on Asn4 and an O-glycosylation (O-glyc) site on Thr37 (Stibbe and Gerlich 1983; Werr and Prange 1998; Schmitt et al. 2004). Capsid interaction sites include residues 96–116 of preS1 and part of a cytosolic loop in the S domain (in red).
Subviral Particles
The envelope proteins also assemble into empty subviral particles (SVPs) at the post-ER/pre-Golgi membrane (Huovila et al. 1992). SVPs are drastically overproduced compared to the number of virions seen in serum of infected patients (Ganem and Prince 2004). Subviral particles are found as spherical structures with a diameter of about 22 nm and filaments of similar diameter but variable length (Gavilanes et al. 1982; Diminsky et al. 1997; Gilbert et al. 2005).
Expression of S is necessary and sufficient for the formation of spherical particles (Liu et al. 1982; Patzer et al. 1986; Bruss and Ganem 1991). During natural infections, however, spherical particles also contain M and trace amounts of L (Heermann et al. 1984). Coassembly of particles with higher amounts of L (ratio of 1:1:4 of L:M:S) leads to the formation of filamentous structures (Heermann et al. 1984; Short et al. 2009). Yeast-expressed recombinant S-HBsAg is the basis of the HBV vaccine (Gerlich and Kann 2010).
Cryo-EM structure determination of filamentous particles reveals tubes with an average diameter of 25 nm with well-defined spike-like features, formed by S dimers, projecting from the membrane surface (Short et al. 2009). The spikes are reminiscent of those seen in a single-particle reconstruction of a virion and consistent with projections observed in the raw data (Dryden et al. 2006). Of note, the ends of filaments appear to be closed off by hemispherical caps corresponding to half of a 22-nm particle (Short et al. 2009).
Release of Viral Particles
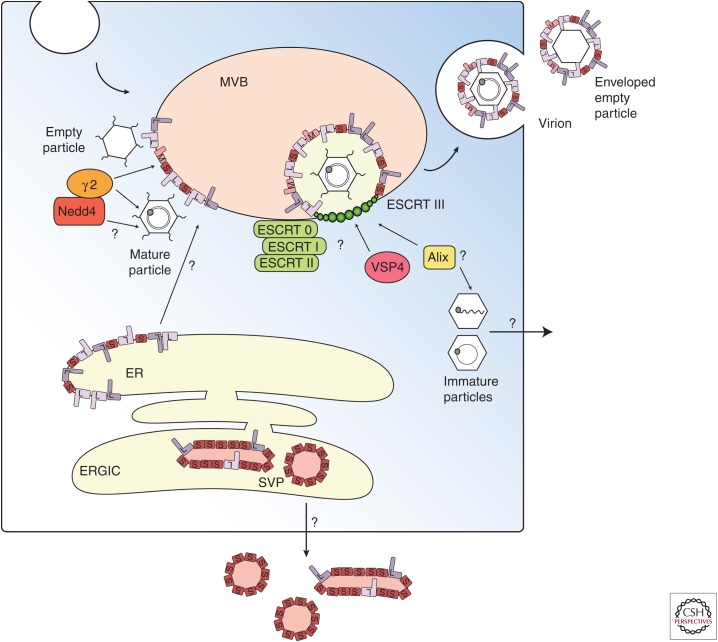
Growing evidence suggests that release of viral particles is mediated by multivesicular bodies (MVBs) of the late endosomal compartment. MVB biogenesis depends on the endosomal sorting complex required for transport (ESCRT) protein complexes (Katzmann et al. 2002; Hanson and Cashikar 2012). Many enveloped RNA viruses, such as retroviruses, filoviruses, rhabdoviruses, and paramyxoviruses, hijack host proteins that are part of the ESCRT machinery to facilitate budding at the plasma membrane (Katzmann et al. 2002; Jasenosky and Kawaoka 2004; Chen and Lamb 2008; Hurley and Hanson 2010; Martin-Serrano and Neil 2011; Hanson and Cashikar 2012). In contrast, it has been suggested that HBV buds into the MVB and exits the cell via an exosomal pathway (Fig. 2) (Lambert et al. 2007). Consistent with this hypothesis, HBV virions have been localized to membranes of the late endosome and large intracellular compartments (Roingeard and Sureau 1998; Falcon et al. 2008). HBV release was shown to be dependent on ESCRT proteins when dominant negative (DN) mutants of ESCRT III, Vsp4, and ALIX inhibited virus assembly and release (Lambert et al. 2007; Watanabe et al. 2007).
Figure 2.
Virions and subviral particles (SVPs) use two different pathways for secretion (adapted from Prange 2012). Capsids are released through budding into multivesicular bodies (MVBs) using endosomal sorting complex required for transport (ESCRT)-0, -I, -II, -III, and Vsp4 protein complexes (Katzmann et al. 2002; Hanson and Cashikar 2012). Envelope proteins facilitate budding of the viral capsids at the MVB membrane; however, it is unclear how envelope proteins are transported from the pre-Golgi/endoplasmic reticulum (ER) membrane to the late endosome. Nonenveloped particles are hypothesized to be released in an ESCRT-independent pathway that uses the apoptosis-linked gene 2 (ALG2)-interacting protein X (Alix), also implicated in the secretion of enveloped particles (Watanabe et al. 2007; Bardens et al. 2011). It is not clear how capsid particles distinguish between pathways of enveloped and nonenveloped release. ERGIC, ER-Golgi intermediate compartment.
It is not clear how HBV recruits the ESCRT machinery. The endosomal protein γ2-adaptin, an adaptor protein that can guide cargo proteins through membrane compartments has been implicated, as down-regulation of γ2-adaptin expression inhibited virion formation and release (Boehm and Bonifacino 2001; Hartmann-Stuhler and Prange 2001; Rost et al. 2006; Lambert et al. 2007). However, interaction with γ2-adaptin may be indirect (Fig. 2) (Garcia et al. 2009). It has been suggested that Nedd4 mediates interactions with γ2-adaptin capsids via the PPAY domain (Rost et al. 2008). However, PPAY is located within the dimer–dimer contact region of the core protein that is essential for capsid assembly (Wynne et al. 1999). Mutation of residue Y132 to alanine interferes with capsid formation (Bourne et al. 2009). Importantly, P130 is not conserved throughout all HBV genotypes (Chain and Myers 2005).
Inhibition of the described host factors (γ2-adaptin and proteins of the ESCRT complex) does not inhibit SVP secretion (Rost et al. 2006; Lambert et al. 2007), indicating that the pathway of virion release differs from the release of SVPs. SVPs are secreted via the general secretory pathway, independent of glycosylation (Patzer et al. 1986; Huovila et al. 1992; Lu et al. 1995; Le Seyec et al. 1999). S-HBsAg assembles first into filamentous particles within the ER that are then transported to the ER-Golgi intermediate compartment (ERGIC). Within the ERGIC lumen, filamentous particles are converted into spherical particles and secreted out of the cell (Patient et al. 2007, 2009).
Capsid and Virion Structure
The structures of T = 4 capsids, formed by the assembly domain, Cp149, have been determined by cryo-EM and X-ray crystallography (Bottcher et al. 1997; Conway et al. 1997; Wynne et al. 1999). Each monomer has five α helices: helices α3 and α4 form a 4-helix bundle with the adjacent monomer, the intradimer interface. Helices α1, α2, and α5 form a domain perpendicular to the 4-helix bundle that modulates dimer–dimer interactions (Fig. 3). This α-helical structure has some similarities to the retroviral Gag protein, but is unrelated to the β-barrel fold found in many unenveloped virus capsids (Zlotnick et al. 1998).
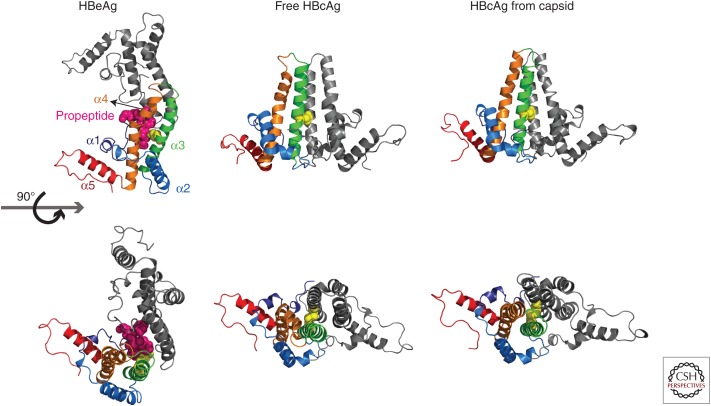
Figure 3.
Structures of hepatitis B core antigen (HBcAg) and hepatitis B e antigen (HBeAg) dimers. HBeAg harbors a 10-amino-acid-long amino-terminal extension (propeptide). The structure of the monomer structure shows modest conformational changes between free HBcAg, HBcAg in capsid, and HBeAg. The largest structural changes are found in helix α3 and α4, located at the intradimer interface (orange) (adapted from Zlotnick et al. 2013). Cysteine-7, located on the amino-terminal extension of HBeAg (3V6Z) (magenta spheres) can form a disulfide with cysteine 61 (yellow spheres). To accommodate this peptide, the monomers are rotated ∼140° about the intradimer interface from their orientation in HBcAg. In HBcAg, the monomers are parallel, and an intradimer Cys61-Cys61 disulfide can form. Structural differences in the spike region as well as the dimer–dimer interface can be observed between free HBcAg dimer (3KXS) and HBcAg from capsid (1QGT) (assembly-inactive and assembled states, respectively).
The intradimer interface is highly dynamic. Its flexibility is evident in the structure of the HBV e antigen (HBeAg). The HBeAg sequence is identical to that of Cp149 but contains a 10-amino-acid amino-terminal extension, the remains of a signal sequence (Fig. 3). HBeAg has an intramonomer disulfide with cysteine-7 of the amino-terminal extension and the highly conserved cysteine 61, located at the intradimer interface (Nassal and Rieger 1993; DiMattia et al. 2013; Zlotnick et al. 2013). This results in a complete reorganization of the dimer interface, rotating one monomer 140° relative to the other (Fig. 3).
During capsid assembly, dimer–dimer contacts are formed by hydrophobic interactions involving helix α5 and the carboxyl terminus with some contribution from the amino-terminus (Wynne et al. 1999; Ceres and Zlotnick 2002). Mutations of highly conserved residues F23 and Y132 to alanine, located at the points of dimer–dimer interactions, inhibit capsid formation (Chain and Myers 2005; Bourne et al. 2009; Packianathan et al. 2009; Alexander et al. 2013).
The CTD, localized to the interior of the capsid, is largely disordered in reconstructions of empty full-length core protein, Cp183 (Wang et al. 2012). However, the 16 arginines of the CTD and seven serines, some of which are able to be phosphorylated, play important roles in packaging nucleic acid and regulating cellular activity. Cp183 shows poor solubility compared with Cp149 (Porterfield et al. 2010). Preventing Cp183 aggregation in vivo may require a host factor.
The core is an active participant in reverse transcription as indicated by mutational studies that show mutants with wild-type-like packing of pgRNA but diminished DNA synthesis (Nassal 1992; Beames and Lanford 1993; Le Pogam et al. 2005; Lewellyn and Loeb 2011a,b; Tan et al. 2013). Also, the capsid surface is perforated by many holes that allow nucleotides to diffuse through during reverse transcription (Wynne et al. 1999).
Although the icosahedral core of HBV is very uniform, the envelopes of secreted virions are diverse in shape and morphology. Virions may have relatively tight-fitting envelopes resulting in an overall diameter of about 42 nm or have filamentous appendages (Fig. 4A) (Stannard and Hodgkiss 1979; Seitz et al. 2007). However, cryo-EM has shown that particles may have tight or gapped envelopes, emphasizing the heterogeneity of this layer (Seitz et al. 2007).
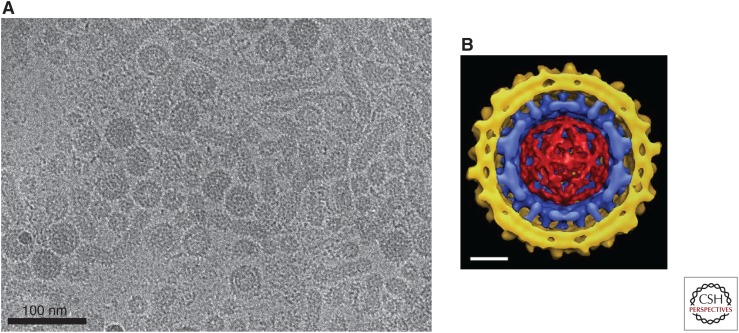
Figure 4.
The hepatitis B virus (HBV) virion. (A) Cryoelectron micrograph (cryo-EM) of virions isolated from chronically infected patients showing virions (Dane particles) containing spherical envelopes and virions with elongated appendages. Also shown are spherical and filamentous subviral particles (provided by Dr. Stefan Seitz). (B) Composite cryoelectron microscopy model of an HBV virion comprising the viral lipid envelope with hepatitis B surface antigen (HBsAg) protrusions (yellow), the icosahedral capsid (blue), and the enclosed double-stranded DNA (dsDNA) (red) (Dryden et al. 2006). The capsid displays icosahedral symmetry with protrusions representing the 4-helix bundles of the hepatitis B core antigen (HBcAg) dimers. Protrusions from the envelope are dimers of predominantly the small (S) form of the envelope protein, S-HBsAg. The viral envelope interacts with the spike tips of the capsid but is not arranged with icosahedral symmetry. Scale bar, 10 nm
In Dane particles, core and envelope proteins form contacts. However, a lack of register between the spikes of the capsid and the envelope proteins results in an ordered, but nonicosahedral envelope (Fig. 4B) (Dryden et al. 2006). This lack of coherence between envelope and capsid is unusual in viruses (Zlotnick and Mukhopadhyay 2011). When the reconstruction was based on the envelope, S dimers formed conspicuous spikes protruding from the envelope; these were spaced about 6 nm apart, reminiscent of those seen in helical reconstructions of SVP filaments (Fig. 4B) (Short et al. 2009). Also visible in the Dane particle reconstruction is density corresponding to dsDNA stretched between CTD clusters underneath fivefold and quasi-sixfold vertices. The organization of nucleic acid in these particles is similar to that of RNA found in unphosphorylated Cp183 capsids that assemble in vitro. (Fig. 4B) (Dryden et al. 2006; Wang et al. 2012). Indeed, the core protein isolated from dsDNA-containing HBV and DHBV cores are largely dephosphorylated (Perlman et al. 2005; Basagoudanavar et al. 2007), which suggests that unphosphorylated core protein packages nucleic acid with this sort of vertex-to-vertex electrostatic organization.
Biochemical studies show that residues on the capsid surface located near the spike tips on the base of the 4-helix bundle, as well as residues of the protruding α-helix 5 are important for envelopment (Bottcher et al. 1998; Ponsel and Bruss 2003). In cryo-EM studies, possibly because of the heterogeneity of core-preS1 interactions, the preS1 domain was not observed (Dryden et al. 2006; Seitz et al. 2007). A further consideration is that residues surrounding the spike tips might not directly be involved in binding envelope proteins but are important for mediating structural changes.
ASSEMBLY IN VITRO
Capsid Assembly Biochemistry
HBV capsid assembly in vitro has been extensively studied using only the assembly domain of the core protein, Cp149. Cp149 dimers expressed in E. coli assemble in response to increasing ionic strength into particles indistinguishable from capsids from cell culture (Kenney et al. 1995; Wingfield et al. 1995; Zlotnick et al. 1996).
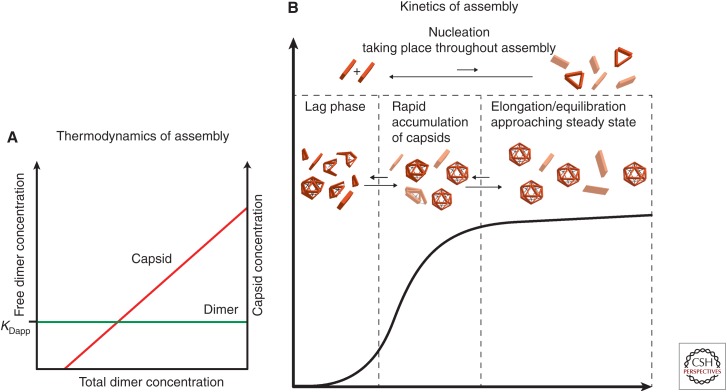
Assembly in vitro has been quantified by inducing assembly of dimer and measuring the yield of capsid and remaining dimer (Zlotnick et al. 1999). When dimer concentration exceeds a pseudocritical concentration, almost all additional free dimer assemble into capsid leaving a nearly constant concentration of free dimer (Fig. 5A) (Katen and Zlotnick 2009). For Cp149 at approximately physiological conditions (150 mM NaCl, 37°C), the pseudocritical concentration is about 5 µM (Ceres and Zlotnick 2002), which is in remarkable agreement with studies of spontaneous assembly of full-length core protein expressed in xenopus oocytes (Seifer et al. 1993).
Figure 5.
Predicted behavior for capsid assembly. (A) Capsid assembly at equilibrium. Very little capsid is observed until the total dimer concentration reaches a pseudocritical concentration of assembly, referred to as KDapp, the apparent dissociation constant. Above KDapp, almost all additional free dimer assemble into capsid (red), leaving a nearly constant concentration of free dimer (green); a slope of almost zero indicates that assembly is very close to equilibrium (Zlotnick 2007; Katen and Zlotnick 2009). (B) Assembly of large populations of particles displays sigmoidal kinetics (Endres and Zlotnick 2002; Hagan and Chandler 2006; Katen and Zlotnick 2009). The observed lag phase of assembly is proportional to the time to establish a steady state of intermediates. Following the lag phase, capsid particles accumulate (Hagan and Chandler 2006; Katen and Zlotnick 2009). As more free subunits are consumed to form capsids, the reaction slowly approaches equilibrium, plateauing when the pseudocritical concentration is reached (Katen and Zlotnick 2009). Nucleation and intermediate formation occurs throughout the entire assembly reaction.
Because assembly appears to equilibrate, it can be described with good approximation by the law of mass action (Zlotnick 1994; Katen and Zlotnick 2009). Assembly of HBV is characterized by multiple weak intersubunit contact energies (about 3–5 kcal/mol [5–8 kT]) equivalent to a millimolar dissociation constant (Ceres and Zlotnick 2002; Mohan et al. 2009). Weak protein–protein interactions support regulated assembly, avoid kinetic traps attributable to errors, and promote the formation of a barely stable dynamic capsid that is primed for uncoating (Katen et al. 2010). Evidence for dynamic changes of capsid was shown by proteolytic cleavage experiments, indicating that Cp149 capsids “breathe” (Hilmer et al. 2008).
The major driving force behind subunit association is entropy, indicated by the temperature dependence of assembly, which is consistent with burial of hydrophobic surface and concomitant release of water (Ceres and Zlotnick 2002). Also, increasing ionic strength increases the yield of capsids (Wingfield et al. 1995; Zlotnick et al. 1999). This has been suggested to arise from conformational changes that respond to ionic strength and divalent cations or ionic shielding of repulsive electrostatic forces (Ceres and Zlotnick 2002; Kegel and van der Schoot 2004; Stray et al. 2004; Choi et al. 2005).
The interdimer contact is a hydrophobic patch with numerous gaps (Wynne et al. 1999; Bourne et al. 2006, 2008). Filling gaps in the hydrophobic interface is predicted to strengthen association energy by increasing buried nonpolar surface (Eisenberg and McLachlan 1986). Indeed, heteroaryldihydropyrimidines (HAPs) and phenylpropenamides (PPAs), two chemical families found to have antiviral activity, act by increasing assembly by filling in the “HAP pocket” at the dimer–dimer interface (Stoltefuss et al. 1999; Delaney et al. 2002; Deres et al. 2003; Bourne et al. 2006, 2008; Stray and Zlotnick 2006; Katen et al. 2010, 2013; Li et al. 2013). Similarly, filling the same pocket with a V124W mutation leads to an assembly hyperactive mutant (Tan et al. 2013).
Although assembly rapidly equilibrates, dissociation in the absence of a catalyst or chaotrope does not. HBV, as well as other viruses, display a high kinetic barrier to dissociation called hysteresis. Dilution experiments and mass spectrometry revealed that capsids were stable even under conditions in which assembly was abrogated (Singh and Zlotnick 2003; Uetrecht et al. 2010). Hysteresis to disassembly has an important biological function. Virions persist even in conditions unfavorable to assembly despite their fragility (Uetrecht et al. 2008).
Kinetics of Assembly
In vivo, virus assembly is dynamic and may not have the time to equilibrate; thus, kinetics may be more relevant to activity in a cell. Cp149 assembly kinetics has been a powerful system for describing the general question of virus assembly as well as the specifics of HBV. Assembly of a single capsid starts with a nucleus followed by “elongation” (i.e., completion of the capsid). Assembly of large populations of particles displays sigmoidal kinetics, which appears similar to nucleated assembly of crystals or filaments (Fig. 5B) (Zlotnick 2005; Hagan and Chandler 2006; Katen and Zlotnick 2009). However, there are striking differences. During polymer assembly, the lag phase corresponds to the formation of a nucleus. The capsid assembly lag phase, however, is proportional to the time required to build intermediates and assemble the first capsid (Hagan and Chandler 2006; Katen and Zlotnick 2009). It has been described as a shock-front of intermediates (Morozov et al. 2009). Intermediates are predicted from graph theory and dynamic simulations to be a compact species (Endres et al. 2005; Moisant et al. 2010; Rapaport 2010; Dykeman et al. 2014). In capsid assembly, the plateau phase has concurrent nucleation and elongation. The reaction slowly approaches equilibrium until it reaches the pseudocritical concentration (Fig. 5B) (Katen and Zlotnick 2009).
High protein concentrations and/or strong subunit association energies between subunits can lead to kinetic trapping of incomplete or incorrect capsids because of depletion of subunits or locking mistakes into growing capsid (Zlotnick et al. 1999). These kinetic traps are local energy minima that persist—they are analogous to misfolded protein. Supporting theoretical predictions, trapped Cp149 assembly intermediates, T = 4 capsids missing 3–16 dimer subunits, have been reported (Pierson et al. 2014).
Allostery
Regulating assembly may be important for preventing misincorporation of the wrong nucleic acid, and minimizing kinetic traps. One mechanism to regulate assembly is allostery. It has been proposed that the capsid proteins undergo a structural transition from an assembly-inactive to an assembly-active conformation, energetically and structurally separating nucleation and elongation (Caspar 1980; Zlotnick 2005). Consistent with this, structural studies reveal differences between free HBV dimers and dimers within capsids (Fig. 3) (Packianathan et al. 2009; DiMattia et al. 2013). This hypothesis is further supported by core protein mutants that alter the assembly behavior. The F97 to leucine mutant, located at the intradimer interface, assembles faster and with stronger association than wild-type protein. Similarly, mutation of L42 to alanine inhibits capsid assembly, although L42 is located at the base of the spike tips far from dimer–dimer contacts (Alexander et al. 2013). Other point mutations that alter assembly unexpectedly alter dimers’ Stokes radius (Bourne et al. 2009; Tan et al. 2013). Another indicator of allostery is the requirement that the disulfide at the intradimer contact be in the reduced state for efficient capsid assembly, suggesting that this region is essential for mediating allosteric changes (Selzer et al. 2014).
The stepwise nature of core protein activity in the HBV lifecycle—packaging nucleic acid, participating in reverse transcription, regulating transport for egress or to the nucleus—all argue that core allostery is critical in vivo.
Assembly around Nucleic Acid
Because of the complexity of virus assembly in vivo, the study of capsid assembly around nucleic acid in vitro has been a valuable tool. Cp183 packages ssRNA with high affinity and high cooperativity, which might be highly important in facilitating specific packaging of viral RNA in vivo (Porterfield et al. 2010). However, assembly lacks specificity, indicating a regulatory step that has yet to be explained (Chen et al. 2011).
CTD phosphorylation appears to be an important regulator of RNA packaging in vivo (Kann and Gerlich 1994; Gazina et al. 2000). Cryo-EM studies of capsids assembled from Cp183 and a phosphorylation mimic, Cp183-EEE, in which three serines are replaced by glutamic acid, show that phosphorylation influenced the organization of the CTD and the packaged pgRNA (Wang et al. 2012). Cp183-EEE CTDs have a more compact organization consistent with inter-CTD salt bridges. In Cp183-EEE capsids, the RNA forms a mesh-like density more consistent with ssRNA. Conversely, the structure of the pgRNA in unphosphorylated capsids closely resembled dsDNA in authentic Dane particles (Dryden et al. 2006; Wang et al. 2012). This structural difference suggests dynamic interactions between CTDs and RNA, which react to changes in phosphorylation (Yu and Summers 1994a,b; Perlman et al. 2005).
Bound nucleic acid is synergistic with viral capsids. An excess of negative charges to positive charges is commonly observed in nucleoprotein complexes with typical values of about 1.6 but going as high as 2.0 (Manning 2001; Belyi and Muthukumar 2006; Hu and Shklovskii 2007; Hagan 2013; Perlmutter et al. 2013). In HBV, the number of positively charged arginines in the CTD correlates with the amount of RNA encapsidated (Zlotnick et al. 1996; Le Pogam et al. 2005; Chua et al. 2009), consistent with a charge neutralization component to encapsidation. However, the change in charge is accounted for by changing the CTD phosphorylation state during the course of reverse transcription. The charge ratio for an immature capsid, assuming a 3500-nt polyadenylated pgRNA and a fully phosphorylated CTD (Perlman et al. 2005), is about 1.5. The same ratio is found for a mature virus with 6300 nt of rcDNA (partially dsDNA) and completely dephosphorylated CTDs.
Cp183 assembles on ssDNA as well as it does on ssRNA, but fails to assemble around dsDNA, suggesting that constraining the less-flexible dsDNA requires stronger dimer–dimer association energies (Dhason et al. 2012; Zlotnick et al. 2012). In vivo, both the increase in internal pressure and also density of negative charge caused by formation of dsDNA might also lead to destabilization. The stiffness of dsDNA in an HBV capsid of only 25 nm internal diameter will substantially affect internal pressure and organization of the nucleic acid (Tzlil et al. 2003; Hagan 2013). Indeed, HBV core particles containing dsDNA appear less stable based on analyses of dsDNA-filled particle structure and measured stability by electrophoresis, velocity sedimentation, and susceptibility to proteolytic cleavage (Zlotnick et al. 2012; Cui et al. 2013).
CONCLUDING REMARKS
Assembly and egress are closely connected activities that are regulated in part by HBV core protein in close concert with other viral and host proteins. Conformational changes of the core protein can modulate assembly, the structure of packaged nucleic acid, and the activity of the packaged polymerase. Interactions of the capsid with the HBV envelope proteins regulate envelopment and envelope organization on the virion. Because of the relative simplicity of this system, both the physics of the virion and its biological regulation are experimentally accessible. Combining in vivo and in vitro studies has shed light on many mechanisms of the HBV life cycle. However, involvement of host proteins during RNA packaging and intracellular trafficking, core protein interactions with the polymerase and RNA during reverse transcription, and the mechanisms of virion envelopment and release are just beginning to be understood.
ACKNOWLEDGMENT
This work is supported by NIH grant R01-AI077688 to A.Z.
Footnotes
Editors: Christoph Seeger and Stephen Locarnini
Additional Perspectives on Hepatitis B and Delta Viruses available at www.perspectivesinmedicine.org
REFERENCES
- Alberti A, Diana S, Scullard GH, Eddleston WF, Williams R. 1978. Full and empty Dane particles in chronic hepatitis B virus infection: Relation to hepatitis B e antigen and presence of liver damage. Gastroenterology 75: 869–874. [PubMed] [Google Scholar]
- Alexander CG, Jurgens MC, Shepherd DA, Freund SM, Ashcroft AE, Ferguson N. 2013. Thermodynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein. Proc Natl Acad Sci 110: E2782–E2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardens A, Doring T, Stieler J, Prange R. 2011. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell Microbiol 13: 602–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Schaller H. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J 11: 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basagoudanavar SH, Perlman DH, Hu J. 2007. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J Virol 81: 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert TF, Rosler C, Malim MH, von Weizsacker F. 2007. Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology 46: 682–689. [DOI] [PubMed] [Google Scholar]
- Beames B, Lanford RE. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology 194: 597–607. [DOI] [PubMed] [Google Scholar]
- Beck J, Nassal M. 2007. Hepatitis B virus replication. World J Gastroenterol 13: 48–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Muthukumar M. 2006. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci 103: 17174–17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum F, Nassal M. 1990. Hepatitis B virus nucleocapsid assembly: Primary structure requirements in the core protein. J Virol 64: 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS. 2001. Adaptins: The final recount. Mol Biol Cell 12: 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher B, Wynne SA, Crowther RA. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386: 88–91. [DOI] [PubMed] [Google Scholar]
- Bottcher B, Tsuji N, Takahashi H, Dyson MR, Zhao S, Crowther RA, Murray K. 1998. Peptides that block hepatitis B virus assembly: Analysis by cryomicroscopy, mutagenesis and transfection. EMBO J 17: 6839–6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne C, Finn MG, Zlotnick A. 2006. Global structural changes in hepatitis B capsids induced by the assembly effector HAP1. J Virol 80: 11055–11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. 2008. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J Virol 82: 10262–10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne CR, Katen SP, Fulz MR, Packianathan C, Zlotnick A. 2009. A mutant hepatitis B virus core protein mimics inhibitors of icosahedral capsid self-assembly. Biochemistry 48: 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol 71: 9350–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss V, Ganem D. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci 88: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL. 1980. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J 32: 103–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceres P, Zlotnick A. 2002. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry 41: 11525–11531. [DOI] [PubMed] [Google Scholar]
- Chain BM, Myers R. 2005. Variability and conservation in hepatitis B virus core protein. BMC Microbiol 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology 372: 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang JC, Zlotnick A. 2011. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog 7: e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gyoo Park S, Yoo JH, Jung G. 2005. Calcium ions affect the hepatitis B virus core assembly. Virology 332: 454–463. [DOI] [PubMed] [Google Scholar]
- Chua PK, Tang FM, Huang JY, Suen CS, Shih C. 2009. Testing the balanced electrostatic interaction hypothesis of hepatitis B virus DNA synthesis by using an in vivo charge re-balance approach. J Virol 84: 2340–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. 1997. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386: 91–94. [DOI] [PubMed] [Google Scholar]
- Cui X, Ludgate L, Ning X, Hu J. 2013. Maturation-associated destabilization of hepatitis B virus nucleocapsid. J Virol 87: 11494–11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J, Ullrich A, Cotten M. 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol 76: 8124–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney WE, Edwards R, Colledge D, Shaw T, Furman P, Painter G, Locarnini S. 2002. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrob Agents Chemother 46: 3057–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, et al. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299: 893–896. [DOI] [PubMed] [Google Scholar]
- Dhason MS, Wang JC, Hagan MF, Zlotnick A. 2012. Differential assembly of hepatitis B virus core protein on single- and double-stranded nucleic acid suggest the dsDNA-filled core is spring-loaded. Virology 430: 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Stahl SJ, Grimes JM, Steven AC, Stuart DI, Wingfield PT. 2013. Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein. Structure 21: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diminsky D, Schirmbeck R, Reimann J, Barenholz Y. 1997. Comparison between hepatitis B surface antigen (HBsAg) particles derived from mammalian cells (CHO) and yeast cells (Hansenula polymorpha): Composition, structure and immunogenicity. Vaccine 15: 637–647. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Wieland SF, Whitten-Bauer C, Gerin JL, Chisari FV, Yeager M. 2006. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell 22: 843–850. [DOI] [PubMed] [Google Scholar]
- Duclos-Vallee JC, Capel F, Mabit H, Petit MA. 1998. Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J Gen Virol 79: 1665–1670. [DOI] [PubMed] [Google Scholar]
- Dykeman EC, Stockley PG, Twarock R. 2014. Solving a Levinthal's paradox for virus assembly identifies a unique antiviral strategy. Proc Natl Acad Sci 111: 5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eble BE, MacRae DR, Lingappa VR, Ganem D. 1987. Multiple topogenic sequences determine the transmembrane orientation of the hepatitis B surface antigen. Mol Cell Biol 7: 3591–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eble BE, Lingappa VR, Ganem D. 1990. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J Virol 64: 1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt SG, Milich DR, McLachlan A. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J Virol 65: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, McLachlan AD. 1986. Solvation energy in protein folding and binding. Nature 319: 199–203. [DOI] [PubMed] [Google Scholar]
- Endres D, Zlotnick A. 2002. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys J 83: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres D, Miyahara M, Moisant P, Zlotnick A. 2005. A reaction landscape identifies the intermediates critical for self-assembly of virus capsids and other polyhedral structures. Prot Sci 14: 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon V, Menéndez I, Acosta-Rivero N, Shibayama M. 2008. Ultrastructural evidences of hepatitis B infection in human liver biopsies disclose complex assembly and morphogenesis pathways for hepatitis B virus. Am J Infect Dis 4: 96–103. [Google Scholar]
- Fallows DA, Goff SP. 1995. Mutations in the ɛ sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J Virol 69: 3067–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernholz D, Galle PR, Stemler M, Brunetto M, Bonino F, Will H. 1993. Infectious hepatitis B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology 194: 137–148. [DOI] [PubMed] [Google Scholar]
- Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S. 2002. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res 30: 4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D, Prince AM. 2004. Hepatitis B virus infection—Natural history and clinical consequences. N Engl J Med 350: 1118–1129. [DOI] [PubMed] [Google Scholar]
- Garcia ML, Byfield R, Robek MD. 2009. Hepatitis B virus replication and release are independent of core lysine ubiquitination. J Virol 83: 4923–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilanes F, Gonzalez-Ros JM, Peterson DL. 1982. Structure of hepatitis B surface antigen. Characterization of the lipid components and their association with the viral proteins. J Biol Chem 257: 7770–7777. [PubMed] [Google Scholar]
- Gazina EV, Fielding JE, Lin B, Anderson DA. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J Virol 74: 4721–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerelsaikhan T, Tavis JE, Bruss V. 1996. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol 70: 4269–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin JL, Ford EC, Purcell RH. 1975. Biochemical characterization of Australia antigen. Evidence for defective particles of hepatitis B virus. Am J Pathol 81: 651–668. [PMC free article] [PubMed] [Google Scholar]
- Gerlich WH, Kann M. 2010. Hepatitis B. In Topley & Wilson's microbiology and microbial infections. Wiley, Hoboken, NJ. [Google Scholar]
- Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, Stuart DI, Rowlands DJ. 2005. Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci 102: 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213: 292–299. [DOI] [PubMed] [Google Scholar]
- Hagan MF. 2013. Modeling viral capsid assembly. Adv Chem Phys 155: 1–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan MF, Chandler D. 2006. Dynamic pathways for viral capsid assembly. Biophys J 91: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A. 2012. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28: 337–362. [DOI] [PubMed] [Google Scholar]
- Hartmann-Stuhler C, Prange R. 2001. Hepatitis B virus large envelope protein interacts with γ2-adaptin, a clathrin adaptor-related protein. J Virol 75: 5343–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. 1984. Large surface proteins of hepatitis B virus containing the pre-S sequence. J Virol 52: 396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilmer JK, Zlotnick A, Bothner B. 2008. Conformational equilibria and rates of localized motion within hepatitis B virus capsids. J Mol Biol 375: 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Boyer M. 2006. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J Virol 80: 2141–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Seeger C. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci 93: 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Shklovskii BI. 2007. Kinetics of viral self-assembly: Role of the single-stranded RNA antenna. Phys Rev E Stat Nonlin Soft Matter Phys 75: 051901. [DOI] [PubMed] [Google Scholar]
- Hu J, Flores D, Toft D, Wang X, Nguyen D. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol 78: 13122–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovila AP, Eder AM, Fuller SD. 1992. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J Cell Biol 118: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. 2010. Membrane budding and scission by the ESCRT machinery: It's all in the neck. Nat Rev Mol Cell Biol 11: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasenosky LD, Kawaoka Y. 2004. Filovirus budding. Virus Res 106: 181–188. [DOI] [PubMed] [Google Scholar]
- Jeong JK, Yoon GS, Ryu WS. 2000. Evidence that the 5′-end cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J Virol 74: 5502–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Cho MH, Park SG, Jung G. 2014. Interaction between nucleophosmin and HBV core protein increases HBV capsid assembly. FEBS Lett 588: 851–858. [DOI] [PubMed] [Google Scholar]
- Junker-Niepmann M, Bartenschlager R, Schaller H. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J 9: 3389–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M, Gerlich WH. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol 68: 7993–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol 145: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan PM, Ford EC, Purcell RH, Gerin JL. 1976. Demonstration of subpopulations of Dane particles. J Virol 17: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katen SP, Zlotnick A. 2009. Thermodynamics of virus capsid assembly. Methods Enzymol 455: 395–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2010. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 5: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2013. Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure. Structure 21: 1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. 2002. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3: 893–905. [DOI] [PubMed] [Google Scholar]
- Kau JH, Ting LP. 1998. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J Virol 72: 3796–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel WK, van der Schoot P. 2004. Competing hydrophobic and screened-coulomb interactions in hepatitis B virus capsid assembly. Biophys J 86: 3905–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney JM, von Bonsdorff CH, Nassal M, Fuller SD. 1995. Evolutionary conservation in the hepatitis B virus core structure: Comparison of human and duck cores. Structure 3: 1009–1019. [DOI] [PubMed] [Google Scholar]
- Kock J, Nassal M, Deres K, Blum HE, von Weizsacker F. 2004. Hepatitis B virus nucleocapsids formed by carboxy-terminally mutated core proteins contain spliced viral genomes but lack full-size DNA. J Virol 78: 13812–13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschel M, Oed D, Gerelsaikhan T, Thomssen R, Bruss V. 2000. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J Virol 74: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Doring T, Prange R. 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and γ2-adaptin. J Virol 81: 9050–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan YT, Li J, Liao W, Ou J. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259: 342–348. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Shim HY, Hsieh A, Min JY, Jung G. 2009. Hepatitis B virus core interacts with the host cell nucleolar protein, nucleophosmin 1. J Microbiol 47: 746–752. [DOI] [PubMed] [Google Scholar]
- Lepere-Douard C, Trotard M, Le Seyec J, Gripon P. 2009. The first transmembrane domain of the hepatitis B virus large envelope protein is crucial for infectivity. J Virol 83: 11819–11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pogam S, Yuan TT, Sahu GK, Chatterjee S, Shih C. 2000. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J Virol 74: 9099–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pogam S, Chua PK, Newman M, Shih C. 2005. Exposure of RNA templates and encapsidation of spliced viral RNA are influenced by the arginine-rich domain of human hepatitis B virus core antigen (HBcAg 165–173). J Virol 79: 1871–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J Virol 72: 5573–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol 73: 2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn EB, Loeb DD. 2011a. The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication. J Virol 85: 1298–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn EB, Loeb DD. 2011b. Serine phosphoacceptor sites within the core protein of hepatitis B virus contribute to genome replication pleiotropically. PloS ONE 6: e17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chirapu SR, Finn MG, Zlotnick A. 2013. Phase diagrams map the properties of antiviral agents directed against hepatitis B virus core assembly. Antimicrob Agents Chemother 57: 1505–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Ou JH. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: Significance of serine in the three repeated SPRRR motifs. J Virol 69: 1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Yansura D, Levinson AD. 1982. Direct expression of hepatitis B surface antigen in monkey cells from an SV40 vector. DNA 1: 213–221. [DOI] [PubMed] [Google Scholar]
- Loffler-Mary H, Dumortier J, Klentsch-Zimmer C, Prange R. 2000. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270: 358–367. [DOI] [PubMed] [Google Scholar]
- Lu X, Mehta A, Dwek R, Butters T, Block T. 1995. Evidence that N-linked glycosylation is necessary for hepatitis B virus secretion. Virology 213: 660–665. [DOI] [PubMed] [Google Scholar]
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, et al. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343: 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludgate L, Ning X, Nguyen DH, Adams C, Mentzer L, Hu J. 2012. Cyclin-dependent kinase 2 phosphorylates s/t-p sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J Virol 86: 12237–12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold CM, Streeck RE. 1993. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol 67: 4588–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold CM, Unckell F, Werr M, Streeck RE. 1995. Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology 211: 535–543. [DOI] [PubMed] [Google Scholar]
- Manning GS. 2001. Counterion condensation on a helical charge lattice. Macromolecules 34: 4650–4655. [Google Scholar]
- Martin-Serrano J, Neil SJ. 2011. Host factors involved in retroviral budding and release. Nat Rev Microbiol 9: 519–531. [DOI] [PubMed] [Google Scholar]
- Mohan S, Kourentzi K, Schick KA, Uehara C, Lipschultz CA, Acchione M, Desantis ME, Smith-Gill SJ, Willson RC. 2009. Association energetics of cross-reactive and specific antibodies. Biochemistry 48: 1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisant P, Neeman H, Zlotnick A. 2010. Exploring the paths of (virus) assembly. Biophys J 99: 1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov AY, Bruinsma RF, Rudnick J. 2009. Assembly of viruses and the pseudo-law of mass action. J Chem Phys 131: 155101. [DOI] [PubMed] [Google Scholar]
- Nassal M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 66: 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M, Rieger A. 1993. An intramolecular disulfide bridge between Cys-7 and Cys-61 determines the structure of the secretory core gene product (e antigen) of hepatitis B virus. J Virol 67: 4307–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Hu J. 2008. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol 82: 6852–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Gummuluru S, Hu J. 2007. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol 81: 4465–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, Hafenstein S, Hu J. 2011. Secretion of genome-free hepatitis B virus—Single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog 7: e1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M. 2008. The structure and functions of NPM1/nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem 143: 441–448. [DOI] [PubMed] [Google Scholar]
- Packianathan C, Katen SP, Zlotnick A. 2009. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol 84: 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient R, Hourioux C, Sizaret PY, Trassard S, Sureau C, Roingeard P. 2007. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J Virol 81: 3842–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient R, Hourioux C, Roingeard P. 2009. Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell Microbiol 11: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer EJ, Nakamura GR, Simonsen CC, Levinson AD, Brands R. 1986. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J Virol 58: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D, Hu J. 2003. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J Virol 77: 2287–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman DH, Berg EA, O’Connor PB, Costello CE, Hu J. 2005. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc Natl Acad Sci 102: 9020–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter JD, Qiao C, Hagan MF. 2013. Viral genome structures are optimal for capsid assembly. eLife 2: e00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DL, Nath N, Gavilanes F. 1982. Structure of hepatitis B surface antigen. Correlation of subtype with amino acid sequence and location of the carbohydrate moiety. J Biol Chem 257: 10414–10420. [PubMed] [Google Scholar]
- Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JC, Zlotnick A, Jarrold MF. 2014. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J Am Chem Soc 136: 3636–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. 1997. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228: 115–120. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Ganem D. 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol 67: 3254–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Ganem D. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol 68: 5579–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsel D, Bruss V. 2003. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol 77: 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porterfield JZ, Dhason MS, Loeb DD, Nassal M, Stray SJ, Zlotnick A. 2010. Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J Virol 84: 7174–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prange R. 2012. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med Microbiol Immunol 201: 449–461. [DOI] [PubMed] [Google Scholar]
- Rabe B, Vlachou A, Pante N, Helenius A, Kann M. 2003. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci 100: 9849–9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe B, Delaleau M, Bischof A, Foss M, Sominskaya I, Pumpens P, Cazenave C, Castroviejo M, Kann M. 2009. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids. PLoS Pathog 5: e1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DC. 2010. Studies of reversible capsid shell growth. J Phys Condens Matter 22: 104115. [DOI] [PubMed] [Google Scholar]
- Roingeard P, Sureau C. 1998. Ultrastructural analysis of hepatitis B virus in HepG2-transfected cells with special emphasis on subviral filament morphogenesis. Hepatology 28: 1128–1133. [DOI] [PubMed] [Google Scholar]
- Rost M, Mann S, Lambert C, Doring T, Thome N, Prange R. 2006. γ-Adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation. J Biol Chem 281: 29297–29308. [DOI] [PubMed] [Google Scholar]
- Rost M, Doring T, Prange R. 2008. γ2-Adaptin, a ubiquitin-interacting adaptor, is a substrate to coupled ubiquitination by the ubiquitin ligase Nedd4 and functions in the endosomal pathway. J Biol Chem 283: 32119–32130. [DOI] [PubMed] [Google Scholar]
- Schädler S, Hildt E. 2009. HBV life cycle: Entry and morphogenesis. Viruses 1: 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht HJ, Kuhn C, Guhr B, Mattaliano RJ, Schaller H. 1987. Biochemical and immunological characterization of the duck hepatitis B virus envelope proteins. J Virol 61: 2280–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Glebe D, Tolle TK, Lochnit G, Linder D, Geyer R, Gerlich WH. 2004. Structure of pre-S2 N- and O-linked glycans in surface proteins from different genotypes of hepatitis B virus. J Gen Virol 853: 2045–2053. [DOI] [PubMed] [Google Scholar]
- Seifer M, Zhou S, Standring DN. 1993. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol 67: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz S, Urban S, Antoni C, Bottcher B. 2007. Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions. EMBO J 26: 4160–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer L, Katen SP, Zlotnick A. 2014. The hepatitis B virus core protein intradimer interface modulates capsid assembly and stability. Biochemistry 53: 5496–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim HY, Quan X, Yi YS, Jung G. 2011. Heat shock protein 90 facilitates formation of the HBV capsid via interacting with the HBV core protein dimers. Virology 410: 161–169. [DOI] [PubMed] [Google Scholar]
- Short JM, Chen S, Roseman AM, Butler PJ, Crowther RA. 2009. Structure of hepatitis B surface antigen from subviral tubes determined by electron cryomicroscopy. J Mol Biol 390: 135–141. [DOI] [PubMed] [Google Scholar]
- Singh S, Zlotnick A. 2003. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J Biol Chem 278: 18249–18255. [DOI] [PubMed] [Google Scholar]
- Stannard LM, Hodgkiss M. 1979. Morphological irregularities in Dane particle cores. J Gen Virol 45: 509–514. [DOI] [PubMed] [Google Scholar]
- Staprans S, Loeb DD, Ganem D. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol 65: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibbe W, Gerlich WH. 1983. Structural relationships between minor and major proteins of hepatitis B surface antigen. J Virol 46: 626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltefuss J, Goldmann S, Krämer T, Schlemmer K-H, Niewöhner U, Paessens A, Lottmann S, Deres K, Weber O. 1999. New dihydropyrimidine derivatives and their corresponding mesomers useful as antiviral agents. Bayer, Leverkusen, Germany: WO patent 9,954,326. [Google Scholar]
- Stray SJ, Zlotnick A. 2006. BAY 41–4109 has multiple effects on hepatitis B virus capsid assembly. J Mol Recognit 19: 542–548. [DOI] [PubMed] [Google Scholar]
- Stray SJ, Ceres P, Zlotnick A. 2004. Zinc ions trigger conformational change and oligomerization of hepatitis B virus capsid protein. Biochemistry 43: 9989–9998. [DOI] [PubMed] [Google Scholar]
- Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B–like virus by reverse transcription of an RNA intermediate. Cell 29: 403–415. [DOI] [PubMed] [Google Scholar]
- Tan Z, Maguire ML, Loeb DD, Zlotnick A. 2013. Genetically altering the thermodynamics and kinetics of hepatitis B virus capsid assembly has profound effects on virus replication in cell culture. J Virol 87: 3208–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303: 1829. [DOI] [PubMed] [Google Scholar]
- Tuttleman JS, Pourcel C, Summers J. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47: 451–460. [DOI] [PubMed] [Google Scholar]
- Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. 2003. Forces and pressures in DNA packaging and release from viral capsids. Biophys J 84: 1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJ, Wingfield PT, Steven AC, Heck AJ. 2008. High-resolution mass spectrometry of viral assemblies: Molecular composition and stability of dimorphic hepatitis B virus capsids. Proc Natl Acad Sci 105: 9216–9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJ. 2010. Subunit exchange rates in hepatitis B virus capsids are geometry- and temperature-dependent. Phys Chem Chem Phys 12: 13368–13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kim S, Ryu WS. 2009. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J Virol 83: 5815–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Dhason MS, Zlotnick A. 2012. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. PLoS Pathog 8: e1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Nickens DG, Lentz TB, Loeb DD, Zlotnick A. 2014. Encapsidated hepatitis B virus reverse transcriptase is poised on an ordered RNA lattice. Proc Natl Acad Sci 111: 11329–11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci 104: 10205–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werr M, Prange R. 1998. Role for calnexin and N-linked glycosylation in the assembly and secretion of hepatitis B virus middle envelope protein particles. J Virol 72: 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Williams RW, Steven AC. 1995. Hepatitis core antigen produced in Escherichia coli: Subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34: 4919–4932. [DOI] [PubMed] [Google Scholar]
- Wittkop L, Schwarz A, Cassany A, Grun-Bernhard S, Delaleau M, Rabe B, Cazenave C, Gerlich W, Glebe D, Kann M. 2010. Inhibition of protein kinase C phosphorylation of hepatitis B virus capsids inhibits virion formation and causes intracellular capsid accumulation. Cell Microbiol 12: 962–975. [DOI] [PubMed] [Google Scholar]
- Wounderlich G, Bruss V. 1996. Characterization of early hepatitis B virus surface protein oligomers. Arch Virol 141: 1191–1205. [DOI] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG. 1999. The crystal structure of the human hepatitis B virus capsid. Mol Cell 3: 771–780. [DOI] [PubMed] [Google Scholar]
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, et al. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1: e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Summers J. 1994a. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol 68: 4341–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Summers J. 1994b. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J Virol 68: 2965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jin L, Jih J, Shih C, Zhou ZH. 2013. 3.5A cryo-EM structure of hepatitis B virus core assembled from full-length core protein. PloS ONE 8: e69729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TT, Sahu GK, Whitehead WE, Greenberg R, Shih C. 1999. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J Virol 73: 5731–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Fu XD, Ou JH. 2005. Suppression of hepatitis B virus replication by SRPK1 and SRPK2 via a pathway independent of the phosphorylation of the viral core protein. Virology 342: 150–158. [DOI] [PubMed] [Google Scholar]
- Zlotnick A. 1994. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J Mol Biol 241: 59–67. [DOI] [PubMed] [Google Scholar]
- Zlotnick A. 2005. Theoretical aspects of virus capsid assembly. J Mol Recognit 18: 479–490. [DOI] [PubMed] [Google Scholar]
- Zlotnick A. 2007. Distinguishing reversible from irreversible virus capsid assembly. J Mol Biol 366: 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Mukhopadhyay S. 2011. Virus assembly, allostery and antivirals. Trends Microbiol 19: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35: 7412–7421. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Stahl SJ, Wingfield PT, Conway JF, Cheng N, Steven AC. 1998. Shared motifs of the capsid proteins of hepadnaviruses and retroviruses suggest a common evolutionary origin. FEBS Lett 431: 301–304. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. 1999. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry 38: 14644–14652. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Suhanovsky MM, Teschke CM. 2012. The energetic contributions of scaffolding and coat proteins to the assembly of bacteriophage procapsids. Virology 428: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Tan Z, Selzer L. 2013. One protein, at least three structures, and many functions. Structure 21: 6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]