Abstract
Since newborn screening (NBS) began in the 1960s, technological advances have enabled its expansion to include an increasing number of disorders. Recent developments now make it possible to sequence an infant’s genome relatively quickly and economically. Clinical application of whole-exome and whole-genome sequencing is expanding at a rapid pace but presents many challenges. Its utility in NBS has yet to be demonstrated and its application in the pediatric population requires examination, not only for potential clinical benefits, but also for the unique ethical challenges it presents.
An infant’s genome can be quickly and economically sequenced with current technologies. However, the clinical value of this approach is limited by practical and ethical issues (e.g., gene selection and parental involvement).
Newborn screening (NBS) in the United States began in the 1960s, shortly after the publication of Robert Guthrie’s paper describing a method for detecting phenylketonuria (PKU) in dried blood spots through a bacterial inhibition assay (Guthrie and Susi 1963). It has been defined as a public health activity aimed at the early identification of infants who are affected by certain genetic, metabolic, or infectious conditions (AAP Committee on Bioethics 2001) for which treatment can prevent unfavorable health outcomes. Millions of lives have been saved and significant morbidities prevented through universal NBS in the United States and other countries. Although a public health success, expansion of NBS has often been driven by technological advances as well as by pressure from the public and special interest groups. Rapid increases in the number of known genetic and metabolic conditions and improved methodologies have led to expansion of candidate disorders for screening. Technological advances have been closely intertwined with the ability to screen for conditions, for example, the adaptation of tandem mass spectrometry (MS/MS) for detection and quantification of multiple analytes in newborns’ blood spot samples (Millington et al. 1989).
Massively parallel sequencing (MPS, also referred to as “next-generation sequencing” or NGS) uses high-throughput sequencers that are able to analyze DNA much more efficiently than previous methods such as Sanger sequencing. This technology provides a new methodology for screening and overcomes one of the major barriers to adding conditions to recommended newborn screening panels. Any number of conditions for which there is a known genetic basis can theoretically be screened for in an individual, potentially including the entire genome. However, screening a newborn with genome-scale sequencing raises significant complexities, including the extent to which parents should be able to learn about genetic predispositions in a newborn, particularly conditions that may not manifest clinically until adulthood, and in many cases that may not have effective preventive strategies. Children lack the autonomy of adults, who can decide for themselves whether to engage in genomic sequencing to determine whether they have a gene mutation that predisposes them to a condition such as cancer. In NBS, these decisions are made by and can directly impact the parents. This represents an additional complication that requires special consideration.
To ultimately determine the clinical utility of genome-scale sequencing in NBS and to evaluate whether such an approach offers added value, it will be imperative to assess the sensitivity and specificity of MPS for currently screened conditions and whether sequencing can provide diagnostic data as accurately as currently used screening methods such as MS/MS. Will these data significantly augment our ability to predict disease prognosis and enable more targeted management? What conditions would then meet the criteria to be added to routine NBS use? To answer these questions, it will be necessary to study the yield of sequencing for common conditions detected by current NBS (e.g., PKU, medium-chain acyl-CoA dehydrogenase deficiency [MCADD], cystic fibrosis [CF] and hearing loss), as well as conditions that meet criteria for NBS but were not possible to detect owing to lack of an adequate screening method (e.g., certain lysosomal storage disorders and primary ciliary dyskinesia). The application of sequencing in NBS will allow not only the delineation of the causative mutation in the proximally causative gene, thus augmenting studies of phenotype-genotype relationships, but it could also create a valuable long-term resource for researchers to investigate how currently unknown loci contribute to clinical heterogeneity.
APPLICATION OF DNA SEQUENCING IN NEWBORN SCREENING
Selection of Genes to Include in Sequencing-Based Newborn Screening
As articulated by Wilson and Jungner: “The central idea of early disease detection and treatment is essentially simple. However, the path to its successful achievement (on the one hand, bringing to treatment those with previously undetected disease, and, on the other, avoiding harm to those persons not in need of treatment) is far from simple though sometimes it may appear deceptively easy” (Wilson and Jungner 1968; Andermann et al. 2008). Introducing genetic testing into screening programs in the past was a relatively slow multistep process with pilot screening programs undertaken after a disease gene or method of identification for a disorder was discovered, and experts agreed was reasonable and efficacious to add to a panel (ACMG Newborn Screening Expert Group 2006; Andermann et al. 2008). Even with multiplex screening methodologies such as MS/MS that can identify analytes associated with multiple inborn errors of metabolism, pilot programs were initiated by states before widespread validation (Frazier et al. 2006).
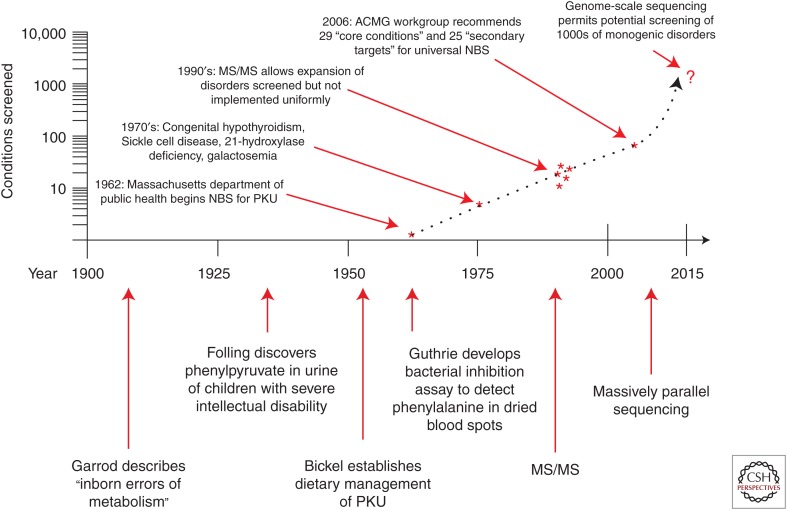
Currently, the Discretionary Advisory Committee on Heritable Disorders in Newborns and Children evaluates conditions nominated for inclusion in NBS programs through a comprehensive systematic evidenced-based review process (see http://www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/index.html; http://www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/nominatecondition/reviews/pompereport2013.pdf) (Kemper et al. 2014). This review process considers not only the magnitude and certainty of net benefit, but also the capability of states to implement comprehensive screening. Twenty-three conditions were considered but not included in the original recommended uniform screening panel (RUSP) because they lack an accurate screening method (ACMG Newborn Screening Expert Group 2006); 21 of these are detectable in some or all cases with molecular genetic analysis and, therefore, have the potential to be added to NBS panels. However, the rate at which new disease genes are being identified outpaces the ability of professionals and policy makers to assess the potential benefits and pitfalls of introducing or expanding genetic screening programs (Andermann et al. 2008), and formal review of the estimated 3260 genes with a human phenotype-causing mutation (see http://omim.org/statistics/geneMap) would be a daunting task. Ultimately, it will be necessary to construct a list of genes associated with conditions that are part of the current RUSP as well as those that are deemed to fulfill criteria for NBS and are detectable by sequencing. This panel would include conditions with onset in childhood in which early identification could allow prevention or amelioration of symptoms. The process will also need to include a mechanism for updating the list to reflect advances in medical genetics. Figure 1 depicts a timeline of advances in technology and newborn screening.
Figure 1.
Milestones in newborn screening are depicted in the lower portion of the figure, with approximate dates on the horizontal axis. The number of conditions screened for (or potentially screened for) is depicted chronologically with asterisks on the vertical axis, plotted on a logarithmic scale. Screening programs have historically differed between states, most dramatically observed after development of tandem mass-spectrometry technology in the 1990s. Adoption of a recommended uniform screening panel in 2005–2006 has gradually led to greater consistency. Development of next-generation sequencing technology in the early 2000s, with subsequent reduction in the cost of genome-scale sequencing, makes it possible to analyze thousands of disease genes. The number of conditions potentially screened for is indicated with a question mark to emphasize the substantial concerns regarding the application of such technology in healthy newborns.
Incorporating Sequencing into Routine NBS Practice
Several challenges must be met to effectively incorporate sequencing into NBS. First, the technical capabilities of MPS need to be evaluated in comparison to standard NBS methods. Next, thoughtful choices must be made about whether to include conditions that are not amenable to current screening methods, yet, would otherwise meet criteria for screening in a public health setting versus others that would be reasonable candidates if accompanied by more rigorous informed parental consent. Finally, any serious proposal to supplement traditional NBS by genome-scale sequencing demands careful consideration of the optimal clinical setting in which parents learn about the series of complex decisions they would need to make to provide informed consent and the methods by which they are guided in this decision making process.
All 50 states in the United States currently screen for a panel of conditions that include hemoglobinopathies, disorders of amino acid, fatty acid and organic acid metabolism, congenital hypothyroidism, congenital adrenal hyperplasia, galactosemia, biotinidase deficiency, and cystic fibrosis. Thirty-four states also mandate newborn hearing screening (see http://genes-r-us.uthscsa.edu/sites/genes-r-us/files/nbsdisorders.pdf). In most states, NBS is mandated by state laws; only a few states consider it voluntary and require specific parental consent (Seashore and Seashore 2005). The public health benefits and importance of NBS must be weighed against the rights of parents to make decisions about their child. Some have argued that preventive programs conducted under public health auspices should be held to the same ethical standards as medical innovations introduced into the private sector (Skrabanek 1990). Some have proposed screening for all but a selected group of conditions, whereas others point out the potential for a “treatment odyssey” undertaken by families whose child is identified with a serious condition through screening for which no effective treatment exists (Baily and Murray 2008; Bailey et al. 2008). Although there is strong support for universal screening for disorders in which early diagnosis and treatment is lifesaving or produces great medical benefit, justifying the omission of explicit informed consent, this will not be the case for many new conditions that will be identifiable through sequencing. Available interventions for these disorders may have varying efficacy, and many disorders could have pleiotropic effects. Incorporation of genetic sequencing panels or even genome-scale sequencing into the NBS paradigm raises significant concerns about the management of such information (Bailey et al. 2008). Conditions that are clinically significant and may benefit patients greatly by surveillance and early diagnosis of complications, such as familial adenomatous polyposis, could have an equivalent impact to conditions that are currently screened. Conditions associated with developmental disability for which there may be no “cure” but for which early intervention and therapy services may be valuable are clearly different than those that have traditionally been included in NBS programs; it would be difficult to justify such screening being “mandatory,” and thus parental informed decision-making will be needed. As public health proponents anticipate the use of NGS to improve health outcomes, care must be taken not to undertake a slippery slope of utilizing this technology without rigorous scientific and ethical examination of its utility, acceptance, and consequences.
Secondary Findings
If genome-scale sequencing ultimately becomes the most cost-effective means of generating sequence data for NBS, the analysis could be focused on a subset of genes through the use of informatics filters. However, whenever genome-scale sequencing is performed, there will inevitably be additional clinically significant variants in genes that may not have been the original intent of the screening. These additional findings could be considered “incidental” or “secondary” findings; whether they should be part of the routine analysis is a subject of intense debate. The American College of Medical Genetics and Genomics (ACMG) recommended that when genome-scale sequencing is performed in a diagnostic setting, known pathogenic or expected pathogenic variants in 56 genes should be reported back to patients unless they opt out of receiving such findings (Green et al. 2013). In general, the ACMG relied on the guiding principle of clinical actionability to generate this list of genes, but the process did not use a systematic approach and the resulting list has been criticized (Burke et al. 2013; Ross et al. 2013a). The list was also not developed specifically for children, and indeed NBS conditions were excluded from consideration. As a result, the list recommended by the ACMG includes some conditions that have onset in adulthood, such as hereditary breast and ovarian cancer susceptibility caused by mutations in BRCA1 and BRCA2. Should these conditions be included as part of NBS when a genome-scale sequencing method is used? Previous recommendations have argued against testing children for adult-onset disorders, preferring to defer such testing until the individual can decide whether he or she wants to have that information (see section on Ethical Considerations).
EXAMPLES OF CONDITIONS AMENABLE TO AUGMENTED NEWBORN SCREENING
Phenylketonuria (PKU)
PKU is one of the most common inborn errors of metabolism detected by NBS. It is a well-characterized amino acid disorder caused by deficiency of the liver enzyme, phenylalanine hydroxylase (PAH), leading to elevated levels of the amino acid, phenylalanine (Phe) in blood and other tissues. It is inherited in an autosomal recessive pattern. Left untreated, PKU causes severe to profound intellectual disability, microcephaly, seizures, and behavior problems. It was the first condition to be screened for in newborns and one in 15,000 infants is born in the United States with classical PKU. Milder variants, known collectively as the hyperphenylalaninemias, result from partial deficiency of the enzyme and occur in approximately one out of 48,000 births. PKU and hyperphenylalanenemia are currently detected through MS/MS of dried blood spots and screen positive cases are confirmed by measuring Phe levels in blood samples. After confirmation, patients are immediately placed on a diet that strictly controls their intake of Phe and their Phe levels are closely monitored. Dietary treatment must begin within the first weeks of life and continue throughout the patient’s lifetime. The amount of protein restriction required to maintain normal blood Phe levels varies among patients. Use of the cofactor tetrahydrobiopterin (BH4; Kuvan) allows ∼50% of patients to increase their protein intake thus approximating a more normal diet. The variable clinical course of PKU is, in part, based on the specific mutations in the gene encoding the l-phenylalanine hydroxylase enzyme and the amount of Phe in the diet. More than 400 mutations have been identified in the PAH gene and show a broad spectrum of types including deletions, insertions, missense, splicing, and nonsense. Although there is a modest correlation between genotype and the distinct phenotypes of classical PKU and hyperphenylalanenemia (Utz et al. 2012), there are other contributing factors as evidenced by sibling discordance, especially in the response to treatment, that is Phe restriction alone, with BH4 or with large neutral amino acids. A “genotype severity” tool has been developed to study the correlation of PAH mutation(s) with responsiveness to BH4 and its use has been recommended to help define which patients will show the best response. However, there is significant overlap between patients of different genotypes and precise predictions cannot be made before a therapeutic trial with the cofactor (Quirk et al. 2012). Genes at other loci may influence Phe transport within the brain as well as play a role in other features of the clinical phenotype; these modifier genes have been hypothesized to exist but have not yet been identified. It has been recommended that all patients with confirmed PKU have mutation analysis for genotype determination (National Institutes of Health Consensus Development Panel 2001). Information about specific PAH mutations as well as variation in other relevant genes, obtained through MPS, may help explain this phenotypic variability. It may also improve treatment outcomes by more targeted intervention and dietary regulation of Phe levels.
Hearing Loss
Hearing loss is the most common birth defect and the most prevalent sensorineural disorder in developed countries (Hilgert et al. 2009). One of every 500 newborns has bilateral permanent sensorineural hearing loss that is greater or equal to a 40 decibel loss. Early detection and appropriate intervention results in improved development of language, cognitive, and social skills (White 2004). In 1993, a National Institutes of Health (NIH) consensus statement recommended that all infants have hearing screening shortly after birth (NIH 1993) and guidelines were established in 2000 (Joint Committee on Infant Hearing 2000). Screening is now performed in all states through Early Hearing Detection and Intervention (EHDI) programs with either otoacoustic emission (OAE) or automated auditory brainstem response (AABR) methodology. In 2009, it was estimated that 96.6% of newborns in the United States had hearing screening (see www.cdc.gov/ncbddd/hearingloss/ehdi-data2012.html).
Hearing loss represents a particularly promising avenue for realizing advances in NBS by using genomic sequencing approaches. Approximately 50% of prelingual deafness has a genetic etiology, with 70% categorized as nonsyndromic of which 75%–85% is autosomal recessive, 15%–24% autosomal dominant, and 1%–2% is X-linked or mitochondrial. Although ∼50% of autosomal recessive nonsyndromic hearing loss is caused by mutations in genes encoding the proteins connexin 26 or 30, 50% is caused by mutations in at least 40 other genes.
Thirty percent of genetic prelingual hearing loss is associated with syndromes (Smith et al. 1993). Syndromic forms can have comorbid conditions including blindness, cardiac arrhythmias, kidney disease, endocrine disorders, and intellectual disability. In newborns, phenotypic features of syndromic hearing loss are usually not yet apparent, making targeted gene testing impossible. Even many of the common syndromic forms of hearing loss show considerable locus heterogeneity. One striking example is Usher syndrome, a condition with early onset deafness and retinitis pigmentosa with progressive loss of vision in later childhood, for which 18 genes or loci have been identified to date (see http://www.omim.org/phenotypicSeries/276900). Testing for mutations in these genes through standard sequencing is prohibitive because of cost and time. Not knowing the etiology may contribute to the stress reported in families after confirmation of hearing loss in their child (Vohr et al. 2008). Additionally, not all infants with significant early-onset hearing loss will have positive newborn screens (Young et al. 2011), thus preventing early identification. Sequencing could potentially detect mutations in all known syndromic and nonsyndromic hearing loss genes, thus providing a specific diagnosis not only in infants with a genetic cause of hearing loss detected through NBS but also in those who would otherwise not be identified through NBHS and thus provides a method to expand the scope of NBS. In addition, its application would increase our knowledge regarding genetic etiologies of hearing loss that are currently poorly understood.
Additional Conditions
Of the 31 conditions in the current RUSP, 27 have identified genetic etiologies, whereas the remaining four (congenital hypothyroidism, hearing loss, critical congenital heart disease, and severe combined immunodeficiencies) are frequently because of identifiable genetic causes (see Table 1). Although other screening methods remain more sensitive and economical, molecular analysis is being increasingly used for confirmatory testing and to determine prognosis and appropriate treatment for many of these conditions as well as for the 26 secondary disorders detected in the differential diagnosis of the core disorders in the RUSP (Carrillo-Carrasco and Venditti 1993; Manoli and Venditti 1993; Bhardwaj et al. 2005; Collins et al. 2010; Bhattacharjee et al. 2014; Kwan et al. 2014).
Table 1.
Recommended uniform screening panel core conditions
| Gene(s) | Metabolic disorder | Endocrine disorder | Hemoglobin disorder | Other disorder | |||
|---|---|---|---|---|---|---|---|
| Organic acid disorder | Fatty acid oxidation disorder | Amino acid disorder | |||||
| Propionic acidemia | PCCB, PCCA | X | |||||
| Methylmalonic academia (methylmalonyl-CoA mutase) | MUT | X | |||||
| Methylmalonic academia (cobalamin disorders) | MMAA, MMAB | X | |||||
| Isovaleric acidemia | IVD | X | |||||
| 3-Methylcrotonyl-CoA carboxylase deficiency | MCCC1, MCCC2 | X | |||||
| 3-Hydroxy-3-methylglutaric aciduria | HMGCL | X | |||||
| Holocarboxylase synthase deficiency | HLCS | X | |||||
| β-Ketothiolase deficiency | ACAT1 | X | |||||
| Glutaric academia type 1 | GCDH | X | |||||
| Carnitine uptake defect/carnitine transport defect | SLC22A5 | X | |||||
| Medium-chain acyl-CoA dehydrogenase deficiency | ACADM | X | |||||
| Very long-chain acyl-CoA dehydrogenase deficiency | ACADVL | X | |||||
| Long-chain l-3 hydroxyacyl-CoA dehydrogenase deficiency | HADHA | X | |||||
| Trifunctional protein deficiency | HADHA, HADHB | X | |||||
| Argininosuccinic aciduria | ASL | X | |||||
| Citrullinemia, type I | ASS1, SLC25A13 | X | |||||
| Maple syrup urine disease | BCKDHA, BCKDHB, DBT | X | |||||
| Homocystinuria | CBS, MTHFR, MTR, MTRR, MMADHC | X | |||||
| Classic phenylketonuria | PAH | X | |||||
| Tyrosinemia, type I | FAH | X | |||||
| Primary congenital hypothyroidism | 1 | X | |||||
| Congenital adrenal hyperplasia | CYP21A2 | X | |||||
| Sickle cell anemia | HBB | X | |||||
| Β-thalassemia | HBB | X | |||||
| S,C disease | HBB | X | |||||
| Biotinidase deficiency | BTD | X | |||||
| Critical congenital heart disease | 2 | X | |||||
| Cystic fibrosis | CFTR | X | |||||
| Classic galactosemia | GALT | X | |||||
| Hearing loss | 3 | X | |||||
| Severe combined immunodeficiencies | 4 | X | |||||
| Conditions Not Included in Original RUSP (2006) because of No Available Screening Method | |||||||
| Hyperbilirubinemia1 | 5 | X | |||||
| Familial hypercholesterolemia1 | 6 | X | |||||
| Carnitine palmitoyltransferase 1B (muscle)1 | CPT1B | X | |||||
| Ornithine transcarbamylase deficiency1 | OTC | X | |||||
| Severe combined immunodeficiencies1 | 4 | X | |||||
| Type 1 diabetes mellitus1 | 7 | ||||||
| Guanidinoacetate methyltransferase deficiency1 | GAMT | ||||||
| Wilson disease1 | ATP7B | X | |||||
| Arginine:glycine amidinotransferase deficiency1 | GATM | X | |||||
| Neuroblastoma1 | 8 | X | |||||
| Turner syndrome1 | 9 | X | |||||
| Carbamylphosphate synthetase deficiency1 | CPS1 | ||||||
| Biliary atresia1 | 10 | X | |||||
| Fragile X syndrome1 | FMR1 | X | |||||
| Congenital disorder of glycosylation type Ib1 | MPI | X | |||||
| Smith–Lemli–Opitz syndrome1 | DHCR7 | X | |||||
| Adrenoleukodystrophy1 | ABCD1 | X | |||||
| Mucopolysaccharidosis 1H1 | IDUA | X | |||||
| Fabry disease1 | GLA | X | |||||
| Creatine transporter defect1 | SLC6A8 | X | |||||
| Lysosomal storage diseases1 | 11 | X | |||||
| Pompe disease1 | GAA | X | |||||
| Krabbe disease1 | GALC | X | |||||
| 1. Most cases are sporadic but 15%–20% are genetic with DUOX2, PAX8, SLC5A5, TG, TPO, TSHB, or TSHR gene mutations | |||||||
| 2. Multifactorial; most genetic causes unknown | |||||||
| 3. Multiple etiologies including genetic and environmental; multiple genes associated with nonsyndromic and syndromic forms | |||||||
| 4. 15 known genes associated; 14% of cases of unknown cause | |||||||
| 5. Most causes are nongenetic | |||||||
| 6. Most common genetic causes include APOB, LDLR, LDLRAP1, and PCSK9 gene mutations. | |||||||
| 7. Most multifactorial with rare single gene causes | |||||||
| 8. Most sporadic; mutations in ALK, KIF1B, and PHOX2B in rare familial cases | |||||||
| 9. Chromosome disorder | |||||||
| 10. Multifactorial, most genetic factors unknown | |||||||
| 11. Large number of disorders under this category with known genetic causes | |||||||
Of the 31 conditions in the current RUSP, 27 have identified genetic etiologies, whereas the remaining four (congenital hypothyroidism, hearing loss, critical congenital heart disease, and severe combined immunodeficiencies) are frequently caused by identifiable genetic defects.
1Adapted from ACMG Newborn Screening Expert Group (2006). Table adapted from www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/recommendedpanel.
There are a number of serious and treatable childhood conditions that are not currently screened for lack of an effective test. MPS has the potential to identify the molecular basis for disorders currently included on NBS panels, but, more importantly, it could significantly expand our ability to detect a much broader range of genetic conditions. Some conditions were considered as candidates for screening by the expert panel convened by the ACMG in 2002 and despite being ranked highly in terms of their clinical significance, availability and efficacy of treatments and potential for avoidance of serious sequelae through early detection, could not be added to the recommended panel due to lack of an available screening test. Many have an underlying genetic etiology, with the causative genes identified and, therefore, could be detected through molecular techniques. In the list of 23 conditions not included in the recommended screening panel because they lack an accurate screening method (ACMG Newborn Screening Expert Group 2006), 21 are detectable in some or all cases with molecular genetic analysis and, therefore, have the potential to be added to NBS panels (see Table 1). Of 52 types of inborn errors of metabolism considered for inclusion in recommended NBS panels, all have the potential for detection with sequencing. These include Fabry disease, familial hypercholesterolemia, Wilson disease and the Creatine Deficiency Syndromes that cannot be detected with standard NBS methods.
Sequencing offers the ability to expand this list beyond inborn errors of metabolism. The list could include, for example, genes associated with early childhood cancer such as multiple endocrine neoplasia type IIB due to mutations in the RET gene, inherited channelopathies that lead to potentially preventable cardiac arrhythmias, and primary ciliary dyskinesia (PCD), a rare, genetically heterogeneous disorder resulting in a range of manifestations including situs inversus, neonatal respiratory distress at full-term birth, recurrent otitis media, chronic sinusitis, chronic bronchitis that may result in bronchiectasis, and male infertility.
Other conditions have not been included in recommended screening panels due to lack of treatment by traditional definitions. Individuals with conditions leading to intellectual disability might derive benefit from early detection and intervention, including earlier enrollment in developmental intervention services and avoidance of the diagnostic odyssey. In 2006, Alexander and van Dyck challenged the traditionally held belief that NBS should only include conditions with effective treatments and broadened the concept to include conditions with benefits to the family for reproductive decision making, the potential to participate in research or innovative therapeutics and avoidance of the diagnostic odyssey (Alexander and van Dyck 2006). On the other hand, genetic information predicting the inevitable development of an incurable genetic disorder may be unwelcome to some parents of otherwise apparently healthy newborns. The practice of mandatory screening for such nonmedically actionable conditions, if widely accepted by public health screening programs, would dramatically alter the nature of the screening program, potentially undermining the currently accepted practice of screening without obtaining explicit consent.
ETHICAL CONSIDERATIONS
Unlike many kinds of medical tests, which provide information of a transient, temporal nature, genetic testing typically can reveal information about an individual’s past, present, and future medical conditions; this information may also have immediate implications for family members. These characteristics, combined with the complexity of genetic information, which ranges from probabilistic to highly deterministic, has led many to view genetic information as somehow different from other kinds of biomedical information (“genetic exceptionalism”). Consideration of the ethical, legal, and social implications of genetic knowledge has been an inherent component of the Human Genome Project and other genomic research efforts (Greely 1998), and inexorable advances in genetic testing have been accompanied by an immense societal discussion about the most appropriate uses of this information in healthcare, in human subjects research, and even in the setting of personal genomic exploration (Bunnik et al. 2011). The thread of an individual’s right to self-determination is woven tightly throughout the ethical considerations of genetic testing (e.g., Nyrhinen et al. 2009; Bunnik et al. 2013). Genetic testing in children raises additional complexities that could potentially alter the dynamic that currently exists in NBS.
Considerations of the benefits and risks of genetic testing are perhaps most acute in children because of their special status as minors, under the guardianship of their parents for a period of time, after which they may achieve independence and acquire their own right to self-determination (Lantos 2010). Although parents are given a significant amount of leeway in their decisions about how to raise their children, there are also limits on this guardianship regarding that child’s future autonomy. Parents have the responsibility to act in their child’s best interests, which is the primary consideration in most approaches to pediatric genetic testing. Further complicating matters, consideration of such testing inevitably occurs in the context of highly variable childhood developmental states and unique family settings (Fanos 1997).
Expert panels have put forth various guidelines to delineate appropriate uses of genetic testing in children (Wertz et al. 1994; ASHG Board of Directors, ACMG Board of Directors 1995; AAP Committee on Bioethics 2001; Ross et al. 2013b). These recommendations have traditionally been grounded in “best interests” being limited strictly to the impact of genetic information for the child in question. For example, it is generally agreed that testing children for adult-onset conditions should be avoided when the information would not directly impact medical management during childhood. This recommendation typically envisions the scenario in which a condition is known to exist in a family and the at-risk child will be able to make an informed decision about genetic testing when they reach adulthood. The recommendation to avoid predictive genetic testing is based in the idea that such testing will not alter medical management of the child, and that there could be psychological harms associated with learning one’s mutation status. The ACMG recommendations regarding the return of adult-onset clinically actionable incidental findings in children (Green et al. 2013) appears to be at odds with these restrictions on testing for adult-onset disorders, except that in the case of a child undergoing diagnostic genome-scale sequencing or NBS, there may be no knowledge in the family about a clinically actionable adult-onset disorder (e.g., a 25-year-old mother with no family history of early-onset breast cancer who inherited a BRCA1 mutation from her father). Such a finding, if not reported, could lead to irreparable harm to the child caused by the early death of a parent from a condition that might have been prevented. This type of incidental finding could therefore have direct psychological benefit to the child and be in the “best interests of the child,” even though the revelation of the information may obviate that child’s “right not to know” later in life. However, opinion seems to be split regarding the justification of revealing information about adult-onset clinically actionable conditions in a child when the benefits are theoretical and less certain to accrue than the discovery of a condition with direct medical implications for the child (e.g., Strong et al. 2014; Yu et al. 2014). Clearly, there is equipoise about the balance of benefits and harms in this situation. There is a view that testing for carrier status for recessive disorders is not likely to benefit the child and should thus be deferred until the individual is considering reproduction. However, it should be noted that substantial counter arguments have been made on behalf of informed decision making by parents, despite these concerns (Pelias 2006; Rhodes 2006).
In the recommendations made by the ACMG and AAP regarding the uniform screening panel (ACMG Newborn Screening Expert Group 2006), the expert group made three recommendations that could have profound implications for NBS via genome-scale sequencing.
First, the expert group recommended that 25 additional “secondary” targets be examined and reported. Although the original intent of NBS was to detect only the specific condition screened for, such as PKU, it has long been recognized that some with positive newborn screens had less medically significant conditions, such as hyperphenylalaninemia, raising concerns that this would cause some children to undergo unnecessary treatments and cause parents undue anxiety (Gurian et al. 2006). These 25 conditions are “clinically significant and revealed by the screening technology but lack an efficacious treatment” and the expert group “thought it was important that such findings be communicated to the health care service community and to families” (ACMG Newborn Screening Expert Group 2006). The direct implication of this recommendation is that any clinically relevant finding from NBS should be reported, which in the case of genome-scale sequencing would include essentially any genetic condition. Presumably, the expert group was not envisioning the use of a genome-scale sequencing test for NBS when they made these recommendations, because many families would refuse such testing if they knew there was a possibility of learning about findings without any related preventive measures (Bombard et al. 2014), thus jeopardizing the immense value of the newborn screen at the population level.
Second, the expert group recommended that states “mandate … reporting of any abnormal results that may be associated with clinically significant conditions, including the definitive identification of carrier status” (ACMG Newborn Screening Expert Group 2006). In practice, carrier results for cystic fibrosis and sickle cell disease are routinely returned as part of NBS. Expanding this recommendation for all conditions detectable by genome-scale sequencing would essentially convert NBS from a program that detects rare, preventable disorders in a small minority of cases into a carrier screening test for all recessive disorders that would yield a handful of findings in every individual screened. Again, the expert group was likely not considering the implications of this recommendation for genome-scale sequencing being used in NBS.
Finally, the expert group recommended that states “consider that the range of benefits realized by NBS includes treatments that go beyond an infant’s mortality and morbidity” (ACMG Newborn Screening Expert Group 2006), which seems to imply that there is value to NBS beyond preventable conditions—That personal utility (or utility as perceived by the family unit) is just as valid a consideration in determining what information to divulge as the traditional values of improving the health of the individual child. This recommendation is somewhat similar to the ACMG incidental findings recommendations, in which the benefit to the child is indirect and related to the overall well-being of the child’s family members. That being said, the expert group’s recommendation, if taken to the logical extreme, could be interpreted as meaning that any genetic information that is desired by the parents is justifiable if considered beneficial by the parents.
Most would likely agree that when genetic information is available, parents should have a reasonable ability to learn such information if desired, and also to refuse information that they do not want. The challenge is in defining what is “reasonable”—Herein lies the equipoise when considering the use of genome-scale sequencing in NBS. It can be argued that parental prerogative is the primary consideration—Parents are responsible for their child’s health care, and learning (or refusing) information about any genetic condition could be considered part of this responsibility. On the other hand, some information could have damaging effects on the child’s own well-being if that knowledge interferes with parental bonding, creates family stress including divorce, or leads to abuse or abandonment. In addition, even the decision making process could lead to strife between parents if they are unable to agree about whether or not to learn such information.
The mainstream consensus of the bioethics community appears to be that adult-onset disorders with no effective prevention or treatment should be off-limits to parents and are most appropriate for informed decision-making by the individual when he or she becomes an adult. That being said, some argue that even these disorders fall within a parent’s responsibility to raise their child to the best of their ability and prepare them for any eventuality, that the theoretical harms are less significant than initially supposed (Malpas 2008) and that parents are in the best position to make decisions relative to their child’s best interests (Robertson and Savulescu 2001). Furthermore, in the case of a disabled child who will likely never be able to make an informed decision, parents could reasonably expect to make such decisions on that child’s behalf.
Clearly, the application of genome-scale sequencing in NBS raises a host of ethical, legal, and social implications (Tarini and Goldenberg 2012). Challenges related to the use of genomic sequencing in newborns, both technical and ethical, will need to be overcome to establish a widely accepted NGS-based platform to augment NBS.
CONCLUSION
The possibility of a significant expansion of NBS raises a number of concerns, including the lack of evidence-based efficacy studies, the need for informed consent, the challenge of providing information and support for families, and the ethical, legal, and social issues associated with such scenarios as disclosure of carrier status or genetic susceptibility to future disease (Taylor and Wilfond 2004; Botkin 2005; Bailey et al. 2006). There are major objectives that need to be addressed to incorporate use of genomics and other technological advances in NBS. Public and professional education will be required, and the expert infrastructure for dealing with children who screen positive will need to be improved significantly (Alexander and van Dyck 2006). Some consideration should be given to whether the long-held criteria for screening should be changed, by broadening the concept of benefit from screening for the child to include the family.
With these challenges in mind, pilot projects to examine these issues are being funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) under the Genomic Sequencing and Newborn Screening Disorders research program. Use of genome-scale sequencing in NBS will require careful consideration and informed decision making by parents and education of providers as they use this technology.
ACKNOWLEDGMENTS
The authors thank other key personnel involved in the research project including Don Bailey, Myra Roche, Laura Milko, Megan Lewis, Christine Rini, Kirk Wilhelmsen, and James P. Evans.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- AAP, Committee on Bioethics. 2001. Ethical issues with genetic testing in pediatrics. Pediatrics 107: 1451–1455. Reaffirmed October 2004. [DOI] [PubMed] [Google Scholar]
- ACMG Newborn Screening Expert Group. 2006. Newborn screening: Toward a uniform screening panel and system—Executive summary. Pediatrics 117: S296–S307. [DOI] [PubMed] [Google Scholar]
- Alexander D, van Dyck PC. 2006. A vision of the future of newborn screening. Pediatrics 117: S350–S354. [DOI] [PubMed] [Google Scholar]
- Andermann A, Blancquaert I, Beauchamp S, Déry V. 2008. Revisiting Wilson and Jungner in the genomic age: A review of screening criteria over the past 40 years. Bull World Health Organ 86: 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHG Board of Directors, ACMG Board of Directors. 1995. Points to consider: Ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am J Hum Genet 57: 1233–1241. [PMC free article] [PubMed] [Google Scholar]
- Baily MA, Murray TH. 2008. Ethics, evidence, and cost in newborn screening. Hastings Center Report 38: 23–31. [DOI] [PubMed] [Google Scholar]
- Bailey DB Jr, Beskow LM, Davis AM, Skinner D. 2006. Changing perspectives on the benefits of newborn screening. Ment Retard Dev Disabil Res Rev 12: 270–279. [DOI] [PubMed] [Google Scholar]
- Bailey DB Jr, Skinner D, Davis AM, Whitmarsh I, Powell C. 2008. Ethical, legal, and social concerns about expanded newborn screening: Fragile X syndrome as a prototype for emerging issues. Pediatrics 121: e693–e704. [DOI] [PubMed] [Google Scholar]
- Bhardwaj U, Zhang YH, Lorey F, McCabe LL, McCabe ER. 2005. Molecular genetic confirmatory testing from newborn screening samples for the common African-American, Asian Indian, Southeast Asian, and Chinese β-thalassemia mutations. Am J Hematol 78: 249–255. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Sokolsky T, Wyman SK, Reese MG, Puffenberger E, Strauss K, Morton H, Parad RB, Naylor EW. 2014. Development of DNA confirmatory and high-risk diagnostic testing for newborns using targeted next-generation DNA sequencing. Genet Med. 10.1038/gim.2014.117. [DOI] [PubMed] [Google Scholar]
- Bombard Y, Miller FA, Hayeems RZ, Barg C, Cressman C, Carroll JC, Wilson BJ, Little J, Avard D, Painter-Main M, et al. 2014. Public views on participating in newborn screening using genome sequencing. Eur J Hum Genet 22: 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botkin JR. 2005. Research for newborn screening: Developing a national framework. Pediatrics 116: 862–871. [DOI] [PubMed] [Google Scholar]
- Bunnik EM, Schermer MH, Janssens AC. 2011. Personal genome testing: Test characteristics to clarify the discourse on ethical, legal and societal issues. BMC Med Ethics 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnik EM, de Jong A, Nijsingh N, de Wert GM. 2013. The new genetics and informed consent: Differentiating choice to preserve autonomy. Bioethics 27: 348–355. [DOI] [PubMed] [Google Scholar]
- Burke W, Matheny Antommaria AH, Bennett R, Botkin J, Clayton EW, Henderson GE, Holm IA, Jarvik GP, Khoury MJ, Knoppers BM, et al. 2013. Recommendations for returning genomic incidental findings? We need to talk! Genet Med 15: 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Carrasco N, Venditti C. 1993. Propionic academia. In GeneReviews (ed. Pagon RA, et al. ) University of Washington, Seattle, WA. [Google Scholar]
- Collins SA, Sinclair G, McIntosh S, Bamforth F, Thompson R, Sobol I, Osborne G, Corriveau A, Santos M, Hanley B, et al. 2010. Carnitine palmitoyltransferase 1A (CPT1A) P479L prevalence in live newborns in Yukon, Northwest Territories, and Nunavut. Mol Genet Metab 101: 200–204. [DOI] [PubMed] [Google Scholar]
- Fanos JH. 1997. Developmental tasks of childhood and adolescence: Implications for genetic testing. Am J Med Genet 71: 22–28. [DOI] [PubMed] [Google Scholar]
- Frazier DM, Millington DS, McCandless SE, Koeberl DD, Weavil SD, Chaing SH, Muenzer J. 2006. The tandem mass spectrometry newborn screening experience in North Carolina: 1997–2005. J Inherit Metab Dis 29: 76–85. [DOI] [PubMed] [Google Scholar]
- Greely HT. 1998. Legal, ethical, social issues in human genome research. Annu Rev Anthropol 27: 473–502. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, et al. 2013. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurian EA, Kinnamon DD, Henry JJ, Waisbren SE. 2006. Expanded newborn screening for biochemical disorders: The effect of a false-positive result. Pediatrics 117: 1915–1921. [DOI] [PubMed] [Google Scholar]
- Guthrie R, Susi A. 1963. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 32: 338–343. [PubMed] [Google Scholar]
- Hilgert N, Smith RJ, Van Camp G. 2009. Function and expression pattern of nonsyndromic deafness genes. Curr Mol Med 9: 546–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint Committee on Infant Hearing; American Academy of Audiology; American Academy of Pediatrics; American Speech-Language-Hearing Association; Directors of Speech and Hearing Programs in State Health and Welfare Agencies. 2000. Principles and guidelines for early hearing detection and intervention programs. Pediatrics 106: 798–817. [DOI] [PubMed] [Google Scholar]
- Kemper AR, Green NS, Calonge N, Lam WK, Comeau AM, Goldenberg AJ, Ojodu J, Prosser LA, Tanksley S, Bocchini JA Jr. 2014. Decision-making process for conditions nominated to the recommended uniform screening panel: Statement of the US Department of Health and Human Services Secretary’s Advisory Committee on Heritable Disorders in Newborns and Children. Genet Med 16: 183–187. [DOI] [PubMed] [Google Scholar]
- Kwan A, Abraham RS, Currier R, Brower A, Andruszewski K, Abbott JK, Baker M, Ballow M, Bartoshesky LE, Bonilla FA, et al. 2014. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA 312: 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantos JD. 2010. Does pediatrics need its own bioethics? Perspect Biol Med 53: 613–624. [DOI] [PubMed] [Google Scholar]
- Malpas PJ. 2008. Predictive genetic testing of children for adult-onset diseases and psychological harm. J Med Ethics 34: 275–278. [DOI] [PubMed] [Google Scholar]
- Manoli I, Venditti CP. 1993. Methylmalonic acidemia. In GeneReviews (ed. Pagon RA, et al. ) University of Washington, Seattle, WA: http://www.ncbi.nlm.nih.gov/books/NBK1116/. [Google Scholar]
- Millington DS, Norwood DL, Kodo N, Roe CR, Inoue F. 1989. Application of fast atom bombardment with tandem mass spectrometry and liquid chromatography/mass spectrometry to the analysis of acylcarnitines in human urine, blood, and tissue. Anal Biochem 180: 331–339. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health Consensus Development Panel. 2001. Phenylketonuria: Screening and management. Pediatrics 108: 972–982. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health Consensus Development Panel. 1993. Early identification of hearing impairment in infants and young children. NIH Consensus Statement 11: 1–24. [PubMed] [Google Scholar]
- Nyrhinen T, Hietala M, Puukka P, Leino-Kilpi H. 2009. Are patient rights to information and self-determination in diagnostic genetic testing upheld? A comparison of patients’ and providers’ perceptions. J Genet Couns 18: 72–81. [DOI] [PubMed] [Google Scholar]
- Pelias MK. 2006. Genetic testing of children for adult-onset diseases: Is testing in the child’s best interests? Mt Sinai J Med 73: 605–608. [PubMed] [Google Scholar]
- Quirk ME, Dobrowolski SF, Nelson BE, Coffee B, Singh RH. 2012. Utility of phenylalanine hydroxylase genotype for tetrahydrobiopterin responsiveness classification in patients with phenylketonuria. Mol Genet Metab 107: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes R. 2006. Why test children for adult-onset genetic diseases? Mt Sinai J Med 73: 609–616. [PubMed] [Google Scholar]
- Robertson S, Savulescu J. 2001. Is there a case in favour of predictive genetic testing in young children? Bioethics 15: 26–49. [DOI] [PubMed] [Google Scholar]
- Ross LF, Rothstein MA, Clayton EW. 2013a. Premature guidance about whole-genome sequencing. Per Med 10 10.2217/pme.13.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross LF, Saal HM, David KL, Anderson RR; American Academy of Pediatrics; American College of Medical Genetics and Genomics. 2013b. Technical report: Ethical and policy issues in genetic testing and screening of children. Genet Med 15: 234–245. [DOI] [PubMed] [Google Scholar]
- Seashore MR, Seashore CJ. 2005. Newborn screening and the pediatric practitioner. Semin Perinatol 29: 182–188. [DOI] [PubMed] [Google Scholar]
- Skrabanek P. 1990. Why is preventive medicine exempted from ethical constraints? J Med Ethics 16: 187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJH, Sheffield AM, Van Camp G. 1993. Nonsyndromic hearing loss and deafness, DFNA3. In GeneReviews (ed. Pagon RA, et al. ) University of Washington, Seattle, WA. [Google Scholar]
- Strong KA, Zusevics KL, Bick D, Veith R. 2014. Views of primary care providers regarding the return of genome sequencing incidental findings. Clin Genet 86: 461–468. [DOI] [PubMed] [Google Scholar]
- Tarini BA, Goldenberg AJ. 2012. Ethical issues with newborn screening in the genomics era. Annu Rev Genomics Hum Genet 13: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HA, Wilfond BS. 2004. Ethical issues in newborn screening research: Lessons from the Wisconsin cystic fibrosis trial. J Peds 145: 292–296. [DOI] [PubMed] [Google Scholar]
- Utz JR, Lorentz CP, Markowitz D, Rudser KD, Diethelm-Okita B, Erickson D, Whitley CB. 2012. START, a double blind, placebo-controlled pharmacogenetic test of responsiveness to sapropterin dihydrochloride in phenylketonuria patients. Molec Genet Metab 105: 193–197. [DOI] [PubMed] [Google Scholar]
- Vohr BR, Jodoin-Krauzyk J, Tucker R, Johnson MJ, Topol D, Ahlgren M. 2008. Results of newborn screening for hearing loss: Effects on the family in the first 2 years of life. Arch Pediatr Adolesc Med 162: 205–211. [DOI] [PubMed] [Google Scholar]
- Wertz DC, Fanos JH, Reilly PR. 1994. Genetic testing for children and adolescents. Who decides? JAMA 272: 875–881. [PubMed] [Google Scholar]
- White KR. 2004. Early hearing detection and intervention programs: Opportunities for genetic services. Am J Med Genet Part A 130A: 29–36. [DOI] [PubMed] [Google Scholar]
- Wilson JMG, Jungner G. 1968. Principles and practice of screening for disease. Public Health Papers (34), World Health Organization. [Google Scholar]
- Young NM, Reilly BK, Burke L. 2011. Limitations of universal newborn hearing screening in early identification of pediatric cochlear implant candidates. Arch Otolaryngol Head Neck Surg 137: 230–234. [DOI] [PubMed] [Google Scholar]
- Yu JH, Harrell TM, Jamal SM, Tabor HK, Bamshad MJ. 2014. Attitudes of genetics professionals toward the return of incidental results from exome and whole-genome sequencing. Am J Hum Genet 95: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]