Abstract
Salmonella enterica is a prevalent food-borne pathogen which can carry multi-drug resistance (MDR) and could pose a threat to human health. Identifying the genetic elements associated with MDR in Salmonella isolated from animals, food, and humans can help determine the sources of MDR in food animals and their impact on human health. Representatives of MDR S. enterica serovars most frequently isolated from healthy animals, retail meat, and human infections in the U.S. and Canada were subjected to detailed genetic analysis (n=56). These included U.S. slaughter (n=12), retail (n=9), and human (9) isolates, and Canadian slaughter (n=9), retail (n=9), and human (n=8) isolates. These isolates were assayed by microarray for antimicrobial resistance and MDR plasmid genes. Genes detected encoded resistance to aminoglycosides (alleles of aac, aad, aph, strA/B); beta-lactams (blaTEM, blaCMY, blaPSE-1); chloramphenicol (cat, flo, cmlA); sulfamethoxazole (sulI); tetracycline (tet(A, B, C, D) and tetR); and trimethoprim (dfrA). Similar resistance genes were detected regardless of serovar, source, or location. Hybridization with IncA/C plasmid gene probes indicated that 27/56 isolates carried a member of this plasmid family; however these plasmids differed in several highly variable regions. Cluster analysis based on genes detected separated most of the isolates into two groups, one with IncA/C plasmids and one without IncA/C plasmids. Other plasmid replicons were detected in all but one isolate, and included I1 (25/56), N (23/56) and FIB (10/56). The presence of different mobile elements along with similar resistance genes suggest that these genetic elements may acquire similar resistance cassettes, and serve as multiple sources for MDR in Salmonella from food animals, retail meat, and human infections.
Keywords: Salmonella, multi-drug resistance, antimicrobial resistance, plasmid
Introduction
Salmonella enterica is a leading cause of food borne illness worldwide.2,5,6,12,29 Salmonella can be transferred to humans through contaminated water and food products, leading to food-borne illness. In most cases, salmonellosis in an otherwise healthy individual results in a self-limiting infection that does not require antimicrobial treatment. Invasive Salmonella infections are more severe and require antimicrobial therapy.2,8 Antimicrobial resistant Salmonella are a concern, as infections caused by these microorganisms may be more difficult to treat compared to their susceptible counterparts.8 Both the U.S. and Canada developed initiatives to monitor the dissemination of antimicrobial resistance in Salmonella and other enteric pathogens isolated from animals, food, and humans.18 The National Antimicrobial Resistance Monitoring System (NARMS) was developed in 1996 to monitor U.S. antimicrobial resistance trends in human diagnostic isolates (Centers for Disease Control and Prevention, CDC), food animals (U.S. Department of Agriculture, Agricultural Research Service, USDA-ARS) and retail meats (U.S. Food and Drug Administration, Center for Veterinary Medicine, FDA-CVM) (http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/default.htm). In Canada, the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) was developed in 2002 to monitor similar antimicrobial resistance trends (http://www.phac-aspc.gc.ca/cipars-picra/index-eng.php). In these programs, isolates are collected and tested for resistance to antimicrobials used in both human and veterinary medicine.
Multi-drug resistant (MDR) Salmonella which are resistant to two or more classes of antimicrobials, are frequently encountered and may reduce the effectiveness of treatments.21 In 2007, 13% of Canadian Salmonella isolated from chickens and swine during slaughter (abattoir) and 3% isolated from retail chicken were resistant to five or more antimicrobials and consequently MDR. Almost 10% of Salmonella isolated in Canada from humans between 2004 and 2006 also exhibited antimicrobial resistance (http://www.phac-aspc.gc.ca/cipars-picra/index-eng.php). In 2007, 10% of Salmonella isolated in the U.S. from chicken, turkey, cattle and swine slaughter isolates were resistant to five or more antimicrobials while 5.6% of retail meat isolates exhibited similar resistances. In U.S. human isolates, 6.9% were resistant to five or more antimicrobials (http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm209340.htm).
Plasmids are often associated with antimicrobial resistance in Salmonella and may also contain additional genes that provide heavy metal resistance, sanitizer resistance, or that aid in virulence and environmental adaptability.8,21,24,29,30 These plasmids are small, circular pieces of DNA that are often self-transmissible to other bacteria through conjugation. The transfer of plasmids from one bacterial cell to another results in the horizontal transfer of genetic material, allowing acquisition of multiple genes by the recipient.10,27,30 Plasmids with the IncA/C replicon have been linked to multiple drug resistance in Salmonella enterica associated with food animals as well as the fish pathogen Yersinia ruckeri and Y. pestis, the causative agent of plague.42 In addition to IncA/C, several other plasmid replicons have been associated with MDR Salmonella, including B/O, HI1, HI2, I1, N, F, and P many of which are found to be co-resident in some MDR Salmonella.37
This study was conducted to identify the antimicrobial resistance genes and genetic elements associated with MDR in Salmonella isolated from humans, animals, and retail meat by NARMS and CIPARS. Genetic analyses included detection of antimicrobial resistance and MDR plasmid genes by DNA microarrays and PCR detection of plasmid replicons.9,23 The detailed analysis of a collection of Salmonella representative of the most frequently isolated MDR serovars, identified antimicrobial resistance and MDR plasmid genes present in U.S. and Canadian isolates. Cluster and linkage analysis of this data was used to identify isolates with significantly similar profiles of antimicrobial resistance and plasmid genes from the different surveillance programs. Several genetic commonalities such as the presence of IncA/C plasmids were detected in MDR Salmonella isolated from different sources including humans, retail meat, and animals at slaughter. Comparable MDR genetics were also identified in isolates from similar animal, meat, and human sources sampled by the U.S. and Canadian surveillance programs.
Materials and Methods
Isolate selection, culture conditions, and antimicrobial susceptibility testing
Salmonella enterica chosen for this study were isolated from various sources by the U.S. Food Safety and Inspection Service (slaughter isolates, n=12), the U.S. Food and Drug Administration, Center for Veterinary Disease (retail isolates, n=9), the Centers for Disease Control and Prevention (human isolates, n=9), the Canadian Integrated Program for Antimicrobial Resistance Surveillance (slaughter isolates, n=9; retail isolates, n=9) and the Public Health Agency of Canada (human isolates, n=8). Isolates were obtained, cultivated, maintained, and stored as frozen stock cultures using standard methods. Culture media was obtained from Difco™ (Becton Dickinson and Company, Sparks, MD). All Salmonella isolates were subjected to testing via the Sensititre™ semi-automated antimicrobial susceptibility system following the manufacturers instructions (TREK Diagnostic Systems, Inc., Westlake, OH) for susceptibility to amikacin, gentamicin, kanamycin, streptomycin, ampicillin, amoxicillin-clavulanic acid, ceftiofur, ceftriaxone, cefoxitin, sulfamethoxazole/sulfisoxazole, trimethoprim-sulfamethoxazole, chloramphenicol, ciprofloxacin, nalidixic acid and tetracycline. Serovars with the highest frequency of MDR for each source agency were determined (MDR defined as resistance to two or more classes of antimicrobials). Isolates where then chosen from these serovars that were resistant to the maximum number of antimicrobial compounds (described below). Prevalence data, detailed isolation and testing methods available in U.S. NARMS reports: http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm209340.htm, and Canadian CIPARS reports: http://www.phac-aspc.gc.ca/cipars-picra/pubs-eng.php
Microarray design and construction
The DNA microarrays and methods used for this analysis have been previously reported and validated.23,31,32 Briefly, the DNA microarrays consisted of 1267 gene probes. Of those probes, 775 were designed to detect antimicrobial resistance genes found in the National Center for Biotechnology Information (NCBI) database. The other 487 gene probes were designed to detect plasmid genes identified in six strains of bacteria.31,32 IncA/C gene probes on the microarray were designed from plasmid sequences identified in Yersinia ruckeri str. YR71 pYR1, Y. pestis biovar Orientalis str. IP275 pIP1202, Photobacterium damselae subsp. piscicida pP99-018, Salmonella enterica subsp. enterica serovar Newport str. SL254 pSN254, Photobacterium damselae subsp. piscicida pP91278 and Escherichia coli p1658/97. Gene sequences from the plasmid pHCM1 found in S. enterica subsp. enterica serovar Typhi str. CT18 were used to design probes for the genes in this IncHI1 plasmid. The microarrays were constructed by spotting probes onto Corning UltraGAPS amino-silane coated slides (Corning Inc., Life Sciences, Acton, MA) with an Qarray mini robot (Genetix, Hampshire, UK) and post processed following the manufacturers recommendations (Corning, Inc.) as previously described.31,32
DNA isolation, labeling, microarray hybridization, and scoring
Total DNA from the isolates was prepared with the GenElute Bacterial Genomic DNA kit (Sigma, St Louis, MO) following instructions for Gram-negative bacteria from 5 mL of overnight cultures grown in LB broth at 37°C with shaking as previously described. Cye dye-labeled dCTP (Amersham, Piscataway, NJ), Klenow fragment (New England Biolabs, Beverly, MA) and random primers were used to label Salmonella genomic DNA overnight in 37°C water bath.23 Labeled DNA was purified using Qiagen PCR clean-up kit (Qiagen, Valencia, CA), and hybridized to the microarray over night at 42°C in a Corning hybridization chamber as previously described. Microarrays were washed following the manufacturers protocol for hybridization with formamide buffer (Corning, Inc.). Slides were scanned using ScanArray Lite microarray analysis system and images were analyzed using the ScanArray Express software version 1.1 (Packard BioChip Technologies, Billerica, MA). Positive hybridizations were determined and scored as previously described.23,32
Plasmid replicon typing
Three panels of multiplex PCR were used to determine the presence of 18 plasmid replicons commonly found in Enterobacteriaceae. PCR primers and parameters were previously described.9,30
Cluster analysis of isolates
Relationships between isolate based on gene content was determined by hierarchical cluster analysis based on antimicrobial resistance and plasmid genes detected by microarray analysis. Open source CLUSTER 3.0 was used for this analysis with Euclidean distance for gene content.13,16 Dendrogram was viewed using Java TreeView version 1.1.4r338 (http://jtreeview.sourceforge.net).
Statistical analysis
Linkage disequilibrium (LD) was calculated as an extension of Fisher’s exact probability test on contingency tables39 as instituted by the program Arlequin17. Standard settings were used, 10,000 steps in the Markov chain and 1,000 dememorization steps; and calculations of D, D′, and r2 coefficients were made with a significance level of 0.05.
Results
MDR in U.S. NARMS and Canadian CIPARS Salmonella enterica isolates selected for the study
The Salmonella enterica serovars chosen for the study were the most frequently isolated MDR serovars found in each source. For U.S. NARMS this was Salmonella serovars Typhimurium, Heidelberg, and Newport for both human (CDC1 – CDC9) and animal (USDA1 – USDA12) isolates. U.S. retail meat isolates (FDA1 – FDA9) were chosen from Salmonella serovars Typhimurium, Heidelberg, and Hadar. Isolates from Canadian CIPARS were serovars Typhimurium, Heidelberg, and Enteritidis from humans (CH1 – CH8), Typhimurium, Heidelberg, Hadar, and Saintpaul, from animals (CA1 – CA9), and serovars Typhimurium, Heidleberg and Hadar from retail meat (CR1 – CR9). Isolates resistant to the greatest number of antimicrobial compounds were chosen from those Salmonella serovars for each agency’s collection from that source. The numbers of antimicrobial compounds the isolates were resistant to range in number from three compounds to 13, with an average of resistance to seven antimicrobials (Table 1 and Figure 1).
Table 1.
Summary of pYR1 and pSN254 IncA/C plasmid genes detected by microarray analysis and replicon types detected by PCR analysis. (Full hybridization data available in supplemental Table S1)
| Year | ID | Serovar | pYR1 | pSN254 | IncA/C Core Regions Detecteda | Plasmid Replicons | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2007 | CDC1 | Typhimurium | 132 | 36 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, I1 |
| 2007 | CDC2 | Typhimurium | 90 | 29 | 1 | 2 | 3 | 4 | 5 | – | – | – | – | 10 | 11 | 12 | A/C, I1 |
| 2007 | CDC3 | Typhimurium | 1 | 12 | – | – | – | – | – | – | – | – | – | – | – | – | FIA, FII |
| 2007 | CDC4 | Heidelberg | 105 | 32 | 1 | 2 | 3 | 4 | 5 | – | – | 8 | 9 | 10 | 11 | 12 | A/C |
| 2007 | CDC5 | Heidelberg | 105 | 31 | 1 | 2 | 3 | 4 | 5 | – | – | 8 | 9 | 10 | 11 | 12 | A/C, N |
| 2007 | CDC6 | Heidelberg | 103 | 32 | 1 | 2 | 3 | 4 | 5 | – | – | 8 | 9 | 10 | 11 | 12 | A/C |
| 2007 | CDC7 | Newport | 90 | 37 | 1 | 2 | 3 | 4 | 5 | – | – | – | – | – | 11 | 12 | A/C, N |
| 2007 | CDC8 | Newport | 131 | 26 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, I1 |
| 2007 | CDC9 | Newport | 136 | 28 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C |
| 2007 | FDA1 | Heidelberg | 15 | 8 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | FDA2 | Typhimurium | 106 | 15 | 1 | 2 | 3 | 4 | 5 | 6 | – | – | – | – | – | – | A/C, I1 |
| 2007 | FDA3 | Heidelberg | 13 | 19 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | FDA4 | Heidelberg | 13 | 8 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | FDA5 | Hadar | 10 | 22 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | FDA6 | Hadar | 7 | 23 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | FDA7 | Hadar | 4 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | N |
| 2007 | FDA8 | Typhimurium | 111 | 22 | 1 | 2 | 3 | 4 | 5 | 6 | – | – | – | – | 11 | 12 | A/C, I1 |
| 2007 | FDA9 | Typhimurium var. 5- | 146 | 19 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | – | 11 | 12 | A/C, N |
| 2007 | USDA1 | Newport | 89 | 30 | 1 | 2 | 3 | 4 | 5 | 6 | – | – | – | 10 | 11 | 12 | A/C, I1 |
| 2007 | USDA2 | Newport | 94 | 32 | 1 | 2 | 3 | 4 | 5 | 6 | – | – | – | 10 | 11 | 12 | A/C, N |
| 2007 | USDA3 | Newport | 82 | 23 | 1 | 2 | 3 | 4 | 5 | 6 | – | – | – | 10 | 11 | 12 | A/C |
| 2007 | USDA4 | Heidelberg | 128 | 36 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, I1 |
| 2007 | USDA5 | Heidelberg | 119 | 33 | – | – | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, N |
| 2007 | USDA6 | Heidelberg | 119 | 33 | – | – | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, N |
| 2007 | USDA7 | Typhimurium | 135 | 36 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, I1 |
| 2007 | USDA8 | Typhimurium | 135 | 40 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, N |
| 2007 | USDA9 | Typhimurium | 127 | 33 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | – | A/C, FIB, I1 |
| 2007 | USDA10 | Heidelberg | 7 | 13 | – | – | – | – | 5 | – | – | – | – | – | – | – | I1 |
| 2007 | USDA11 | Heidelberg | 14 | 5 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | USDA12 | Heidelberg | 144 | 36 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, I1 |
| 2004 | CH1 | Heidelberg | 44 | 8 | – | 2 | 3 | – | 5 | – | – | – | – | 10 | – | 12 | A/C |
| 2005 | CH2 | Typhimurium | 133 | 29 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, I1 |
| 2007 | CH3 | Typhimurium | 96 | 13 | 1 | 2 | 3 | 4 | 5 | 6 | – | – | – | 10 | 11 | 12 | A/C, N |
| 2005 | CH4 | Heidelberg | 80 | 19 | 1 | 2 | 3 | 4 | 5 | – | – | – | – | 10 | 11 | 12 | A/C, N |
| 2005 | CH5 | Heidelberg | 130 | 29 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, N |
| 2005 | CH6 | Enteritidis | 134 | 31 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | A/C, FIB, N |
| 2006 | CH7 | Typhimurium | 1 | 4 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2006 | CH8 | Enteritidis | 1 | 4 | – | – | – | – | – | – | – | – | – | – | – | – | FIB, N |
| 2007 | CR1 | Typhimurium | 17 | 22 | – | 2 | – | – | – | – | – | – | – | – | – | – | FIB, N |
| 2007 | CR2 | Heidelberg | 9 | 6 | – | – | – | – | – | – | – | – | – | – | – | – | HI1 |
| 2007 | CR3 | Typhimurium | 1 | 6 | – | – | – | – | – | – | – | – | – | – | – | – | FIB, N |
| 2007 | CR4 | Hadar | 9 | 22 | – | – | – | – | – | – | – | – | – | – | – | – | I1, N |
| 2007 | CR5 | Typhimurium | 1 | 6 | – | – | – | – | – | – | – | – | – | – | – | – | FIB, N |
| 2007 | CR6 | Hadar | 4 | 4 | – | – | – | – | – | – | – | – | – | – | – | – | N |
| 2007 | CR7 | Heidelberg | 0 | 14 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | CR8 | Heidelberg | 0 | 19 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | CR9 | Typhimurium | 0 | 8 | – | – | – | – | – | – | – | – | – | – | – | – | N |
| 2005 | CA1 | Saintpaul | 9 | 27 | – | – | 3 | – | – | – | – | – | – | – | – | – | P, N |
| 2007 | CA2 | Heidelberg | 28 | 24 | – | 2 | – | – | – | – | – | – | 9 | – | – | – | I1 |
| 2007 | CA3 | Typhimurium | 2 | 10 | – | – | – | – | – | – | – | – | – | – | – | – | FIB, FII, N, F |
| 2007 | CA4 | Hadar | 4 | 7 | – | – | – | – | – | – | – | – | – | – | – | – | nd |
| 2007 | CA5 | Heidelberg | 8 | 4 | – | – | – | – | – | – | – | – | – | – | – | – | FII, I1, F |
| 2007 | CA6 | Typhimurium var. 5- | 2 | 7 | – | – | – | – | – | – | – | – | – | – | – | – | FIB |
| 2007 | CA7 | Typhimurium | 3 | 7 | – | – | – | – | – | – | – | – | – | – | – | – | I1 |
| 2007 | CA8 | Typhimurium | 1 | 10 | – | – | – | – | – | – | – | – | – | – | – | – | FIB, N |
| 2007 | CA9 | Typhimurium | 3 | 9 | – | – | – | – | – | – | – | – | – | – | – | – | FIB, N |
|
| |||||||||||||||||
| Total on array | 180 | 47 | |||||||||||||||
IncA/C plasmid core regions are based upon the assay by Welch et al. 2007. Core regions are numbered in order 1 to 12 around the plus strand of the IncA/C plasmid sequence. Regions are scored as present if all genes within them were detected by microarray analysis of the isolate. These regions are identified with the full hybridization data in supplemental Table S1.
nd: none detected
Figure 1.

Cluster analysis of Salmonella enterica isolates based on results of microarray detection of antimicrobial resistance and plasmid genes. The IncA/C positive cluster, group A, and IncA/C negative cluster, group B are labeled and indicated by brackets. Complete cluster analysis data is available in Supplemental Figure S1. Resistance profiles of each isolate is listed with the following abbreviations for the antimicrobials: AMC, amoxicillin-clavulanic acid; AMK, amikacin; AMP, ampicillin; CHL, chloramphenicol; CIP, ciprofloxacin; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; SMX, sulfamethoxazole; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TCY, tetracycline; TIO, ceftiofur. Antimicrobial resistance genes detected are summarized for each isolate. Genes and gene families with multiple probes are summarized and presented only once with different genes and alleles of related genes separated by a comma; full hybridization data are available in Supplementary Table S1. Replicons detected in each isolate by PCR analysis are also listed with (−) indicating that none of the replicons assayed were detected.
Detection of antimicrobial resistance and plasmid genes by microarray analysis
Resistance genes consistent with the observed phenotypes were detected in the isolates. A summary of the genes detected most frequently in the isolates is in Figure 1 (complete hybridization data available in supplemental table S1); an exception was isolate CH1, where only a strA/B gene was detected. The most prevalent aminoglycoside resistance genes detected were alleles of aac(3) (24/56), aac(6′) (22/56), aadA (34/56), aadB (27/56), aadE (14/56), aph (21/56), and strAB (39/56). Detected β-lactam resistance associated genes included blaTEM (32/56), blaCMY-2 (30/56), blaPSE-1 (36/56), and the β-lactamase resistance transcriptional regulatory gene ampR (5/56). Chloramphenicol resistance genes included cat (36/56), floR (27/56) and cmlA (7/56). Sulfamethoxazole resistance genes in the isolates include sulI (26/56) and sulII (23/56). Frequently detected tetracycline resistance associated genes were tet(A) (45/56), tet(B) (8/56), tet(C) (8/56), tet(D) (7/56) and the regulatory gene tetR (50/56). Also detected in several isolates were alleles of trimethoprim resistance genes, such as dfr (18/56).
Many of the isolates hybridized to multiple probes for IncA/C and HI1 plasmid genes. Twenty-seven isolates hybridized to a large number of IncA/C plasmid genes including genes in core regions used to define the IncA/C plasmid backbone (Table 1).31,42 Detection of these core regions, based upon sequence data from the pYR1 and pSN254 plasmids42, indicate that IncA/C plasmids are likely present in these 27 isolates. Eight of nine U.S. human isolates had IncA/C plasmid genes detected, three of which contained genes in all 12 core regions. The remaining isolates were missing genes in core regions 6–7, 6–9 or 6–10, which have been identified as a region of insertions and deletions (indels) in the family of IncA/C plasmids.24,31,32,42 Three of nine U.S. retail meat isolates contained IncA/C plasmid genes but none of the isolates contained genes in all 12 core regions. One isolate, FDA9, is missing genes in region 10, another, FDA8, is missing regions 7–10 and another isolate, FDA2, is missing genes in regions 7–12. Four of the 10 U.S. animal isolates that contain IncA/C plasmids hybridized to genes in all 12 core regions. The remaining six isolates are missing genes in either regions 1–2, 7–9, or region 10–12. Five of 8 Canadian isolates from humans likely contain IncA/C plasmids. Three of the Canadian IncA/C positive human isolates contained genes in all 12 core regions and two isolates were missing genes in the indel at regions 6–9 or 7–9. Canadian Human Isolate CH1 had a different pattern of core IncA/C genes detected than all isolates in that group, with IncA/C regions 1, 4, 6–9, and 11 being missing. None of the Canadian retail meat or animal isolates had IncA/C genes detected in a large enough number to indicate the presence of IncA/C plasmids. However, one of the Canadian retail meat isolates, CR2, was the only isolate in the study with numerous IncHI1 plasmid genes detected (183/206), most likely indicating the presence of an HI1 plasmid.24
Plasmid replicon typing
Replicons were detected in all isolates except Canadian animal isolate CA4. The IncA/C (27/56), I1 (25/56), N (23/56) and FIB (10/56) were the most prevalent replicons detected among all isolates (Figure 1 and Table 1). The IncA/C replicon was detected in all of the 27 isolates where large numbers IncA/C genes (>52) were detected by microarray analysis, confirming the presence of IncA/C plasmids in these isolates. These included U.S. human, retail and slaughter isolates, as well as Canadian human isolates. However, the IncA/C replicon was not detected in Canadian retail or animal slaughter isolates. Plasmid replicons I1 and N were widespread and detected in isolates from all sources in both countries. Replicons FII, FIA, P, F, and HI1 were present in fewer isolates. Most isolates (n=51) contained one or two replicon types, three isolates were positive for three replicon types and a single isolate was positive for four replicons. Overall, half of these 18 replicon types associated with Enterobacteriaceae were detected in at least one of the isolates.9
Cluster and LD analysis
Almost all of the Salmonella isolates fell into two major groups based upon the antimicrobial resistance and plasmid genes detected. These were group A (n=27) with numerous IncA/C plasmid genes detected and group B (n=27) with very few IncA/C genes detected (Figure 1 and Table 1). Two isolates, CR2 and CA2, were an exception to this and did not group together or with any of the other isolates. For CR2, this was due to the detection of an almost complete HI1 plasmid making its genes detected unique from all other isolates in the study. While isolate CA2 had several IncA/C plasmid genes detected, it did not have enough to cluster with group A, and also was not PCR positive for the IncA/C replicon. However, CA2 had too many IncA/C genes detected to cluster with group B and thus was a lone isolate. All 27 isolates of group A were IncA/C positive via microarray analysis as well as plasmid replicon typing. CH1, an isolate from a human in Canada, was different from other isolates in group A. This isolate did not hybridize with as many IncA/C gene probes as other isolates in group A, but five of the core regions were detected (2, 3, 5, 10, and 12) as was the IncA/C replicon by PCR analysis (Table 1). Group A consists mostly of U.S. slaughter, U.S. human and Canadian human isolates, although some U.S. retail meat isolates were present in this cluster. In addition to IncA/C, replicons I1, N and FIB were detected in group A. Cluster B contained 27 isolates in which IncA/C was not detected by microarray analysis or replicon typing. This cluster consists mostly of Canadian retail, Canadian animal slaughter and U.S. retail meat isolates. However, U.S. slaughter, U.S. human and Canadian human isolates were present in lower numbers. Replicons I1, FIB, N, P, FIA, and FII were also frequently detected in this cluster.
Linkage disequilibrium (LD) analysis detected significant linkage between the isolates’ plasmid replicon, serovar, source agency, and animal source (cattle, chicken, swine, or turkey and their retail meat products, and also human isolates) (Figure 2). Significant linkage often corresponded with the separation of isolates into groups A and B by the cluster analysis of the microarray data. Most notably IncA/C had significant linkage with serovar, agency, and animal source, which are reflected in clusters A and B. All six Salmonella Newport isolates were in group A and contained IncA/C; while none of the six Salmonella Hadar isolates contained IncA/C and thus were in group B, leading to LD between IncA/C and these serovars. Significant positive LD was also detected for IncA/C to human isolates, with 14 out of 17 human isolates in group A; likewise, IncA/C was linked to cattle isolates, with all five in group A. The animal or animal meat product from which the isolate was collected also demonstrated a negative association with IncA/C. Only one of the 12 swine isolates and one of nine turkey isolates contained IncA/C plasmids and were in group A, while 10 of the remaining swine and all eight remaining turkey isolates were found in group B. These linkages suggest that the groups formed by clustering isolates based upon antimicrobial and plasmid genes are significantly different from each other and reflect gene content associated with specific replicons, serovars, agencies, and animal sources.
Figure 2.
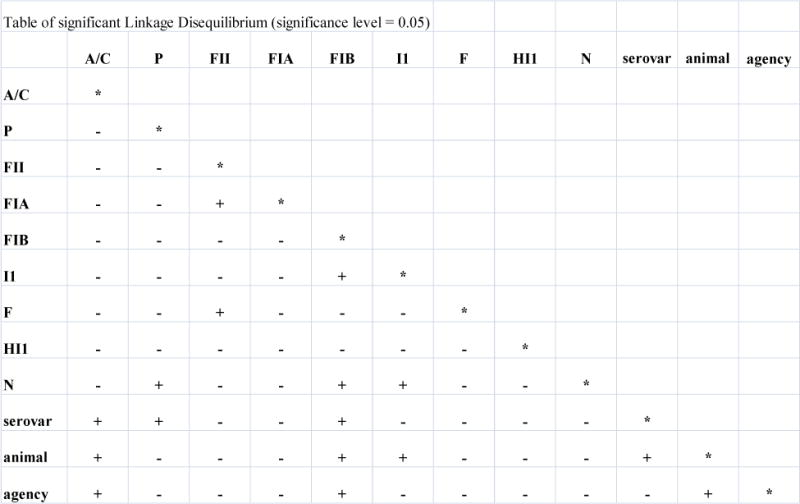
Pairwise linkage disequilibrium based on nine plasmid replicon groups, serovar, phage type and animal source of Salmonella Typhimurium strains. (+) indicates a P value of ≤0.05 or less and significant linkage disequilibrium; and a (−) indicates a P value of greater than >0.05 and no significant linkage disequilibrium. Replicon groups B/O, FIC, T, K, W, Y, FIA, X, L/M were not present in any isolate and were subsequently removed from the analysis.
Animal and IncI1 were linked due to eight out of nine turkey isolates having the IncI1 replicon detected; conversely, only four out of 12 swine isolates had the IncI1 replicon. IncI1 also had a negative relationship with IncN attributable to only one of 23 IncN positive isolates also had the IncI1 replicon detected. Replicon FIB showed linkage with serovar, agency, and animal due to eight isolates being Typhimurium, only one isolate was from a U.S. agency (USDA), and six were from swine. Replicon FIB was also detected in eight of the isolates found in group B, and thus had a negative association with IncA/C and group A. However, some linkages are less clear due to the small number of isolates with the replicon detected. Linkages with IncF, FIA, FII, and P were detected, but are suspect due to only one or two of these replicons being detected in all of the isolates. For example, IncP is in disequilibrium with serovar and IncN, but only one P replicon was detected which is not a sampling large enough to determine if this association is representative of the bacterial population. Animal and agency also showed significant LD, which is likely an artifact due to no Canadian isolates were available from beef or cattle for this study.
Discussion
To investigate MDR in Salmonella enterica isolated from animals, retail meat, and infected humans, a small sampling of the most frequently isolated MDR serovars in the U.S. and Canada were analyzed to determine genetic mechanisms associated with their antimicrobial resistance. Overall, many of the antimicrobial resistance genes detected in the study have been previously found in MDR isolates from the U.S. and Canada in other studies using similar methods or other complementary methods.24,30,31,42 The large number of genes detected in this study and their combination with plasmid genes and plasmid replicon typing allows a better understanding of MDR in Salmonella isolates from these sources. Cluster analysis based on the genes detected divided the isolates into two large clusters: A, with IncA/C plasmids and B, without IncA/C plasmids. This reveals the impact of IncA/C plasmids on MDR in U.S and Canadian isolates, while also implicating other genetic elements that are also associated with MDR in Salmonella.
The resistance genes detected within group A and group B were shared at a high level, indicating the presence of common mechanisms. Antimicrobial resistance genes usually found in IncA/C plasmids (e.g. blaCMY-2, floR, aac(3), aadA, aphA1, strA/B, sulI, sulII, dfrA, tet(A), tet(B), tet(C), tet(D), and tetR) were detected in most isolates of group A.24,30,31,42 Isolates in group B had resistance genes indicative of integrons like SGI-1 or other MDR plasmids (e.g. blaPSE-1, blaTEM, floR, aac(3), aadA, sulI, dfrA, tet(A/G), and tetR).24,30,35 In Addition, many of the resistance genes found in isolates from both group A and B were also similar, in spite of the different genetic vehicles for these MDR genes.24,30,35 This may reflect the cassette nature of many of these resistance genes, where the genes encoding MDR can be transferred to plasmids, to integrons, and/or to the chromosome resulting in different genetic elements carrying similar resistance genes. For example, many IncA/C plasmids and a variety of Salmonella Genomic Island-1 (SGI-1) sequences have been shown to share resistance genes cassettes.24,30 In addition, IncA/C plasmids have been shown to mobilize genetic elements such as SGI-1 and antimicrobial resistance gene cassettes in-trans.14 This type of movement could explain many of the genes found in common throughout these MDR isolates.
Certain resistance genes were also found to be widespread among these isolates and between the two clusters. For example, the blaCMY-2 gene that encodes resistance to extended spectrum cephalosporins and other beta-lactams was found in isolates from both cluster A and B.11,21,22 MDR IncA/C plasmids that carry the blaCMY-2 gene have been reported and characterized as a major contributor to MDR in Salmonella from the U.S. and Canada.3,11,21,22 In IncA/C positive group A, 23/27 isolates have the blaCMY-2 gene detected. However, 7/27 isolates in group B also have the blaCMY-2 gene detected. Five of these are Salmonella Heidelberg isolates which also have the IncI1 plasmid replicon detected. IncI1 plasmids carrying a blaCMY-2 gene have recently been found in Salmonella Heidelberg human isolates in the U.S.19,20 Therefore, it is likely that there are two or more genetic elements carrying the blaCMY-2 genes in MDR Salmonella from in these isolates. The prevalence of MDR Salmonella resistant to cephalosporins due to blaCMY-2 encoding IncA/C plasmids has declined recently in animal isolates; however, the spread of a blaCMY-2 gene by other genetic elements, such as IncI1, could cause increased prevalence of Salmonella resistant to extended spectrum cephalosporins in the future.
A large amount of variability was found in the core backbone regions of the IncA/C plasmids detected in group A. Most of the regions missing are previously described indels within regions 6–10, 1–2 and 7–12.24,31,32,42 The genes encoded in these indels have been observed to have effects on a great diversity of plasmid functions. Some of these include: transmissibility of the plasmids, mobilization of other genetic elements, phenotypes conferred by the plasmids, response to selective pressures including heavy metals, chemical sanitizers, and traditional antimicrobials, as well as phylogenetic implications.24,31,32,42 These aspects of the IncA/C plasmid family have been well characterized in other studies of Salmonella as well as other bacteria containing IncA/C plasmids.24,31–33,41,42 Interestingly, while IncA/C is associated with almost half of the MDR Salmonella isolates in this study, there is a large amount of diversity within the structure of the plasmids in these isolates, possibly indicating several different phylogenetic origins of the IncA/C plasmids in these Salmonella isolates and even within a single serovar of Salmonella. This suggests that there are multiple sources of MDR IncA/C Salmonella in these isolates rather than the spread of a single IncA/C plasmid among the different MDR Salmonella found in animals, retail meat, and human infections. Alternatively, the plasmids may have spread some time ago and then undergone mutations and deletions within certain serovars or in certain environments resulting in the diversity we have observed.
Other plasmids that could potentially carry MDR genes were detected in each of the large clusters, including IncI1 which has been previously shown to carry blaCMY-2 or MDR in bacteria isolated from humans and animals.19,20,25,26 IncN, IncFIB, and other plasmid replicons were also detected demonstrating that the genetic elements carrying MDR, while dominantly associated with IncA/C and I1, are complex and could be associated with other plasmids and genetic elements such as integrons like Salmonella Genomic Island 1 (SGI1).24,30,35 This observation is also supported by the two lone MDR isolates that are genetically distinct from the two large clusters, including one carrying an IncHI1 plasmid which is often associated with MDR in Typhoid and Paratyphoid serovars of Salmonella.36,40
Within both large clusters, isolates which grouped together into sub-clusters also had other similarities. The isolates were often the same serovar when isolated from the same type of animal or animal meat, or when isolated by the same agency or in the same country. This was especially true for human isolates which clustered most closely to other human isolates and in some cases human isolates from the U.S. and Canada clustered together. However, human isolates did not clustered with animal or meat isolates. The clustering did show that 14 out of 17 human isolates were associated with IncA/C, a relationship between source and IncA/C that exhibits significant LD. This likely reflects an association between MDR human salmonellosis and Salmonella serovars, such as Newport, Heidelberg, Typhimurium, and others that carry a plasmid from the MDR IncA/C family.19,20,24 This association has been described in several reports on MDR human isolates collected during this time period (2005–2007), and emphasizes the impact of IncA/C plasmids on U.S. and Canadian human MDR Salmonella infections.4,15,19,20,24 U.S. animal MDR Salmonella isolates were also found to be mostly IncA/C positive (10/12), as were three of the U.S. retail meat isolates. In contrast all Canadian animal and retail meat isolates lacked the IncA/C plasmid7,21,22,24,28,30. Canadian monitoring of cattle and beef had previously detected MDR Salmonella with the IncA/C plasmid, however sampling of cattle and beef in Canada was discontinued before this study.
Human infections caused by MDR Salmonella are usually thought to be foodborne, with animals serving as reservoirs of resistance and retail meats acting as a vector for human disease.1,2,43 Agencies in the U.S. and Canada have developed programs to monitor the levels of antimicrobial resistance in Salmonella found in food animals, retail meats, and humans. This study was designed to investigate MDR Salmonella collected by these programs and to determine the genetic elements associated with antimicrobial resistance. Genetic elements, such as IncA/C and IncI1 plasmids were found in most of the isolates, but other plasmids and genetic elements were also associated with MDR in many of these Salmonella. It was also observed that IncA/C was found in most human MDR isolates from both the U.S. and Canada. These results suggest that in addition to phenotypic monitoring of antimicrobial resistance in Salmonella, investigations of the genetic elements causing resistance is necessary to understand the cause, prevalence, and spread of MDR and determine the impact of food borne Salmonella on human health.
Supplementary Material
Figure S1. Cluster analysis of Salmonella Typhimurium isolates based on results of microarray detection of antimicrobial resistance and plasmid genes. Probes with positive hybridizations are indicated with a black block; those with negative hybridizations are indicated with a gray block.
Acknowledgments
The authors thank Jennifer Turpin, Georgina Hidalgo, Jovita Haro, Benny Barrett, and Takiyah Ball at USDA for technical assistance. From CIPARS we thank Lucie Dutil for isolate selection, and Laura Martin and Emily Weir for DNA isolation. We would also like to dedicate this manuscript in memory of our colleague, Lucie Dutil, whom we had the privilege to know and work with through the NARMS and CIPARS collaborations.
Footnotes
Note: The mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, CDC, FDA, or PHAC.
Reference List
- 1.Alcaine SD, Soyer Y, Warnick LD, Su WL, Sukhnanand S, Richards J, Fortes ED, McDonough P, Root TP, Dumas NB, Grohn Y, Wiedmann M. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl Environ Microbiol. 2006;72:7575–7585. doi: 10.1128/AEM.01174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaine SD, Warnick LD, Wiedmann M. Antimicrobial resistance in nontyphoidal Salmonella. J Food Prot. 2007;70:780–790. doi: 10.4315/0362-028x-70.3.780. [DOI] [PubMed] [Google Scholar]
- 3.Allen KJ, Poppe C. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by beta-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can J Vet Res. 2002;66:137–144. [PMC free article] [PubMed] [Google Scholar]
- 4.Andrysiak AK, Olson AB, Tracz DM, Dore K, Irwin R, Ng LK, Gilmour MW. Genetic characterization of clinical and agri-food isolates of multi drug resistant Salmonella enterica serovar Heidelberg from Canada. BMC Microbiol. 2008;8:89. doi: 10.1186/1471-2180-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob Agents Chemother. 2005;49:836–839. doi: 10.1128/AAC.49.2.836-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butaye P, Michael GB, Schwarz S, Barrett TJ, Brisabois A, White DG. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect. 2006;8:1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 7.Call DR, Kang MS, Daniels J, Besser TE. Assessing genetic diversity in plasmids from Escherichia coli and Salmonella enterica using a mixed-plasmid microarray. J Appl Microbiol. 2006;100:15–28. doi: 10.1111/j.1365-2672.2005.02775.x. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr Issues Mol Biol. 2003;5:113–122. [PubMed] [Google Scholar]
- 9.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Carattoli A, Miriagou V, Bertini A, Loli A, Colinon C, Villa L, Whichard JM, Rossolini GM. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg Infect Dis. 2006;12:1145–1148. doi: 10.3201/eid1207.051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A, Tosini F, Giles WP, Rupp ME, Hinrichs SH, Angulo FJ, Barrett TJ, Fey PD. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob Agents Chemother. 2002;46:1269–1272. doi: 10.1128/AAC.46.5.1269-1272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Aoust JY. Salmonella Species. In: Doyle MP, Beauchat LR, Montville TJ, editors. Food Microbiology: Fundamentals and Frontiers. ASM Press; Washington D.C: 1997. pp. 129–158. [Google Scholar]
- 13.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 14.Douard G, Praud K, Cloeckaert A, Doublet B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS ONE. 2010;5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutil L, Irwin R, Finley R, Ng LK, Avery B, Boerlin P, Bourgault AM, Cole L, Daignault D, Desruisseau A, Demczuk W, Hoang L, Horsman GB, Ismail J, Jamieson F, Maki A, Pacagnella A, Pillai DR. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg Infect Dis. 2010;16:48–54. doi: 10.3201/eid1601.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorka-Cray PJ, Dargatz D, Petersen K, Tollefson L. (FDA CVM. USDA. 8-16-2005. USDA, ARS).National antimicrobial resistance monitoring system. Veterinary strains. [Google Scholar]
- 19.Folster JP, Pecic G, Bolcen S, Theobald L, Hise K, Carattoli A, Zhao S, McDermott PF, Whichard JM. Characterization of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from humans in the United States. Foodborne Pathog Dis. 2010;7:181–187. doi: 10.1089/fpd.2009.0376. [DOI] [PubMed] [Google Scholar]
- 20.Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM. Characterization of bla(CMY)-Encoding Plasmids Among Salmonella Isolated in the United States in 2007. Foodborne Pathog Dis. 2011;8:1289–1294. doi: 10.1089/fpd.2011.0944. [DOI] [PubMed] [Google Scholar]
- 21.Frye JG, Fedorka-Cray PJ. Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int J Antimicrob Agents. 2007;30:134–142. doi: 10.1016/j.ijantimicag.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Frye JG, Fedorka-Cray PJ, Jackson CR, Rose M. Analysis of Salmonella enterica with Reduced Susceptibility to the Third-Generation Cephalosporin Ceftriaxone Isolated from U.S. Cattle During 2000–2004. Microb Drug Resist. 2008;14:251–258. doi: 10.1089/mdr.2008.0844. [DOI] [PubMed] [Google Scholar]
- 23.Frye JG, Lindsey RL, Rondeau G, Porwollik S, Long F, McClelland M, Jackson CR, Englen MD, Meinersmann RJ, Berrang ME, Davis JA, Barrett JB, Turpin JB, Thitaram SN, Fedorka-Cray PJ. Development of a DNA Microarray to Detect Antimicrobial Resistance Genes Identified in the National Center for Biotechnology Information Database. Microb Drug Resist. 2009;16:9–19. doi: 10.1089/mdr.2009.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glenn LM, Lindsey RL, Frank JF, Meinersmann RJ, Englen MD, Fedorka-Cray PJ, Frye JG. Analysis of Antimicrobial Resistance Genes Detected in Multidrug-Resistant Salmonella enterica Serovar Typhimurium Isolated from Food Animals. Microb Drug Resist. 2011 doi: 10.1089/mdr.2010.0189. [DOI] [PubMed] [Google Scholar]
- 25.Johnson TJ, Jordan D, Kariyawasam S, Stell AL, Bell NP, Wannemuehler YM, Alarcon CF, Li G, Tivendale KA, Logue CM, Nolan LK. Sequence analysis and characterization of a transferable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli. Infect Immun. 2010;78:1931–1942. doi: 10.1128/IAI.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson TJ, Shepard SM, Rivet B, Danzeisen JL, Carattoli A. Comparative genomics and phylogeny of the IncI1 plasmids: A common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid. 2011;66:144–151. doi: 10.1016/j.plasmid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol. 2007;73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang MS, Besser TE, Hancock DD, Porwollik S, McClelland M, Call DR. Identification of specific gene sequences conserved in contemporary epidemic strains of Salmonella enterica. Appl Environ Microbiol. 2006;72:6938–6947. doi: 10.1128/AEM.01368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavigne JP, Blanc-Potard AB. Molecular evolution of Salmonella enterica serovar Typhimurium and pathogenic Escherichia coli: from pathogenesis to therapeutics. Infect Genet Evol. 2008;8:217–226. doi: 10.1016/j.meegid.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Lindsey RL, Fedorka-Cray PJ, Frye JG, Meinersmann RJ. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl Environ Microbiol. 2009;75:1908–1915. doi: 10.1128/AEM.02228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsey RL, Frye JG, Fedorka-Cray PJ, Meinersmann RJ. Microarray-based analysis of IncA/C plasmid-associated genes from multidrug-resistant Salmonella enterica. Appl Environ Microbiol. 2011;77:6991–6999. doi: 10.1128/AEM.00567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindsey RL, Frye JG, Fedorka-Cray PJ, Welch TJ, Meinersmann RJ. An oligonucleotide microarray to characterize multidrug resistant plasmids. J Microbiol Methods. 2010;81:96–100. doi: 10.1016/j.mimet.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 33.Lindsey RL, Frye JG, Thitaram SN, Meinersmann RJ, Fedorka-Cray PJ, Englen MD. Characterization of multidrug-resistant Escherichia coli by antimicrobial resistance profiles, plasmid replicon typing, and pulsed-field gel electrophoresis. Microb Drug Resist. 2011;17:157–163. doi: 10.1089/mdr.2010.0148. [DOI] [PubMed] [Google Scholar]
- 34.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, Harkins CR, Wang C, Nguyen C, Berghoff A, Elliott G, Kohlberg S, Strong C, Du F, Carter J, Kremizki C, Layman D, Leonard S, Sun H, Fulton L, Nash W, Miner T, Minx P, Delehaunty K, Fronick C, Magrini V, Nhan M, Warren W, Florea L, Spieth J, Wilson RK. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 35.Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006;8:1915–1922. doi: 10.1016/j.micinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 36.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O’Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–52. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 37.Poole TL, Edrington TS, Brichta-Harhay DM, Carattoli A, Anderson RC, Nisbet DJ. Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack bla(CMY) in the A/C plasmid backbone. Foodborne Pathog Dis. 2009;6:1185–1194. doi: 10.1089/fpd.2009.0316. [DOI] [PubMed] [Google Scholar]
- 38.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 39.Slatkin M. Linkage disequilibrium in growing and stable populations. Genetics. 1994;137:331–336. doi: 10.1093/genetics/137.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wain J, Diem Nga LT, Kidgell C, James K, Fortune S, Song DT, Ali T, Gaora O, Parry C, Parkhill J, Farrar J, White NJ, Dougan G. Molecular analysis of incHI1 antimicrobial resistance plasmids from Salmonella serovar Typhi strains associated with typhoid fever. Antimicrob Agents Chemother. 2003;47:2732–2739. doi: 10.1128/AAC.47.9.2732-2739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welch TJ, Evenhuis J, White DG, McDermott PF, Harbottle H, Miller RA, Griffin M, Wise D. IncA/C plasmid-mediated florfenicol resistance in the catfish pathogen Edwardsiella ictaluri. Antimicrob Agents Chemother. 2009;53:845–846. doi: 10.1128/AAC.01312-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, LeClerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE. 2007;2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao S, Qaiyumi S, Friedman S, Singh R, Foley SL, White DG, McDermott PF, Donkar T, Bolin C, Munro S, Baron EJ, Walker RD. Characterization of Salmonella enterica serotype newport isolated from humans and food animals. J Clin Microbiol. 2003;41:5366–5371. doi: 10.1128/JCM.41.12.5366-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cluster analysis of Salmonella Typhimurium isolates based on results of microarray detection of antimicrobial resistance and plasmid genes. Probes with positive hybridizations are indicated with a black block; those with negative hybridizations are indicated with a gray block.


