Abstract
BK polyomavirus (BKV) is important pathogen for kidney transplant recipients, as it is frequently re-activated, leading to nephropathy. The aim of this study was to investigate the phylogenetic reconstruction and polymorphism of the VP2 gene in BKV isolated from Chinese kidney transplant recipients. Phylogenetic analysis was carried out in the VP2 region from 135 BKV-positive samples and 28 reference strains retrieved from GenBank. The unweighted pair-group method with arithmetic mean (UPGMA) grouped all strains into subtypes, but failed to subdivide strains into subgroups. Among the plasma and urine samples, all plasma (23/23) and 82 urine samples (82/95) were identified to contain subtype I; the other 10 urine samples contained subtype IV. A 86-bp fragment was identified as a highly conserved sequence. Following alignment with 36 published BKV sequences from China, 92 sites of polymorphism were identified, including 11 single nucleotide polymorphisms (SNPs) prevalent in Chinese individuals and 30 SNPs that were specific to the two predominant subtypes I and IV. The limitations of the VP2 gene segment in subgrouping were confirmed by phylogenetic analysis. The conserved sequence and polymorphism identified in this study may be helpful in the detection and genotyping of BKV.
Keywords: BK polyomavirus, VP2 gene, genotypes, polymorphism, Chinese
Introduction
BK polyomavirus (BKV), which belongs to the genus Polyomavirus of the family Polyomaviridae, is a ubiquitous double-stranded DNA virus prevalent in humans. It remains latent in the urogenital tract or mononuclear cells following a primary infection during childhood. Its serum positive rate reaches up to 90% in adults (1–3).
BKV rarely activates in immune-competent individuals, while it reactivates frequently in renal transplant recipients under immunosuppressive conditions. At an early stage, 10–60% of transplant recipients exhibit asymptomatic viruria. Viruria can also be observed at a late stage (4). As BKV DNA is detected in peripheral blood, it reflects the continuous replication of BKV and the destruction of targeted cells. A high number of copies of BKV DNA in the blood is associated with an increased incidence of BKV-associated nephropathy (BKVN), ureteral stenosis and hemorrhagic cystitis. Among these, BKVN is the most common. It mainly manifests as renal tubular necrosis or interstitial nephritis. Eventually, it results in the loss of graft (5–7).
The BKV genome is a double-stranded DNA that comprises ~5 kb. It is generally divided functionally into three parts: An early encoding region that encodes small and large T-antigens, a late encoding region that encodes the structural proteins VP1–3, as well as one that encodes agnoprotein (8,9). BKV can be genotyped by molecular phylogenetic analysis and serological typing. Based on restriction fragment length polymorphism and comparisons of partial VP1 sequences, a classification system comprising four subtypes I–IV has been introduced (10). This system corresponds to a previous serological typing scheme (11). According to mutation of the VP1 sequence, subtypes I and IV are divided further into four subgroups (I/a, I/b-1, I/b-2, I/c) and six subgroups (IV/a-1, IV/a-2, IV/b-1, IV/b-2, IV/c-1, IV/c-2), respectively (12,13). BKV subtypes show a specific geographic distribution worldwide. It is reported that subtype I is prevalent throughout the world, while subtype IV is prevalent mostly in East Asia (14). There are numerous mutation sites in the BKV genome, and the most polymorphic coding region in the viral genome is VP1. Significant variation is also present in the large T-antigen gene, wherein polymorphisms are found in 11.39% of all nucleotide sites (15). BKV polymorphism loci can also be obtained through comparisons of non-coding control region (NCCR) regions (16). Studies focusing on the polymorphism of the VP1 gene and mutations in amino acid residues encoded by the VP1 gene have developed quickly (17). In the majority of cases, researchers have opted to reconstruct a phylogenetic tree on the basis of the VP1 gene (18,19). However, investigation of the BKV VP2 gene has been limited. In 2009, Luo et al conducted the first genotyping of BKV using the BKV VP2 gene (20). However, considering the variable genetic background of BKV in that study, whether the conclusions can be applied to the Chinese population merits investigation. Studies of single nucleotide polymorphisms (SNPs) and BKV prevalence in the Chinese population are scarce, and the BKV genotyping and polymorphism analysis of Chinese samples has rarely been reported. The principal aims of the present study were to subtype BKV from a phylogenetic standpoint, identify the conserved sequence, and determine SNPs in the VP2 gene. Additionally, SNP and genotyping analysis were conducted specifically on samples from a Chinese population.
Materials and methods
Patients and samples
A total of 135 adult renal transplant recipients (69 males and 66 females) from the Transplantation Center of the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between April 2010 and November 2012 were included. The clinical samples consisted of 95 urine samples, 23 plasma samples and 17 other samples. They were collected after explaining the purpose of the research and obtaining written informed consent from the patients. None of the patients developed nephropathy or renal transplantation dysfunction during the study. This research was approved by the Ethical Decision Committee of the Research Administration at Wenzhou Medical University.
DNA extraction
BKV DNA was extracted from samples using the DNA Fast 2000 DNA Extraction kit (Shanghai Fastagen Biotechnology Co., Ltd., Shanghai, China) according to the manufacturer's specifications. Centrifugation at 800 × g for 10 min was conducted to separate 100 µl urinary sediment from 10 ml urine. In addition, 100 µl plasma with leukocytes was extracted from 2 ml whole blood after standing for 5–15 min. A 100-µl volume was used for other samples. DNA was then extracted from pretreated samples following the respective instructions.
Nested polymerase chain reaction (PCR) and sequencing
Amplification of the VP2 gene fragment was conducted by nested PCR, using TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China). Two pairs of specific primers were designed using Primer Premier 5 software (Premier Biosoft International, Palo Alto, CA, USA) to select a highly conserved region of the BKV (BK polyomavirus DNA, complete genome, isolate: TW-2, GenBank accession number: AB213487.1). The pair of outer primers was as follows: Upstream primer, 5′-TGGGACCTAGTTGCCAGTGTATC-3′ (676–697) and downstream primer, 5′-AAGAGCAGGTGTTACAGTCCC-3′ (1557–1577). The pair of inner primers was as follows: Upstream primer, 5′-AAGTTCAAATTGCATCCCT-3′ (766–784) and downstream primer, 5′-TTCTTTGATTAGCACCTCCTG-3′ (1495–1515). The specificity of these primers was tested using the Primer-BLAST tool provided by the National Center for Biotechnology Information (NCBI: http://www.ncbi.nlm.nih.gov/), using default parameters. DNA extracted from clinical specimens was amplified by nested PCR with the aforementioned primers. Agarose gel electrophoresis was performed for the relatively qualitative screening of BKV-positive samples and to judge the credibility of the PCR product. BKV-positive PCR products were sequenced by Sun Biotech Co. Ltd. (Beijing, China). The polymorphic sites of a 684-bp sequence in the VP2 gene were selected for analysis. The sequence analysis utilized BioEdit version 7.0.2 (Ibis Biosciences, Carlsbad, CA, USA). DNAMAN version 5.2.10 (Lynnon Biosoft, San Ramon, CA, USA) and DNAStar bioinformatics software version 7.1 (DNASTAR Inc., Madison, WI, USA).
Extracted reference sequence of BKV strains for phylogenetic analysis
Twenty-eight BKV whole-genome sequences, the majority of which were from China, were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The subtypes and accession numbers of the 28 virus strains used for the sequence analysis are shown in Table I. Corresponding fragments of reference strains in the VP2 gene were sorted using DNAMAN software.
Table I.
Reference BKV strains in the phylogenetic analysis.
| Virus isolate | Subtype/subgroup | Geographical origin | GenBank accession |
|---|---|---|---|
| DUN | I/a | USA | NC_001538 |
| WW | I/b-1 | South Africa | AB211371 |
| DIK | I/b-1 | Netherlands | AB211369 |
| FNL-12 | I/b-2 | Finland | AB263918 |
| JL | I/b-2 | Netherlands | AB211370 |
| SHA-62 | I/c | China | AB365175 |
| SHA-70 | I/c | China | AB365176 |
| FUJ-4 | I/c | China | AB369095 |
| FUJ-6 | I/c | China | AB269826 |
| GBR-12 | II | England | AB263920 |
| ETH-3 | II | Ethiopia | AB263916 |
| AS | III | England | M23122 |
| SEC-3 | IV/a-1 | China | AB269860 |
| SHA-43 | IV/a-1 | China | AB365171 |
| RYU-3 | IV/a-2 | Japan | AB211389 |
| FUJ-13 | IV/a-2 | China | AB269826 |
| THK-8 | IV/b-1 | Japan | AB211390 |
| TW-3 | IV/b-1 | Japan | AB211391 |
| KOM-2 | IV/b-2 | Japan | AB211387 |
| JPN-15 | IV/b-2 | Japan | AB269834 |
| FUJ-32 | IV/c-1 | China | AB269828 |
| SHA-78 | IV/c-1 | China | AB365178 |
| NEC-4 | IV/c-1 | China | AB269854 |
| NWC-8 | IV/c-1 | China | AB269858 |
| NWC-14 | IV/c-1 | China | AB269855 |
| SWC-1 | IV/c-1 | China | AB269863 |
| SWC-2 | IV/c-1 | China | AB269864 |
| SWC-4 | IV/c-1 | China | AB269865 |
BKV, BK polyomavirus.
Phylogenetic analysis
Using phylogenetic analysis, VP2 sequences of BKV from the 135 specimens were aligned with the 28 recently published sequences of reference strains using NCBI BLAST, DNAMAN and ClustalX version 2.1 software (University College Dublin, Dublin, Ireland). Following the alignment, sequence analysis was performed with ClustalX software. Neighbor joining (NJ) and an un-weighted pair-group method with arithmetic means (UPGMA) trees were constructed using MEGA software, version −5.03 (Mega Software, Philadelphia, PA, USA). To estimate the confidence level of the branching patterns of the tree, bootstrap resampling tests were performed 1,000 times.
SNP analysis of urine and plasma samples in a Chinese population
For SNP analysis, 36 reference strains isolated from a population in China were extracted from GenBank. They included 18 sequences of subtype I and 18 sequences of subtype IV. The 118 specimen strains from plasma or urine (samples from other sources were excluded) together with 36 reference strains were analyzed to determine the SNP positions using DNAMAN software. These positions can be used to distinguish between subtypes I and IV in China. The subtypes and accession numbers of the 36 reference strains are shown in Table II.
Table II.
Extracted reference BKV sequences from China.
| Strain | Subtype | Sample source | GenBank accession |
|---|---|---|---|
| FUJ-13 | IV | Urine | AB269826 |
| NEC-15 | IV | Urine | AB269856 |
| NEC-7 | I | Urine | AB365144 |
| NEC-12 | I | Urine | AB263930 |
| NWC-7 | IV | Urine | AB269857 |
| NWC-14 | IV | Urine | AB269855 |
| SEC-3 | IV | Urine | AB269860 |
| SEC-21 | IV | Urine | AB269862 |
| SHA-7 | I | Urine | AB365159 |
| SHA-10 | I | Urine | AB365161 |
| SHA-22 | I | Urine | AB365164 |
| SHA-25 | I | Urine | AB365166 |
| SHA-30 | IV | Urine | AB365168 |
| SHA-41 | I | Urine | AB365170 |
| SHA-47 | IV | Urine | AB365172 |
| SHA-56 | I | Urine | AB365174 |
| SHA-72 | I | Urine | AB365177 |
| SWC-2 | IV | Urine | AB269864 |
| NEC-4 | I | Urine | AB269854 |
| NEC-22 | IV | Urine | AB365146 |
| NEC-8 | I | Urine | AB263931 |
| NEC-14 | IV | Urine | AB269852 |
| NWC-8 | IV | Urine | AB269858 |
| NWC-15 | IV | Urine | AB269856 |
| SEC-6 | IV | Urine | AB269861 |
| SHA-4 | I | Urine | AB365158 |
| SHA-13 | I | Urine | AB365162 |
| SHA-19 | I | Urine | AB365163 |
| SHA-23 | I | Urine | AB365165 |
| SHA-28 | IV | Urine | AB365167 |
| SHA-40 | I | Urine | AB365169 |
| SHA-43 | IV | Urine | AB365171 |
| SHA-55 | IV | Urine | AB365173 |
| SHA-62 | I | Urine | AB365175 |
| SWC-1 | IV | Urine | AB269863 |
| SWC-4 | IV | Urine | AB269865 |
BKV, BK polyomavirus.
Statistical analysis
All statistical analyses were conducted using SPSS software, version 18.0 (IBM SPSS, Armonk, NY, USA). Differences in the viral subtypes obtained from plasma and urine samples were compared using Fisher's exact test. P<0.05 was considered to indicate a statistically significant result.
Results
Amplification using nested PCR
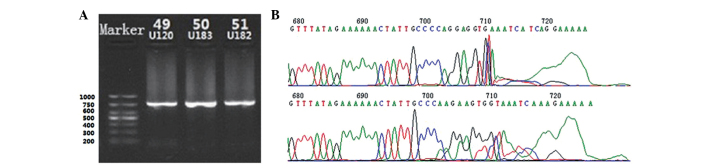
Following amplification by nested PCR with an inner and outer primer, corresponding BKV VP2 gene products with a total length of 750 bp were obtained. The products were verified by agarose gel electrophoresis (Fig. 1A). DNA sequencing results confirmed that the products were the target gene (Fig. 1B).
Figure 1.
(A) Electrophoresis of the nested PCR products of clinically isolated BKV VP2 DNA fragments showed that the PCR products were specific 750-bp DNA segments of the VP2 gene (inner product). (B) PCR product sequences were confirmed as the VP2 gene by ABI chromatogram (terminal part of the sequencing). BKV, BK polyomavirus; PCR, polymerase chain reaction.
Phylogenetic analysis
UPMGA method
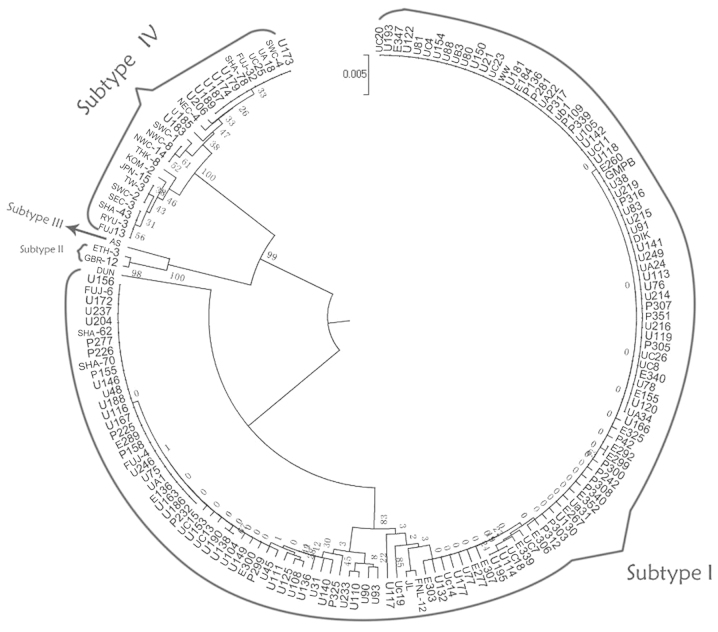
A UPGMA phylogenetic tree (Fig. 2) was constructed for partial segments of the VP2 gene and the 163 strains (135 sample strains and 28 reference strains) were grouped into three major branches. Two primary branches had high bootstrap probabilities (BPs), both reaching 99%. Based on the locations of reference strains, the majority of the samples belonged to the largest branch on the phylogenetic tree (subtype I), whereas the rest were ascribed to the other branch (subtype IV). The results were confirmed to have high reliability and were consistent with serology and the genotyping consequences of other gene segments.
Figure 2.
Phylogenetic analysis of the VP2 gene segment by the UPMGA method Strains that were grouped together were labeled with their corresponding genotypes. Three phylogenetic clustering patterns were reproduced by the VP2 gene. The maximal one was subtype I, containing reference strains DIK, WW, JL and DUN. The minimal one included subtype II and III, comprising three strains. The other one belonged to subtype IV. The confidence level between the sequenced VP2 PCR products was expressed by bootstrap probabilities in percentage (%BPs). UPMGA, unweighted pair-group method with arithmetic mean; PCR, polymerase chain reaction.
The largest branch included the reference strains DIK, WW, JL, FNL-12, FUJ-4, FUJ-6, SHA-62, SHA-70 and DUN, which have conventionally been classified as subtype I. DUN belongs to subgroup Ia, as reported previously (19). Subtype Ia was distant from the other clusters at the evolutionary distance shown in Fig. 2. These strains could then be subdivided into corresponding subgroups, for example, DIK and WW belonging to subgroup Ib-1. However, certain clinical isolated strains, for example U93 and U90, could not be classified within any subgroup; the BP values of certain strains, such as UC20 and U193, were as low as 0. These strains indicate the poor credibility of the subgroups when divided.
Another large branch contained reference strains such as FUJ-13 and RYU-3, which were attributed to subtype IV. The bootstrap value of each clade ranged from 25 to 61%, and typing credibility was low. SHA-78 and SWC-4 (IV/c-1) were subdivided into one group, but with a low BP value (32%). By analyzing tiny arborizations of this branch, it was found that it was not possible to precisely classify many of the identified reference strains into correct clades. For example, SEC-3 (IV/a-1), SHA-43 (IV/a-1), RYU-3 (IV/a-2) and FUJ-13 (IV/a-2) clustered in one clade. NEC-4 and NWC-14 (IV/c-1) did not cluster together although they belonged to the same subgroup.
The small cluster in the subtype IV branch included two more detailed clades: One comprised strains GBR-13 and ETH-3, and the other was AS. The former belonged to subtype II. The latter belonged to subtype III. According to the constructed phylogenetic tree, there is a tight evolutionary link between subtype II and III. There were, however, no clinical isolated strains in the cluster.
In the genotyping results, all 23 plasma samples belonged to subtype I. There were 82 samples of subtype I and 13 of subtype IV in 95 urine samples. Thus, there were certain statistical differences between the subtype distribution in the blood and urine samples (P=0.082).
NJ method
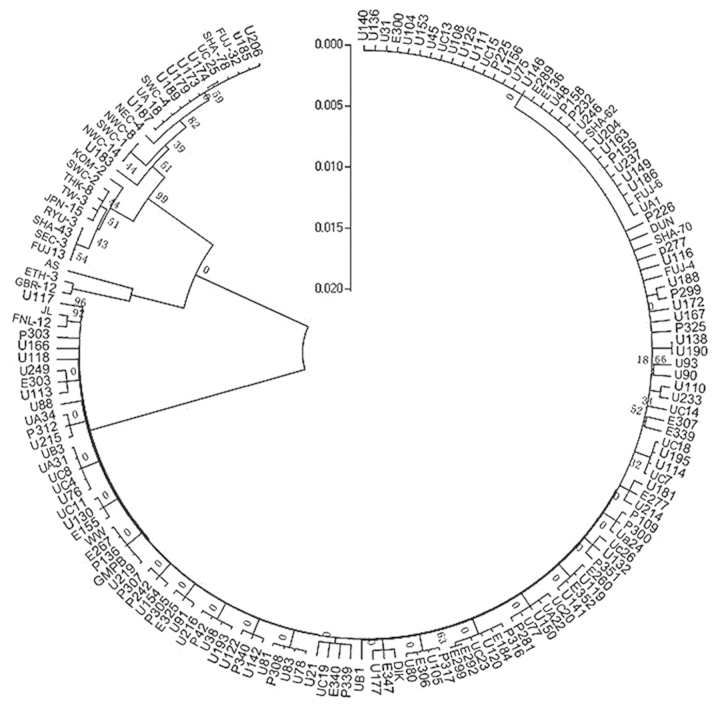
A phylogenetic tree (Fig. 3) was constructed with the NJ method for partial segments of the VP2 gene. Isolates that were analyzed could be divided into two main clades by this tree. However, the BPs values of major branches ranged from 0 to 17%. This indicated the low reliability of the construction results. The confidence of subgroups in the first branch was verified. The bootstrap value was 0. Among three subclusters from the second branch, the third bootstrap value was 99%. Other grouped isolates shared BP values from 7 to 82%. In addition, reference strains of the same subgroup could not be distributed into the same branch, whereas different ones were distributed into the same cluster.
Figure 3.
Phylogenetic analysis of VP2 gene segment by NJ method two major branches were evidently shown. Whereas the BPs values of major branches ranged from 0 to 17%. And reference strains of the same subgroup couldn't be distributed into the same branch, while different ones were distributed into the same cluster.
Application of SNP in specimen strains derived from plasma and urine in genotyping
SNP sites and ubiquitous mutations in Chinese strains
BKV sequences from plasma and urine samples were aligned with 36 reported viral strains obtained from Chinese sources, and 92 SNP sites were identified in total (Table III). In addition, there were 11 SNPs (Table IV) existing in all of them as hot spots. This indicates that these sites may be prevalent in a Chinese population. Among these common SNP sites, G889C, A922G, T1146G, C1272G, G930A and G931A could lead to changes in amino acid residues. The specific transformations are as follows: Serine to threonine (TAG→TAC), lysine to arginine (AAG→AGG), serine to alanine (TCT→GCT), glutamine to glutamic acid (CAA→GAA), and aspartic acid to serine (GAT→AGT). All the aforementioned changes are sense mutations. A905G, G1160A, C1023T, T1049A and G1169A are synonymous mutations.
Table III.
Nucleotide polymorphisms of the VP2 gene segment.
| Position | Subtype I | Subtype IV |
|---|---|---|
| 790 | A | G/A |
| 795 | A | T/A |
| 811 | T | C/T |
| 848 | T | A/T |
| 856 | T | G/T |
| 889 | G | C |
| 894 | A | G/A |
| 899 | T | G/T |
| 902 | C | T/C |
| 905 | A | G |
| 908 | T | C/T |
| 922 | A | G |
| 926 | C | T/C |
| 930 | G | A |
| 931 | A | G |
| 935 | T | C/T |
| 965 | C | G/T/C |
| 970 | A | G/A |
| 975 | T | C/T |
| 986 | T | A/T |
| 989 | G | A/G |
| 990 | G | A/G |
| 996 | T | G/T |
| 1015 | A | G/A |
| 1023 | C | T |
| 1027 | T | C/T |
| 1043 | T | A/T |
| 1049 | T | A |
| 1067 | T | A/T |
| 1068 | G | A/G |
| 1078 | A | G/A |
| 1079 | T | G/A |
| 1083 | G | A/G |
| 1091 | T | C/T |
| 1146 | T | G |
| 1154 | C | A/T/C |
| 1160 | G | A |
| 1166 | G | A/G |
| 1169 | G | G |
| 1174 | G | A/G |
| 1181 | A | G/A |
| 1182 | A | T/A |
| 1187 | T | C/T |
| 1189 | T | C/T |
| 1191 | A | G/A |
| 1200 | T | C/T |
| 1208 | A | G/A |
| 1217 | G | A/G |
| 1226 | C | T/C |
| 1248 | A | G/A |
| 1271 | A | G/A |
| 1272 | C | G |
| 1274 | A | T/A |
| 1284 | G | A/G |
| 1287 | C | T/C |
| 1289 | T | G/T |
| 1295 | T | C/T |
| 1300 | G | A/G |
| 1304 | C | T/C |
| 1316 | A | G/A |
| 1322 | A | G/T/A |
| 1337 | T | A/ |
| 1342 | G | A/G |
| 1343 | T | G/T |
| 1345 | T | C/T |
| 1347 | C | A/C |
| 1361 | T | C/T |
| 1364 | T | C/T |
| 1367 | T | A/T |
| 1385 | T | G/T |
| 1388 | A | G/A |
| 1389 | G | C/G |
| 1392 | G | A/G |
| 1400 | A | C/A |
| 1407 | A | T/A |
| 1408 | T | G/T |
| 1412 | C | G/C |
| 1422 | C | A/C |
| 1425 | C | G/C |
| 1427 | A | G/A |
| 1429 | G | A/C/G |
| 1431 | G | A/G |
| 1440 | G | T/G |
| 1441 | G | C/G |
| 1446 | T | A/T |
| 1449 | A | T/A |
| 1450 | T | A/C/T |
| 1451 | A | G/A |
| 1452 | G | A/G |
| 1457 | A | G/A |
| 1458 | A | G/A |
| 1465 | C | T/C |
DUN was selected as the standard strain.
Table IV.
Prevalent SNP sites in Chinese viral strains.
| Position | 889 | 905 | 922 | 930 | 931 | 1023 | 1049 | 1146 | 1160 | 1169 | 1272 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard strain | G | A | A | G | A | C | T | T | G | G | C |
| Specimen | C | G | G | A | G | T | A | G | A | A | G |
DUN was selected as the standard strain.
SNP sites for distinguishing subtype I and IV
On the basis of the phylogenetic tree, alignment on a partial fragment of the VP2 gene at the nucleotide level was conducted with the aforementioned 154 strains. Thirty SNP sites reflected the difference between subtype I and IV in a Chinese population. These sites are clearly able to distinguish BKV subtypes I and IV (Table V).
Table V.
Thirty sites to distinguish subtypes I and IV.
| Position | Subtype I | Subtype IV |
|---|---|---|
| 848 | T | C |
| 902 | C | T |
| 965 | C | T |
| 986 | T | A |
| 1067 | A | A |
| 1154 | C | A |
| 1181 | A | G |
| 1187 | T | C |
| 1217 | G | A |
| 1248 | A | G |
| 1274 | A | T |
| 1284 | G | A |
| 1287 | C | T |
| 1316 | C | T |
| 1304 | A | G |
| 1322 | G | T |
| 1337 | A/G | A |
| 1342 | T | A |
| 1343 | G | G |
| 1347 | T | A |
| 1361 | C | C |
| 1364 | T | C |
| 1367 | T | A |
| 1389 | G | C |
| 1400 | A | C |
| 1412 | C | G |
| 1422 | C | A |
| 1425 | C | G |
| 1427 | A | G |
| 1429 | G/C | A |
DUN was selected as the standard strain.
Discussion
BKV is the only polyomavirus reported to be divided into four subtypes on the basis of serological and genotyping methods. Four subgroups within subtype I (I/a, I/b-1, I/b-2, I/c) and six subgroups within subtype IV can be further distinguished with nucleotide variation (10,21). Previous studies describe basically four genotyping schemes associated with the phylogenetic construction, where the genotyping is: i) based on the whole BKV genome; ii) based on the complete BKV VP1 gene; iii) based on a part of the BKV VP1 gene; and iv) based on the complete BKV large T antigen (LTA) gene (16,22,23). However, the contributory role of the VP2 gene in genotyping has not been well examined. According to the phylogenetic tree in the present study, the VP2 gene was able to resolve the subtypes, but not the subgroups.
The 940-bp VP2 gene belongs to the late coding region. It encodes viral capsid protein VP2 (9). A complete VP2 gene was first used for genotyping by Luo et al in 2009 (20), and was demonstrated to enable the four subtypes of BKV to be classified, but not their subdivision into subgroups. The results of the present study, using a VP2 gene fragment, are in accordance with those of the study by Luo et al (20). Similar phylogenetic analysis on other regions beyond the VP2 gene or genetic polymorphism analysis is required to differentiate subgroups from subtypes. In the process of phylogenetic analysis, the present study found that the NJ method was not able to produce a proper phylogenetic tree, and indicates that the UPMGA method is better for genotyping. The NJ method is commonly adopted for the VP1 gene (18,19). Therefore, the analysis conducted in the present study indicates that various means of construction should be put into use according to the target gene. It is notable that genotyping based on the VP1 gene yields a clearer definition of subgroups and shorter gene fragments (~300 bp) (22,24). This indicates that VP1 carries more information than the VP2 gene. The VP2 gene failed to subdivide strains into subgroups in the present study of Chinese samples, restricting its further application in BKV genotyping.
Numerous researchers have shed light on the association between the distribution of BKV subtypes and patient populations (14,25,26). The results of these studies have indicated that subtype I predominates in all geographical regions, while subtype IV occurs at lower rates and mostly in East Asia, and subtypes II and III rarely occur. These results are consistent with the observations of the present study. The distribution pattern of BKV subtypes clarified in a study conducted by Zhong et al showed that the frequency of subtype IV was variable among populations, and in northeast USA, Finland, Japan and China, and its frequency ranged between 17 and 36% (27). In the present study, subtype IV accounted for only 7% of the samples, and the study included a greater number of clinical samples than most previous research. The small proportion of subtype IV identified in the present study is inconsistent with the co-migration hypothesis of Zhong et al which suggests that BKV migrates with human race and the proportion of subtype IV in East Asia is relatively large.
With the evident regional characteristics of BKV gene polymorphism, its patterns of mutations differ from place to place. In order to reveal the unique patterns at the nucleic acid level in China, a large-sample investigation among the Chinese population would be of tremendous significance. Therefore, to further this effort, using the phylogenetic tree with UPGMA method, the present polymorphism study targeting BKV strains in China was performed.
A total of 11 SNP sites that are prevalent in BKV isolated from Chinese individuals were identified. Only four sites, 1023, 1146, 1169 and 1272, correspond with those in the global range outcomes reported by Luo et al (20). Specific SNPs for the identification of subtype I or IV in a Chinese population were also analyzed. The specific SNPs that were identified include many previously reported SNPs, but not all of them. Based on this finding, it is notable that BKV in the Chinese population may evolve and mutate in a particular pattern. With an appropriately chosen SNP as a target, defining subgroups would not require sequencing of the entire gene, and would be more simple and rapid, providing an alternative to BKV genotyping. Ultimately, the reliability of phylogenetic reconstruction depends on the accuracy of the algorithm, and no matter what method is adopted, deviation exists (28). The application of SNP genotyping can avoid this problem. The present study of the features and genotyping of BKV in China included a greater number of clinical samples than were included in the majority of studies in this field, and so should be reliable. However, limitations also exist, as the samples were collected from a localized area. Additional sequences from other areas in China are required to confirm the results.
Precise and accurate detection of BKV offers technical support for the monitoring of BKV infection status and the prophylaxis of BKVN in renal transplant recipients. With regard to mutations of the BKV genome, nucleotide-based testing would not detect all BKV DNA. The gold standard of quantitative PCR and a standard substance remain unavailable. The separated conserved region of the VP2 gene may be useful as a new target for PCR or a specific probe assay.
Since the discovery of BKV in 1971 (29), its pathogenicity, virulence, and the mechanism of reactivation have been actively studied. The initial manifestation of BKV reactivation following renal transplantation is asymptomatic viruria. The subsequent viremia and overt nephropathy may culminate in graft loss (30). In earlier studies, researchers conjectured that the severity of infection and viral spread had a close correlation with certain mutants in the amino acid residues of the protein (31). In the present study, it was found that with the exception of a few positions, the majority of the SNPs did not result in variations in amino acids. Whether these mutants influence the expression level of the VP2 protein and cause the change in pathogenicity remains to be elucidated in further research. In the present study, the BKV isolated from blood samples was all of subtype I, while subtype IV was only extracted from urine. It is conceivable that the imbalance in subtypes may have a close affinity to viral virulence or can be accounted for by different stages of infection. Another theory suggests that the intensity of immunosuppression and genetic susceptibility of the host immune system are the principal determinants of BKV-mediated tissue injury in the kidney (32). To verify the validity of these assumptions, an innovative experimental design and new experimental methods are urgently required.
In summary, the construction of a phylogenetic tree based on the VP2 gene of BKV was able to divide BKV into four subtypes, but not into subgroups. Certain prevalent SNPs of BKV in a Chinese population were isolated. The specific positions at which SNPs occur could be used for the identification of subtypes I and IV. A conserved sequence within the VP2 gene was identified. Further studies involving novel genotyping methods may lead to accurate identification of BKV subtypes and elucidate the genetic diversity of BKV.
Acknowledgements
The authors thank Hui-Ying Xu for editing the manuscript. The study was supported by a grant for surgical nutrition from the Top Key Discipline and Technology Foundation of Zhejiang Province (No. 2013C37046). The study was also supported by the Zhejiang Provincial Undergraduate Scientific and Technological Innovation Project and Fresh Talent Program. (No. 2014R413014).
References
- 1.Martini F, Iaccheri L, Lazzarin L, et al. SV40 early region and large T antigen in human brain tumors, peripheral blood cells and sperm fluids from healthy individuals. Cancer Res. 1996;56:4820–4825. [PubMed] [Google Scholar]
- 2.De Mattei M, Martini F, Corallini A, et al. High incidence of bk virus large-T-antigen-coding sequences in normal human tissues and tumors of different histotypes. Int J Cancer. 1995;61:756–760. doi: 10.1002/ijc.2910610603. [DOI] [PubMed] [Google Scholar]
- 3.Bechert CJ, Schnadig VJ, Payne DA, Dong J. Monitoring of BK viral load in renal allograft recipients by real-time PCR assays. Am J Clin Pathol. 2010;133:242–250. doi: 10.1309/AJCP63VDFCKCRUUL. [DOI] [PubMed] [Google Scholar]
- 4.Randhawa P, Brennan D. BK virus infection in transplant recipients: an overview and update. Am J Transplant. 2006;6:2000–2005. doi: 10.1111/j.1600-6143.2006.01403.x. [DOI] [PubMed] [Google Scholar]
- 5.Drew RJ, Walsh A, Laoi Ní B, Conneally E, Crowley B. BK virus (BKV) plasma dynamics in patients with BKV-associated hemorrhagic cystitis following allogeneic stem cell transplantation. Transpl Infect Dis. 2013;15:276–282. doi: 10.1111/tid.12066. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: Interdisciplinary analyses and recommendations. Transplantation. 2005;79:1277–1286. doi: 10.1097/01.TP.0000156165.83160.09. [DOI] [PubMed] [Google Scholar]
- 7.Moens U, Rekvig OP. Molecular biology of BK virus and clinical and basic aspects of BK virus renal infection. In: Khalili K, Stoner GL, editors. Human polyomaviruses: molecular and clinical perspectives. John Wiley & Sons; New York, NY: 2001. pp. 359–408. [DOI] [Google Scholar]
- 8.Cole CN, Conzen SD. Polyomaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th. Philadelphia, PA: 2001. pp. 2141–2174. (Lippincott Williams and Wilkins). [Google Scholar]
- 9.Seif I, Khoury G, Dhar R. The genome of human papovavirus BKV. Cell. 1979;18:963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- 10.Jin L. Rapid genomic typing of BK virus directly from clinical specimens. Mol Cell Probes. 1993;7:331–334. doi: 10.1006/mcpr.1993.1047. [DOI] [PubMed] [Google Scholar]
- 11.Knowles WA, Gibson PE, Gardner SD. Serological typing scheme for BK-like isolates of human polyomavirus. J Med Virol. 1989;28:118–123. doi: 10.1002/jmv.1890280212. [DOI] [PubMed] [Google Scholar]
- 12.Stoner GL, Alappan R, Jobes DV, Ryschkewitsch CF, Landry ML. BK virus regulatory region rearrangements in brain and cerebrospinal fluid from a leukemia patient with tubulointerstitial nephritis and meningoencephalitis. Am J Kidney Dis. 2002;39:1102–1112. doi: 10.1053/ajkd.2002.32795. [DOI] [PubMed] [Google Scholar]
- 13.Zheng HY, Nishimoto Y, Chen Q, et al. Relationships between BK virus lineages and human populations. Microbes Infect. 2007;9:204–213. doi: 10.1016/j.micinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Nishimoto Y, Zheng HY, Zhong S, et al. An Asian origin for subtype IV BK virus based on phylogenetic analysis. J Mol Evol. 2007;65:103–111. doi: 10.1007/s00239-006-0269-6. [DOI] [PubMed] [Google Scholar]
- 15.Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. Phylogenetic analysis of polyomavirus BK sequences. J Virol. 2006;80:8869–8879. doi: 10.1128/JVI.02178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drew RJ, Walsh A, Laoi BN, Crowley B. Phylogenetic analysis of the complete genome of 11 BKV isolates obtained from allogenic stem cell transplant recipients in Ireland. J Med Virol. 2012;84:1037–1048. doi: 10.1002/jmv.23240. [DOI] [PubMed] [Google Scholar]
- 17.Luo C, Hirsch HH, Kant J, Randhawa P. VP-1 quasispecies in human infection with polyomavirus BK. J Med Virol. 2012;84:152–161. doi: 10.1002/jmv.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boukoum H, Nahdi I, Foulongne V, et al. Distribution of BK polyomavirus genotypes in Tunisian renal transplant recipients. J Med Virol. 2011;83:725–730. doi: 10.1002/jmv.22035. [DOI] [PubMed] [Google Scholar]
- 19.Slavov S, Tsekov I, Kalvatchev Z. Sequence variations of the VP1 gene of Polyomavirus hominis 1 among Bulgarians. J Med Virol. 2010;82:325–330. doi: 10.1002/jmv.21684. [DOI] [PubMed] [Google Scholar]
- 20.Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol. 2009;83:2285–2297. doi: 10.1128/JVI.02180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikegaya H, Saukko PJ, Tertti R, et al. Identification of a genomic subgroup of BK polyomavirus spread in European populations. J Gen Virol. 2006;87:3201–3208. doi: 10.1099/vir.0.82266-0. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Gibson PE. Genomic function and variation of human polyomavirus BK (BKV) Rev Med Virol. 1996;6:201–214. doi: 10.1002/(SICI)1099-1654(199612)6:4<201::AID-RMV177>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 23.Smith RD, Galla JH, Skahan K, et al. Tubulointerstitial nephritis due to a mutant polyomavirus BK virus strain, BKV (Cin), causing end-stage renal disease. J Clin Microbiol. 1998;36:1660–1665. doi: 10.1128/jcm.36.6.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takasaka T, Goya N, Tokumoto T, et al. Subtypes of BK virus prevalent in Japan and variation in their transcriptional control region. J Gen Virol. 2004;85:2821–2827. doi: 10.1099/vir.0.80363-0. [DOI] [PubMed] [Google Scholar]
- 25.Ledesma J, Bouza E, González-Nicolás M, de Viedma DG, Rodríguez-Sánchez B, Muñoz P. BK polyomavirus genotyping at inter-and intra-patient level in Spain. J Med Virol. 2013;85:1402–1408. doi: 10.1002/jmv.23612. [DOI] [PubMed] [Google Scholar]
- 26.Yogo Y, Zhong S, Xu Y, et al. Conserved archetypal configuration of the transcriptional control region during the course of BK polyomavirus evolution. J Gen Virol. 2008;89:1849–1856. doi: 10.1099/vir.0.2008/000836-0. [DOI] [PubMed] [Google Scholar]
- 27.Zhong S, Randhawa PS, Ikegaya H, et al. Distribution patterns of BK polyomavirus (BKV) subtypes and subgroups in American, European and Asian populations suggest co-migration of BKV and the human race. J Gen Virol. 2009;90:144–152. doi: 10.1099/vir.0.83611-0. [DOI] [PubMed] [Google Scholar]
- 28.Li WH. Proc Natl Acad Sci. Vol. 78. USA: 1981. Simple method for constructing phylogenetic trees from distance matrices; pp. 1085–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (BK) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/S0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 30.Chehadeh W, Nampoory MR. Genotypic diversity of polyomaviruses circulating among kidney transplant recipients in Kuwait. J Med Virol. 2013;85:1624–1631. doi: 10.1002/jmv.23639. [DOI] [PubMed] [Google Scholar]
- 31.Randhawa P, Kant J, Shapiro R, Tan H, Basu A, Luo C. Impact of genomic sequence variability on quantitative PCR assays for diagnosis of polyomavirus BK infection. J Clin Microbiol. 2011;49:4072–4076. doi: 10.1128/JCM.01230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carr MJ, McCormack GP, Mutton KJ, Crowley B. Unique BK virus non-coding control region (NCCR) variants in hematopoietic stem cell transplant recipients with and without hemorrhagic cystitis. J Med Virol. 2006;78:485–493. doi: 10.1002/jmv.20566. [DOI] [PubMed] [Google Scholar]