Abstract
In order to estimate the effects of small interfering RNA (siRNA) targeting retinoblastoma binding protein 2 (RBP2) on the proliferation, expression, invasion, migration and tumorigenicity abilities of papillary thyroid carcinoma K1 cells, siRNA targeting RBP2 (RBP2-siRNA) and negative control siRNA were transfected into K1 cells. The mRNA levels of RBP2 in the transfected cells were estimated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and the protein levels of RBP2 in these cells were evaluated by western blot analysis and immunocytochemical (ICC) analyses. The growth, tumorigenicity, migration and invasion abilities of the transfected cells were measured by Cell Counting Kit-8 (CCK-8), soft agar colony formation and transwell chamber assay, respectively. The ICC results demonstrated that the protein expression levels of RBP2 were lower in the RBP2-siRNA-transfected cells than in the blank and control cells (analysis of variance, F=26.754, P<0.01). RBP2-siRNA downregulated RBP2 at the mRNA (t=8.869) and protein level (F=60.835) (P=0.000 vs. control cells). In addition, the transfection of RBP2-siRNA into K1 cells also suppressed cell proliferation at 24, 48 and 72 h post-transfection (t=7.650, P<0.01; t=2.606, P=0.016; and t=2.377, P=0.027, respectively). Compared with the control group, the number of invasive and migrated cells were significantly reduced in the RBP2-siRNA-transfected group (t=4.774 and t=6.366, respectively; P<0.01). Furthermore, the tumorigenic potential of the cells transfected with RBP2-siRNA was markedly reduced, as indicated by the soft agar formation assay (t=2.749, P=0.014 vs. control cells). In conclusion, the transfection of RBP2-siRNA into papillary thyroid carcinoma K1 cells suppressed the expression of RBP2 in these cells, and reduced their proliferation, invasion, migration and tumorigenic potential. Therefore, targeting RBP2 may be an efficient approach to control thyroid carcinoma.
Keywords: papillary thyroid carcinoma, retinoblastoma binding protein 2, siRNA, transwell chamber assay, soft agar colony formation assay
Introduction
Thyroid cancer is the most common type of cancer of the endocrine system, and its incidence has increased in recent years (1–3). Although a large proportion of patients with thyroid cancer tend to present a good outcome, ~10% of patients, who do not receive radioiodine, may experience recurrence of the disease and mortality (4). Therefore, sensitive markers for the diagnosis of thyroid cancer, and efficient target genes for the treatment of this disease, are required. Despite previous studies on the occurrence of thyroid carcinoma have been published, the molecular mechanism of this type of cancer remains to be elucidated (5–7).
Genetic mechanisms are not the only path to gene disruption in cancer. Pathological epigenetic changes, non-sequence-based alterations that are inherited through cell division, are increasingly being considered as alternatives to mutations and chromosomal alterations in disrupting gene function (8). Certain histone demethylases have been proved to be involved in tumor metastasis (9). Previous studies have demonstrated that KDM5B histone demethylase controlled epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family (10), and histone demethylase KDM6B promoted epithelialmesenchymal transition (11). Retinoblastoma binding protein 2 (RBP2) was initially identified as a retinoblastoma binding protein that exhibited histone 3 lysine 4 (H3K4) demethylation activity (12,13). It is a member of the Jumonji/AT-rich interactive domain (JARID) protein family, an important epigenetic molecule, has been implicated in cancer and other diseases (14,15). Although it has been shown that RBP2 is highly expressed in a number of types of cancer (16–19) and that the deregulation of RBP2 may result in human diseases, especially developmental disorders (14), there remains a lack of studies into the pathological roles of RBP2 in thyroid carcinoma. Therefore, the present study aimed to characterize the role of RBP2 in thyroid tumors.
Materials and methods
Cell culture
Papillary thyroid carcinoma K1 cells were purchased from The European Collection of Cell Cultures (Salisbury, UK). The cells were cultured in Dulbecco's Modified Eagle's medium (DMEM) (Gibco Life Technologies, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (FBS) (GE Healthcare Life Sciences, Logan, CA, USA), glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 mg/ml), and incubated at 37°C with 5% CO2.
Transfection of small interfering RNA (siRNA)
The sequences of the siRNA targeting RBP2 (RBP2-siRNA) and the non-targeting scrambled siRNA (Shanghai GenePharma Co., Ltd., Shanghai, China), used as a negative control, were as follows: 5′-CCA GCA CCA CCU CCU UCC UUC AUAA-3′ and 5′-UUC UCC GAA CGU GUC ACG UTT-3′, respectively. K1 cells (1.8×105) were seeded in 6-well plates, and incubated for 18 h, prior to being transfected with the RBP2-siRNA (siRNA group) or negative control siRNA, using HiPerFect Transfection Reagent (Qiagen GmbH, Hilden, Germany). Cells without transfecting were regarded as blank group.
Immunocytochemistry (ICC) staining of cell slides
Protein expression of RBP2 was detected by ICC staining of K1 cells seeded on cover slips. For the experiment, ~2×105 K1 cells were added to each cover slip in a 6-well plate, and incubated for 4 h, to allow the cells to adhere to the cover slip in situ. Next, 3–4 ml complete culture medium (DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin) was added to each well, and the cells were cultured for additional 36 h. The cells were then fixed with 4% (v/v) paraformaldehyde for ≥20 min. The ICC was performed as previously described by Kong et al (20) using the following antibodies: Monoclonal rabbit-anti-human-RBP2, (Cell Signaling Technology, Danvers, MA, USA) and horseradish peroxidase-conjugated anti-rabbit secondary antibody (ZSGD-BIO, Peking, China). The brown color observed in the cells nuclei was regarded as positive expression of RBP2. Positive cells were counted using an eyepiece graticule at ×400 magnification (Olympus, Tokyo, Japan). For each slide, ≥10 views were counted. Each experiment was performed in triplicate.
Total RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RT-qPCR analysis was performed at 48 h post-transfection. TRIzol reagent (Invitrogen Life Technologies) was used to extract the total RNA content from the transfected cells, and 1.5 µg RNA was used for the synthesis of the first strand of cDNA with a reverse transcriptase (Promega Corporation, Madison, WI, USA). The relative mRNA levels of RBP2 were normalized to the levels of β-actin, and normalized using the formula 2−ΔΔCT with SYBR Green qPCR Kit (Roche Diagnostics, Basel, Switzerland). The cycling conditions were as follows: 95°C, 5 min, 1 cycle; followed by 35 cycles of 95°C for 5 sec, 50°C for 30 sec and 72°C for 32 sec. Specific primer pairs (Shanghai GenePharma Co., Ltd., Shanghai, China) were used for RBP2 (forward, 5′-GCT GCT GCA GCC AAA GTTG-3′; and reverse, 5′-AGC ATC TGC TAA CTG GTC-3′) and β-actin (forward, 5′-GAG CAA GAG AGG CAT CCTCA-3′; and reverse, 5′-AGC CTG GAT AGC AAC GTACA-3′).
Western blot analysis
Cells were harvested 48 h post-transfection, and lysed with RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China). The protein concentration in the cell lysates was quantified by BCA assay. The proteins were separated by 10% SDS-PAGE, and subsequently transferred onto polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc., Hercules, USA). The membranes were incubated with the monoclonal rabbit-anti-human primary antibodies (dilution 1:100; Cell Signaling Technology, Inc., Danvers, USA) at 4°C for 24 h. Next, the membranes were washed with Tris-buffered saline and Tween 20 (TBST) for three times, and incubated with goat anti-rabbit HRP-conjugated secondary antibody (Gene Tech Co., Ltd., Shanghai, China). Proteins were detected using ECL Plus (Boster, Wuhan, China).
Cell counting Kit-8 (CCK-8) assay
CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was performed to evaluate the effect of RBP2-siRNA on cell proliferation. K1 cells transfected with RBP2-siRNA or neagtive control siRNA were seeded into 96-well plates at a density of 3.8×103 cells/well, and cultured for 24, 48 and 72 h. At the time points indicated above, 90 µl DMEM and 10 µl CCK-8 were added to each well, and the cells were incubated for additional 2 h. Subsequently, the supernatant was removed, and the absorbance at 450 nm wavelength was recorded using a microplate reader (Bio-Rad Laboratories, Inc.). Those wells containing 10% CCK-8 (DMEM 90 µl and 10 µl CCK-8) were regarded as blanks. The experiment was performed with 6 repeated measures of each experimental value, and repeated 3 times.
Invasion and migration assays
The invasion ability of K1 cells was evaluated by Transwell chamber assay (Corning Life Sciences, New York, USA). The chamber was covered with 50 µl Matrigel Basement Membrane Matrix (2 mg/ml; BD Biosciences, Franklin Lakes, NJ, USA). Serum-free DMEM (200 µl) containing 2×104 cells was added to the upper chamber, while the lower chamber was filled with 600 µl DMEM supplemented with 15% FBS. Following incubation for 20 h, the cells on the upper chamber were removed, and the remaining cells were fixed with methanol for 20 min, prior to being dyed with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The migration assay was conducted following the aforementioned steps, except for the absence of the Matrigel Basement Membrane Matrix layer, and ~3.5×104 cells were added to the upper chamber. Each experiment was performed in triplicate and repeated 3 times.
Soft agar colony formation assay
The capacity of the K1 cells to form colonies was assessed by soft agar colony formation assay conducted in 6-well plates. In each well, a base layer was created with 0.7% soft agar, and a top layer was subsequently added, which contained 0.35% soft agar and 103 suspended cells. The cells were cultured for ~14 days, until the newly formed colonies were visible. The experiment was performed in triplicate and repeated 3 times.
Statistical analysis
The data were evaluated by the Student's t-test for 2 groups, or analysis of variance for ≥3 groups, using SPSS software, version 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of RBP2 by RBP2-siRNA
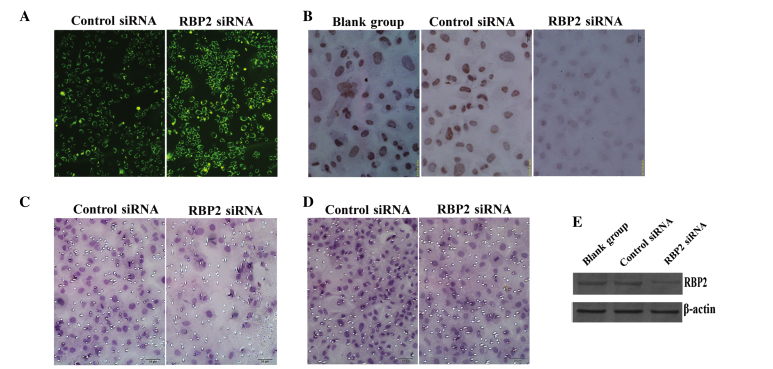
RBP2-siRNA and negative control siRNA were transfected into thyroid carcinoma K1 cells. The fluorescence emission of green fluorescent protein in the cytoplasm of the transfected cells was recorded and photographed (Fig. 1A). The protein expression levels of RBP2 were detected by ICC analysis in the blank (cells without transfecting), RBP2-siRNA-transfected and negative control siRNA-transfected cells. The results indicated that the transfection of RBP2-siRNA into K1 cells suppressed the expression of RBP2 in these cells (Fig. 2A). The expression levels of RBP2 in the nuclei of the RBP2-siRNA-transfected cells were lower than in the blank and negative control cells (F=26.754, P<0.01; Fig. 1B). No significant difference was observed between the blank and control groups (P>0.05).
Figure 1.
(A) The fluorescence emission of thyroid carcinoma K1 cells transfected with control siRNA and RBP2-siRNA was recorded (magnification, ×200). (B) The protein expression levels of RBP2 were lower in the RBP2-siRNA-transfected cells than in the blank and negative control cells, according to the results of immunocytochemical analysis (magnification, ×400). (C) Transwell assay revealed the presence of invasive cells through the membrane in the control and RBP2-siRNA groups (magnification, ×200). (D) The cell migration ability was lower in the RBP2-siRNA-transfected cells, compared with the control cells, as evaluated by transwell assay (magnification, ×200). (E) Western blot analysis demonstrated that the protein expression levels of RBP2 were markedly reduced in the RBP2-siRNA group, compared with the blank and negative control groups. β-actin was used as reference. siRNA, small interfering RNA; RBP2, retinoblastoma binding protein 2; RBP2-siRNA, siRNA targeting RBP2; blank-group, non transfected cells.
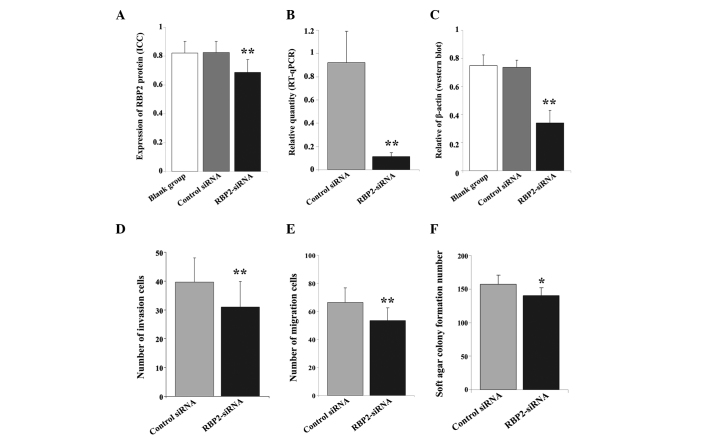
Figure 2.
(A) Immunocytochemical and (B) RT-qPCR analyses revealed a marked difference in the (A) protein and (B) mRNA expression levels of RBP2 in thyroid carcinoma K1 cells transfected with RBP2-siRNA, compared with the control cells. (C) Western blot analysis demonstrated a significant difference in the protein levels of RBP2 across the 3 groups of cells. β-actin was used as reference. Transwell assay identified a lower number of (D) invasive and (E) migrated cells in the RBP2-siRNA-transfected cells, compared with the control group. (F) The transfection with RBP2-siRNA affected the tumorigenicity ability of K1 cells, since a lower number of newly formed colonies were observed in the RBP2-siRNA-transfected group, compared with the control group. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; RBP2, retinoblastoma binding protein 2; siRNA, small interfering RNA; RBP2-siRNA, siRNA targeting RBP2; control siRNA, negative control siRNA targeting RBP2; blank group, non transfected cells. **P<0.01 vs. blank group, negative control; *P<0.05 vs. negative control.
To confirm the above data, the mRNA levels of RBP2 were quantified by RT-qPCR. The transfection of RBP2-siRNA into K1 cells significantly reduced the mRNA expression levels of RBP2 in these cells (F=31.385, P<0.01; Fig. 3). Compared with the control group, RBP2-siRNA notably downregulated the mRNA levels of RBP2 (t=8.869, P<0.01; Fig. 2B). The protein levels of RBP2 were measured by western blot analysis, using β-actin as reference. The results presented in Fig. 1E indicated that compared with the blank cells, the expression of RBP2 was notably inhibited in the RBP2-siRNA-transfected cells (F=60.835, P=0.000; Fig. 2C).
Figure 3.
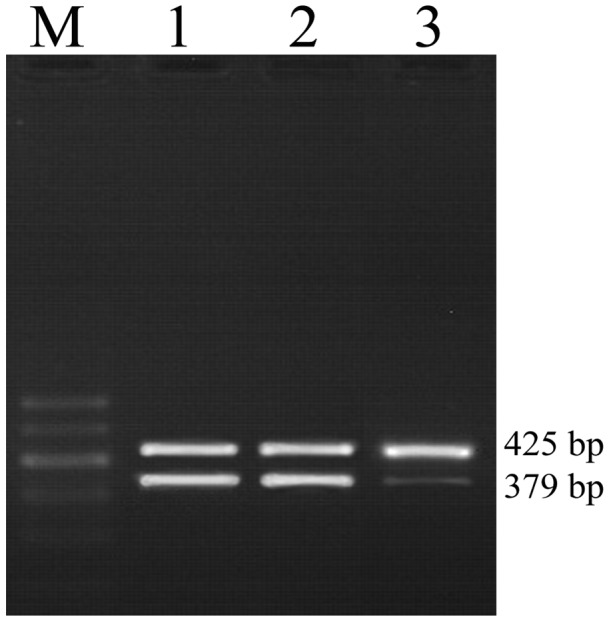
RT-PCR analysis was conducted to evaluate the mRNA expression levels of RBP2 in thyroid carcinoma K1 cells transfected with RBP2-siRNA or control siRNA. The results revealed that K1 cells transfected with RBP2-siRNA (lane 3) presented lower mRNA levels of RBP2 than the blank (lane 1) and control cells (lane 2). The quantification of mRNA levels of RBP2 were normalized to levels of β-actin, and normalized using the formula 2−ΔΔCt with SYBR Green qPCR kit [Fig. 2B; (t=8.869; P=0.000)]. RBP2, retinoblastoma binding protein 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; RBP2-siRNA, siRNA targeting RBP2.
Downregulation of RBP2 suppressed the proliferation ability of K1 cells
The effect of RBP2 on cell proliferation ability was estimated by CCK-8 assay. The data revealed that, compared with the control cells, the optical density of the cells transfected with RBP2-siRNA was significantly reduced at 24, 48 and 72 h post-transfection (OD=0.325±0.067, t=7.650, P<0.01; OD=0.605±0.118, t=2.606, P=0.016; and OD=0.906±0.070, t=2.377, P=0.027, respectively; Table I).
Table I.
Optical density values (mean ± standard deviation) of Cell Counting kit-8 assay.
| Cell groups | 24 h | 48 h | 72 h |
|---|---|---|---|
| Control siRNA | 0.507±0.048 | 0.714±0.083 | 0.978±0.078 |
| RBP2-siRNA | 0.325±0.067 | 0.605±0.118 | 0.906±0.070 |
| t/p | 7.650/0.000 | 2.606/0.016 | 2.377/0.027 |
Thyroid carcinoma K1 cells were transfected with RBP2-siRNA or control siRNA, and subjected to Cell Counting Kit-8 assay at 24, 48 and 72 h post-transfection, in order to assess their proliferation ability. RBP2, retinoblastoma binding protein 2; siRNA, small interfering RNA; RBP2-siRNA, siRNA targeting RBP2; t/p, ratio of t- vs. P-value.
Downregulation of RBP2 reduced the invasion and migration ability of K1 cells in vitro
The invasion ability of K1 cells was evaluated by transwell assay. Compared with the siRNA control group, the number of invasive cells on the RBP2-siRNA-transfected group was significantly reduced (39.67±8.36 vs. 30.96±8.94; t=4.774, P=0.000) as indicated in Figs. 1C and 2D. Similarly, the number of migrated cells was also notably reduced (66.44±10.30 vs. 53.53±8.89; t=6.366, P=0.000), as indicated in Figs. 1D and 2E.
Transfection of RBP2-siRNA inhibited the colony formation ability of K1 cells
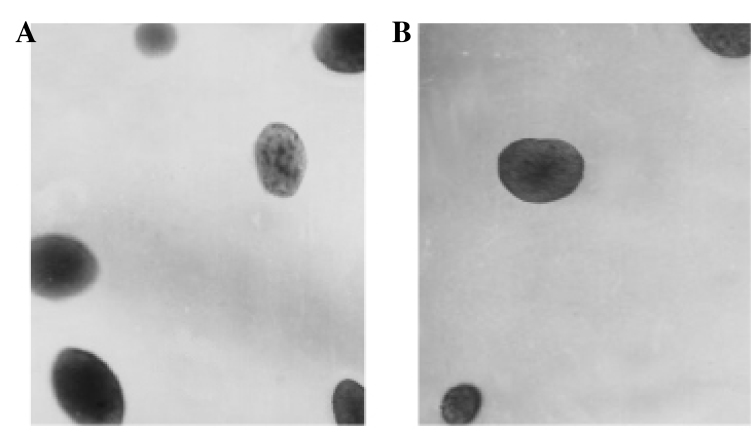
The in vitro tumorigenicity ability of K1 cells transfected with RBP2-siRNA was estimated by soft agar colony formation assay. Compared with the control group (Fig. 4A), the number of newly formed colonies observed in the RBP2-siRNA-transfected group was reduced (t=2.749, P=0.014; Figs. 2F and 4B).
Figure 4.
Effect of RBP2-siRNA on the tumorigenicity ability of thyroid carcinoma K1 cells, as evaluated by soft agar formation assay. (A) Control siRNA-transfected cells. (B) RBP2-siRNA-transfected cells. The ability of the K1 cells to form colonies was significantly inhibited following transfection with RBP2-siRNA (P<0.01 vs. control cells). RBP2, retinoblastoma binding protein 2; siRNA, small interfering RNA; RBP2-siRNA, siRNA targeting RBP2.
Discussion
Genetic and epigenetic modifications have been demonstrated to be involved in the tumorigenesis and development of cancer (21). Therefore, the stable inheritance of epigenetic states is required for the cells and tissues in the body to maintain their specific functions (21). The alteration of epigenetic modifications has been mainly associated with the methylation of DNA and modifications of histones (22,23). RBP2 has been recently identified as a histone demethylase, which regulates gene expression by demethylating H3K4me3/2 (24,25). RBP2 influences cell cycle withdrawal and cell differentiation via retinoblastoma protein (pRB) (13). Previous studies have demonstrated that RBP2 represses the transcription of its target genes, which are activated by pRB, by associating with the promoters of these actively transcribed genes (14,26). Previous studies have observed that RBP2 was preferentially bound to the proximal promoter regions of p27, cyclin D1, and integrin β1, whereby RBP2 repressed the expression of p27, but acted as an activator of the gene expression for cyclin D1 and integrin β1 (27). In agreement with these results, previous studies have reported that knocking down RBP2 led to overexpression of p27 (12). Since RBP2 occupies and regulates the promoters of multiple genes containing H3K4me3, RBP2 is likely to be involved in cancer progression (15). Previous studies have revealed that RBP2 was overexpressed in numerous types of cancer, including hepatocellular, non-small cell lung, breast and gastric cancer (16–19). Previous studies have demonstrated that RBP2 was overexpressed in hepatocellular carcinoma cells, which was downregulated by homo sapiens microRNA-212 (hsa-miR-212). In addition to inhibiting the expression of RBP2, the overexpression of hsa-miR-212 in hepatocellular carcinoma cells reduced the cell proliferation ability and induced cellular senescence (16). Furthermore, Wang et al (17) observed that RBP2 was also overexpressed in non-small cell lung cancer, whereby regulated the expression of E- and N-cadherin via the activation of Akt signaling. Previous studies also reported the overexpression of RBP2 in human lung cancer, and revealed that RBP2 promoted proliferation, motility, migration, invasion and metastasis in lung cancer cells; whereas depletion of RBP2 inhibited proliferation, motility, migration, invasion and metastasis in these cells (17). Similarly, previous studies observed that RBP2 was critical for the progression and metastasis of breast cancer, since RBP2 acted as a pleiotropic positive regulator of several genes involved in metastasis (18,27). In gastric cancer, RBP2 was also observed to be overexpressed, and directly regulated by miR-212 (19). Collectively, these findings indicate that RBP2 may participate in tumorigenesis, although its mechanism of action remains unclear (28).
Due to the increasing incidence in thyroid cancer in recent years (29), clinical markers for the diagnosis of this condition are urgently required. Previous studies have indicated that RBP2 was overexpressed in thyroid carcinoma, and therefore, RBP2 may be considered as a clinical marker for thyroid cancer (20). In the present study, the effects of RBP2-siRNA on the biological behavior of thyroid carcinoma cells, including cell proliferation, invasion and colony formation abilities, were investigated in vitro. The ICC results indicated that RBP2-siRNA significantly suppressed the expression of RBP2 in RBP2-siRNA-transfected cells. Similarly, the mRNA and protein levels of RBP2 were notably reduced in these cells. The transfection with RBP2-siRNA also reduced the proliferation, migration, invasion and tumorigenicity abilities of the transfected cells. In summary, downregulation of RBP2 via RBP2-siRNA reduced the expression levels of RBP2, and inhibited the proliferation, invasion and tumorigenicity abilities of thyroid carcinoma cells. These data indicate that RBP2 may be a therapeutic target for the inhibition of tumor progression and metastasis in thyroid cancer.
In conclusion, the results of the present study indicate that downregulation of RBP2 notably suppressed the biological behavior of thyroid carcinoma K1 cells. Knocking down RBP2 markedly affected the mRNA expression levels of RBP2 in RBP2-siRNA-transfected K1 cells, and inhibited the proliferation, migration, invasion and colony formation abilities of these cells. In summary, RBP2 appears to be essential for thyroid carcinoma cells to maintain their malignant behavior. Therefore, targeting RBP2 may be an efficient approach to control thyroid carcinoma.
Acknowledgements
The present study was supported by the Natural Science Foundation of Shandong (Shandong, China) (grant nos. ZR2011HM057 and ZR2009CM070).
References
- 1.Sherman SI. Thyroid carcinoma. Lancet. 2003;361:501–511. doi: 10.1016/S0140-6736(03)12488-9. [DOI] [PubMed] [Google Scholar]
- 2.Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014;40:317–322. doi: 10.1001/jamaoto.2014.1. [DOI] [PubMed] [Google Scholar]
- 3.Moura MA, Bergmann A, Aguiar SS, Thuler LC. The magnitude of the association between smoking and the risk of developing cancer in Brazil: A multicenter study. BMJ Open. 2014;4:e003736. doi: 10.1136/bmjopen-2013-003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilli T, Prasad KV, Jayarama S, Pacini F, Prabhakar BS. Potential utility and limitations of thyroid cancer cell lines as models for studying thyroid cancer. Thyroid. 2009;19:1333–1342. doi: 10.1089/thy.2009.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medici M, Porcu E, Pistis G, Teumer A, Brown SJ, Jensen RA, Rawal R, Roef GL, Plantinga TS, Vermeulen SH, et al. Identification of novel genetic loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS Genet. 2014;10:e1004123. doi: 10.1371/journal.pgen.1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akeno N, Miller AL, Ma X, Wikenheiser-Brokamp KA. p53 suppresses carcinoma progression by inhibiting mTOR pathway activation. Oncogene. 2015;34:589–99. doi: 10.1038/onc.2013.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moretti S, Menicali E, Voce P, Morelli S, Cantarelli S, Sponziello M, Colella R, Fallarino F, Orabona C, Alunno A, et al. Indoleamine 2,3-dioxygenase 1 (IDO1) is up-regulated in thyroid carcinoma and drives the development of an immunosuppressant tumor microenvironment. J Clin Endocrinol Metab. 2014;99:E832–E840. doi: 10.1210/jc.2013-3351. [DOI] [PubMed] [Google Scholar]
- 8.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 9.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schübeler D, van Nimwegen E, Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Enkhbaatar Z, Terashima M, Oktyabri D, Tange S, Ishimura A, Yano S, Suzuki T. KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle. 2013;12:2100–2112. doi: 10.4161/cc.25142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, Zeng J, Wang L, Fang M, Wang Q, Zhao M, Xu X, Liu Z, Li W, Liu S, et al. Histone demethylase retinoblastoma binding protein 2 is overexpressed in hepatocellular carcinoma and negatively regulated by hsa-miR-212. PLoS One. 2013;8:e69784. doi: 10.1371/journal.pone.0069784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Wang Y, Wu H, Hu L. RBP2 induces epithelial-mesenchymal transition in non-small cell lung cancer. PLoS One. 2013;8:e84735. doi: 10.1371/journal.pone.0084735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J, Liu Z, Cheung WK, Zhao M, Chen SY, Chan SW, Booth CJ, Nguyen DX, Yan Q. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Reports. 2014;6:868–877. doi: 10.1016/j.celrep.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiping Z, Ming F, Lixiang W, Xiuming L, Yuqun S, Han Y, Zhifang L, Yundong S, Shili L, Chunyan C, Jihui J. MicroRNA-212 inhibits proliferation of gastric cancer by directly repressing retinoblastoma binding protein 2. J Cell Biochem. 2013;114:2666–2672. doi: 10.1002/jcb.24613. [DOI] [PubMed] [Google Scholar]
- 20.Kong L, Zhang G, Wang X, Zhou J, Hou S, Cui W. Immunohistochemical expression of RBP2 and LSD1 in papillary thyroid carcinoma. Rom J Morphol Embryol. 2013;54:499–503. [PubMed] [Google Scholar]
- 21.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 22.Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Fang Y, Xiao F, An Z, Hao L. Systematic review on the relationship between genetic polymorphisms of methylenetetrahydrofolate reductase and esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2011;12:1861–1866. [PubMed] [Google Scholar]
- 24.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Defeo-Jones D, Huang PS, Jones RE, Haskell KM, Vuocolo GA, Hanobik MG, Huber HE, Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- 27.Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW, Chan MY, Lin FM, Huang HD, Chen YT, Jeng YM, et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013;73:4711–4721. doi: 10.1158/0008-5472.CAN-12-3165. [DOI] [PubMed] [Google Scholar]
- 28.Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic regulation by lysine demethylase 5 (kdm5) enzymes in cancer. Cancers (Basel) 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin N, Du XL, Reitzel LR, Xu L, Strugis EM. impact of enhanced detection on the increase in thyroid cancer incidence in the United States: Review of incidence trends by socioeconomic status within the surveillance, epidemiology, and end results registry, 1980–2008. Thyroid. 2013;23:103–110. doi: 10.1089/thy.2012.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]