Abstract
Thyroid transcription factor-1 (TTF-1) reportedly possesses oncogenic and suppressive roles within the same tumor type and may play a dual function in the progression of lung cancer. Immunohistochemistry (IHC) and mRNA in situ hybridization (ISH) are commonly used methods for detecting protein or mRNA expression. The present study compared the concordance rate of the two methods in the evaluation of thyroid transcription factor-1 (TTF-1) expression in non-small cell lung carcinoma (NSCLC) using tissue microarray-based IHC and mRNA ISH. TTF-1 protein and mRNA expression levels were examined in 196 cases of NSCLC. The IHC and mRNA ISH agreement was 91.3% (179/196), and near-perfect agreement was observed between the two methods (κ-coefficient, 0.848). There was no significant difference between IHC and mRNA ISH, as analyzed by the McNemar-Bowker test (P=0.219). The present findings proved that IHC is comparable to mRNA ISH for evaluating TTF-1 expression in NSCLC. These two methods can be used to detect TTF-1 expression in future studies.
Keywords: tissue microarray, immunohistochemistry, mRNA in situ hybridization, non-small cell lung cancer, thyroid transcription factor-1
Introduction
As the primary cause of cancer-associated mortality globally, lung cancer is an important public health problem. In men, lung cancer is ranked first among the commonly observed malignant tumors, while in females, the disease is ranked second or third in the majority of developed countries. The 5-year survival rate is ~15% and the cure rate remains low (1). Therefore, the elucidation of the tumorigenesis of lung carcinoma is of paramount importance for the improvement of public health.
Thyroid transcription factor-1 (TTF-1), also known as NKX2-1 or TITF-1, is mapped to chromosome 14q13 and the protein a 38-kDa transcription factor that is normally expressed in the thyroid gland, lungs and brain. This type of exhibits significant functions in the development, cell growth and differentiation processes. TTF-1 reportedly possesses oncogenic and suppressive functions within the same tumor type and may play a dual function in the progression of lung cancer (2). TTF-1 is also used in the differentiation between primary and metastatic lung tumors (3,4).
Immunohistochemistry (IHC) has been used (5–7) to detect TTF-1 expression in lung carcinomas. TTF-1 expression differs among the subtypes of lung carcinoma, with strong expression in lung adenocarcinoma and small cell lung carcinoma (SCLC), and weak or no expression in squamous cell lung carcinoma and large cell lung carcinoma. In our previous studies, mRNA in situ hybridization (ISH) was used to detect TTF-1 mRNA expression in different subtypes of lung cancer. The TTF-1 protein expression level detected by IHC is consistent with that detected by mRNA ISH (8). However, no comparison of the statistical significance of the two methods has been performed.
In the present study, TTF-1 expression was investigated using IHC and mRNA ISH, and the concordance rate of the two methods was compared for detecting TTF-1 expression in non-SCLC (NSCLC).
Materials and methods
Patients
A total of 196 patients with NSCLC who were treated in Nanfang Hospital (Southern Medical University and Guangdong Provincial Institute of Nephrology, Guangzhou, Guangdong, China) between January 2000 and December 2010 were selected for this study. The diagnosis of NSCLC mainly included adenocarcinoma and squamous cell carcinoma based on the pathological classification of the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (9). The inclusion criteria for this study were a diagnosis of NSCLC, no radiotherapy or chemotherapy, and the availability of an adequate paraffin block for the analysis. Specimens from these patients were obtained from the Departments of Pathology and Thoracic Surgery of Nanfang Hospital. The specimens included tumor tissues from 92 primary lung adenocarcinomas and 104 primary squamous cell lung carcinomas.
Written informed consent was obtained from all patients according to the protocols of the Southern Medical University Ethics Committee, and the study was approved by the ethics committee of South Hospital of Southern Medical University.
IHC and ISH in the tissue microarray (TMA)
A TMA was constructed from the 196 paraffin-embedded blocks according to a previously described procedure (10–12). Four tissue cores selected from each sample were used to construct a 30×30 matrix microarray. In the final row of the microarray, 16 cores were used as sample location indicators. Sections (4-µm thick) were cut from the TMA blocks and used for the IHC and mRNA ISH labeling.
For IHC, TMA sections (4-µm thick) were stained with mouse anti-human TTF-1 monoclonal antibody (clone 8G7G3/1; Dako, Carpinteria, CA, USA) using the standard streptavidin-biotin complex method according to the manufacturer's instructions.
For mRNA ISH, a TTF-1 mRNA–specific oligonucleotide probe designed by Bioasia Co., Ltd., (Shanghai, China) was used to detect TTF-1 mRNA expression in the TMA samples. The sequence of the probe was 5′-GCCGACAGGTACTTCTGTTGCTTGAAGCGT-3′. The probe was labeled with digoxin on the 3′ and 5′ ends. TMA sections (4-µm thick) were deparaffinized, rehydrated, rinsed in phosphate-buffered saline (PBS) and digested with pepsin at 37°C for 20 min. The slides were hybridized with the riboprobe at 38°C for 16 h. The hybridization samples were then washed sequentially in 2X sodium chloride and sodium citrate (SSC) at 37°C for 5 min and in 0.5X SSC at 37°C for 30 min. The slides were treated with 3% hydrogen peroxide to inactivate the endogenous peroxidase. Pepsin diluted with 3% citric acid was subsequently used to expose mRNA fragments. The sections were hybridized with a probe diluted with hybridization solution for 16 h at 38°C. The hybridization samples were sequentially washed three times in 2X SSC for 5 min at 37°C, twice in 0.5X SSC for 5 min at 37°C and once in 0.2X SSC for 15 min at 37°C.
The hybridized probe was detected using an in situ hybridization detection kit (Wuhan Boster Biological Technology, Ltd., Wuhan, China) according to the manufacturer's instructions (13). Briefly, the slides were incubated with blocking reagent for 30 min at 37°C, and then with biotin-labeled sheep antidigoxigenin for 1 h at 37°C. The slides were sequentially washed with 0.5 M PBS at 37°C for 5 min, and then incubated with streptavidin-biotin complex for 30 min and washed four times in 0.5 M PBS for 5 min. Finally, the slides were incubated with biotin peroxidase for 30 min at 37°C and washed four times in 0.5 M PBS for 5 min. The peroxidase reaction was enhanced using 3,3′-diaminobenzidine. The IHC and ISH slides were counterstained with hematoxylin and cover-slipped.
Two cases of normal lung tissue were used as internal positive controls for IHC and ISH. PBS or hybridization solution was used to replace the primary antibody or the probe in order to serve as negative controls.
Analysis of IHC and mRNA ISH slides
The samples were analyzed under a 40X objective lens using a BX51 light microscope (Olympus, Tokyo, Japan). A total of 20 fields were randomly selected from four randomly chosen representative tissue cores of a specimen on the TMA. In total, 200 tumor cells were observed. Those with dark brown particles in the nuclei or cytoplasm were considered to demonstrate positive protein or mRNA expression. Two independent analysts assessed IHC and ISH staining effects.
TTF-1 IHC and ISH staining was recorded as no expression (absent staining, −), low expression (≥30% of tumor cells with weak staining intensity, +) or high expression (≥30% of tumor cells with strong staining intensity, ++). No expression was considered as negative, while low or high expression was considered as positive.
Statistical analysis
Comparisons between IHC and mRNA ISH methods were analyzed using the McNemar-Bowker test. Concordance data obtained from IHC and mRNA ISH were determined. The κ statistic method was used to measure the agreement of positive ratios between the two assays. The κ statistic evaluates the level of agreement following adjustment for agreement expected to occur by chance alone, with a κ-coefficient of >0.80 indicating near-perfect agreement, 0.61–0.80 indicating substantial agreement, 0.41–0.60 indicating moderate agreement, 0.21–0.40 indicating fair agreement, >0–0.20 indicating slight agreement and 0 indicating no agreement or a random association (14). SPSS 13.0 (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
In the 196 cases of NSCLC detected by IHC and mRNA ISH using TMAs, the two techniques agreed that 109 were TTF-1-negative (−), 27 exhibited low expression (+) and 43 exhibited high expression (++). Representative IHC images are shown in Fig. 1 and mRNA ISH images are shown in Fig. 2. In the 46 cases detected with ++ TTF-1 protein expression on IHC, 43 were ++ and 3 were +, as detected by mRNA ISH. No cases were negative when detected by mRNA ISH. Of the 50 cases detected with ++ TTF-1 mRNA expression on ISH, 43 were ++, 4 were + and 3 were negative, as detected by IHC. The agreement between IHC and mRNA ISH was near-perfect at 91.3% (179/196), with a κ-coefficient of 0.848 (Table I). There was no significant difference between the two methods on the McNemar-Bowker test (P=0.219) (Table I).
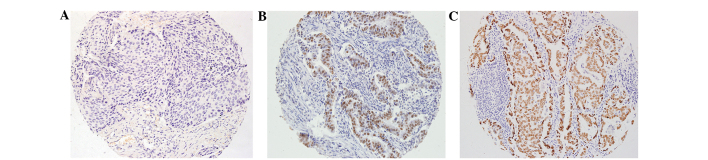
Figure 1.
Immunohistochemistry for detecting thyroid transcription factor-1 protein expression in non-small cell lung cancer (×200 magnification). (A) No expression (absent staining, −). (B) Low expression (≥30% of tumor cells with weak staining intensity, +). (C) High expression (≥30% of tumor cells with strong staining intensity, ++).
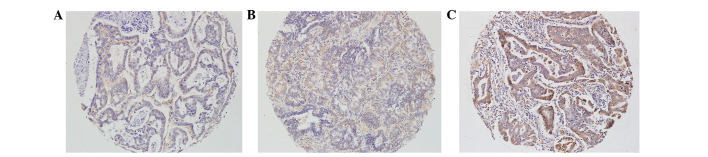
Figure 2.
mRNA in situ hybridization (ISH) for detecting thyroid transcription factor-1 mRNA expression in non-small cell lung cancer (ISH; ×200 magnification). (A) No expression (absent staining, −). (B) Low expression (≥30% of tumor cells with weak staining intensity, +). (C) High expression (≥30% of tumor cells with strong staining intensity, ++).
Table I.
Comparison of IHC vs. mRNA ISH for detecting TTF-1 expression in non-small cell lung cancer.
| mRNA ISH | ||||
|---|---|---|---|---|
| IHC | − | + | ++ | Total |
| − | 109 | 5 | 3 | 117 |
| + | 2 | 27 | 4 | 33 |
| ++ | 0 | 3 | 43 | 46 |
| Total | 111 | 35 | 50 | 196 |
Overall agreement, 0.913; κ-coefficient, 0.848. −, Absent staining; +, ≥30% of tumor cells with weak staining intensity; ++, ≥30% of tumor cells with strong staining intensity. TTF-1, thyroid transcription factor-1; IHC, immunohistochemistry; ISH, in situ hybridization.
Discussion
TMAs, also known as tissue chips, are a novel technology that were invented in 1998 by Konenen et al (15) based on cDNA microarrays. This technique has a high throughput and is considered to be a resource-conserving technology in which tens of thousands of typical minute cylindrical tissue samples or cells from numerous tumor types are transferred to a new paraffin block. TMA can be used to detect DNA, RNA or proteins in a range of clinical or basic studies (16–18).
In the present study, IHC and mRNA ISH were used to detect TTF-1 gene expression in NSCLC. The McNemar-Bowker test demonstrated that there was no significant difference between the two methods (P=0.219), and the κ-test revealed near-perfect agreement between them (κ-coefficient, 0.848). This finding shows that the highly conserved transcription factor TTF-1 is crucial at the protein and mRNA levels in lung cancer development and progression.
With the development and progression of scientific technology of targeted agents in NSCLC, the majority of patients are not considered suitable for molecularly targeted therapy, as they do not express any presently known genetic alterations that make them suitable candidates for such treatment (19).
According to the central dogma of genetics, genes are first transcribed into mRNA and then translated into proteins. In the present study, the TTF-1 gene status was detected and analyzed at the transcriptional level using mRNA ISH and at the translational level using IHC. The concordance rate of the two methods was near-perfect (91.3%; κ-coefficient, 0.848; P=0.219). However, of the 50 cases with ++ TTF-1 mRNA expression, 3 cases were negative on IHC. Of the 46 cases with ++ TTF-1 protein expression, no cases were negative on mRNA ISH. This finding indicates that the transcriptional step is the crucial process prior to translation. Similar to TTF-1 protein expression, positive TTF-1 mRNA expression in NSCLC indicates a good prognosis in lung cancer patients. The expression patterns of TTF-1 protein and mRNA in the present study are consistent with those of our previous studies (10,11). TTF-1 and p63 immunostaining has been utilized successfully in previous studies (20,21) to facilitate pathological differentation between small cell carcinoma and poorly differentiated pulmonary SCC using cytological samples. Kapila et al (20) were able to catergorize NSCLC samples using a restricted panel of antibodies. The authors were also able to differentiate between adenocarcinoma and SCC samples. Furthermore, Kalhor et al (21) employed TTF-1 and p63 immunostaining to identify primary lung tumors, which could be further catergorized as SCCs.
During the construction of the TMAs, representative tumor areas were carefully chosen from the donor blocks. The four cores biopsied in each donor block highly represented the characteristics of each lung cancer. All 196 samples were detected simultaneously on a single glass slide to ensure a comparable and consistent data analysis.
Furthermore, additional studies are required to authenticate the possible diagnostic value of the current findings. For example, future studies should include intraobserver and interobserver variability in their evaluation; sensitivity, specificity, predictive values for the presence of TTF-1 positivity; and possibly a correlation between the different growth patterns, TTF-1 gene expression and TTF-1 amplification.
In conclusion, TTF-1 expression, as detected by IHC and mRNA ISH using TMA technology, could reveal the biological features of samples and detect gene expression.
Acknowledgements
This study was supported by a grant (no. 81100496) from the National Natural Science Foundation of China.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra M, Morgan V, Hamati AK, Al-Abbadi M. Carcinoma of unknown primary: Check the liver… thanks to TTF-1. Tenn Med. 2012;105:35–36. [PubMed] [Google Scholar]
- 4.Ye J, Findeis-Hosey JJ, Yang Q, McMahon LA, Yao JL, Li F, Xu H. Combination of napsin A and TTF-1 immunohistochemistry helps in differentiating primary lung adenocarcinoma from metastatic carcinoma in the lung. Appl Immunohistochem Mol Morphol. 2011;19:313–317. doi: 10.1097/PAI.0b013e318205b059. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura N, Miyagi E, Murata S, Kawaoi A, Katoh R. Expression of thyroid transcription factor-1 in normal and neoplastic lung tissues. Mod Pathol. 2002;15:1058–1067. doi: 10.1097/01.MP.0000028572.44247.CF. [DOI] [PubMed] [Google Scholar]
- 6.Tang X, Kadara H, Behrens C, Liu DD, Xiao Y, Rice D, Gazdar AF, Fujimoto J, Moran C, Varella-Garcia M, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: Implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011;17:2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 8.Bai XY, Shen H. Quantitative analysis of thyroid transcription factor-1 mRNA expressions in primary lung cancer and its metastatic foci. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28:20–25. (In Chinese) [PubMed] [Google Scholar]
- 9.Feng RE. IASLC/ATS/ERS international multidisciplinary new classification of lung adenocarcinoma and its clinical significance. Zhonghua Jie He He Hu Xi Za Zhi. 2012;35:95–96. (In Chinese) [PubMed] [Google Scholar]
- 10.Li X, Wan L, Shen H, Geng J, Nie J, Wang G, Jia N, Dai M, Bai X. Thyroid transcription factor-1 amplification and expressions in lung adenocarcinoma tissues and pleural effusions predict patient survival and prognosis. J Thorac Oncol. 2012;7:76–84. doi: 10.1097/JTO.0b013e318232b98a. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Wan L, Geng J, Wu CL, Bai X. Aldehyde dehydrogenase 1A1 possesses stem-like properties and predicts lung cancer patient outcome. J Thorac Oncol. 2012;7:1235–1245. doi: 10.1097/JTO.0b013e318257cc6d. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Bai X, Zhang C, Zhang X. Evaluation of Her2 gene amplification in breast cancer using nuclei microarray in situ hybridization. Int J Mol Sci. 2012;13:5519–5527. doi: 10.3390/ijms13055519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H, Zhang X, Zhang X, Yang D, Meng L, Zhang Y, Zhou S. The effect of esculentoside A on lupus nephritis-prone BXSB mice. Arch Med Sci. 2013;9:354–360. doi: 10.5114/aoms.2012.31439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landis JR, Koch GG. A one-way components of variance model for categorical data. Biometric. 1977;33:671–679. doi: 10.2307/2529465. [DOI] [Google Scholar]
- 15.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 16.Rimm DL, Camp RL, Charette LA, Olsen DA, Provost E. Amplification of tissue by construction of tissue microarrays. Exp Mol Pathol. 2001;70:255–264. doi: 10.1006/exmp.2001.2363. [DOI] [PubMed] [Google Scholar]
- 17.von Wasielewski R, Mengel M, Wiese B, Rüdiger T, Müller-Hermelink HK, Kreipe H. Tissue array technology for testing interlaboratory and interobserver reproducibility of immunohistochemical estrogen receptor analysis in a large multicenter trial. Am J Clin Pathol. 2002;118:675–682. doi: 10.1309/URLK-6AVK-331U-0V5P. [DOI] [PubMed] [Google Scholar]
- 18.Braunschweig T, Chung JY, Hewitt SM. Tissue microarrays: Bridging the gap between research and the clinic. Expert Rev Proteomics. 2005;2:325–336. doi: 10.1586/14789450.2.3.325. [DOI] [PubMed] [Google Scholar]
- 19.Fennell DA, Myrand SP, Nguyen TS, Ferry D, Kerr KM, Maxwell P, Moore SD, Visseren-Grul C, Das M, Nicolson MC. Association between gene expression profiles and clinical outcome of pemetrexed-based treatment in patients with advanced non-squamous non-small cell lung cancer: exploratory results from a phase II study. PLoS One. 2014;9:e107455. doi: 10.1371/journal.pone.0107455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapila K, Al-Ayadhy B, Francis IM, George SS, Al-Jassar A. Subclassification of pulmonary non-small cell lung carcinoma in fine needle aspirates using a limited immunohistochemistry panel. J Cytol. 2013;30:223–225. doi: 10.4103/0970-9371.126640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalhor N, Zander DS, Liu J. TTF-1 and p63 for distinguishing pulmonary small-cell carcinoma from poorly differentiated squamous cell carcinoma in previously pap-stained cytologic material. Mod Pathol. 2006;19:1117–1123. doi: 10.1038/modpathol.3800629. [DOI] [PubMed] [Google Scholar]