ABSTRACT
Triggers and regulatory pathways that effectively link human cytomegalovirus (HCMV) major immediate early (MIE) latent-lytic switch activation with progeny production are incompletely understood. In the quiescently infected human NTera2 cell model of primitive neural stem cells, we found that costimulation with vasoactive intestinal peptide (V) and phorbol ester (P) synergistically activated viral infection, but this effect waned over time. Coupling retinoic acid (R), an inducer of neuronal differentiation, to VP pulse stimulation attenuated the decline in viral activity and promoted the spread of the active infection through concentric layers of neighboring cells as cellular differentiation progressed. R stimulation alone was unable to activate the infection. The MIE enhancer cis-regulatory mechanisms responsible for this result were characterized by a strategy of combinatorial mutagenesis of five cis-acting element types (retinoic acid receptor binding elements [RARE], cyclic AMP [cAMP] response elements [CRE], NF-κB binding sites [kB], serum response element, and ETS/ELK-1 binding site) and multiple methods of assessment. We found that the CRE and kB combination sets the preinduction enhancer tone, is the major initiator and amplifier of RVP-induced MIE gene expression, and cooperates with RARE during cellular differentiation to enhance viral spread. In predifferentiated NTera2, we also found that the CRE-kB combination functions as initiator and amplifier of unstimulated HCMV MIE gene expression and cooperatively interacts with RARE to enhance viral spread. We conclude that RVP-stimulated signaling cascades and cellular differentiation operate through the enhancer CRE-kB-RARE core in strengthening induction of HCMV MIE gene expression in linkage with viral propagation.
IMPORTANCE Cytomegalovirus-seropositive persons commonly lack detectable levels of cytomegalovirus replication, even when profoundly immunocompromised. In a human NTera2 cell model of primitive neural stem cells carrying resting cytomegalovirus genomes, we show that costimulation of protein kinase A and C-delta signaling cascades in conjunction with retinoic acid-induced neuronal differentiation brings about progeny virus propagation. Iterated DNA binding sites for retinoic acid receptor, CREB, and NF-κB family members in the cytomegalovirus major enhancer are at the crux in the pathway to HCMV activation. The stimulated CREB and NF-κB binding site combination vigorously initiates and amplifies the active cytomegalovirus infection and cooperates with activated retinoic acid receptor binding sites to further promote viral proliferation and spread between differentiated cells. These results support a paradigm in which a specific combination of stimuli coupled with cellular differentiation satisfies a core cis-activating code that unlocks enhancer silence to repower the cycle of cytomegalovirus propagation.
INTRODUCTION
The majority of the world's population is carrier to the human cytomegalovirus (HCMV) (1). Like other Herpesviridae family members, HCMV dwells in its host in a quasi-inactive state of viral latency until prompted by a specific set of stimuli or conditions to produce viral progeny. HCMV inhabits virtually all types of human tissues, a feature that has long complicated efforts in transplantation. Shedding of virus into saliva, urine, semen, and cervicovaginal secretions is the usual means by which HCMV is passed to susceptible individuals (2). Among the HCMV-seropositive population, episodic shedding of infectious virus into bodily fluids or molecular traces of viral lytic cycle activity in tissues are seldom detected in healthy persons but are commonly detected in persons with severe cellular immune deficiency or inflammatory conditions (e.g., septic shock or burns). However, many HCMV-seropositive persons with the same predisposing conditions do not experience the outcome of productive HCMV replication or viral shedding (3–5). Interhost differences in factors that drive HCMV latent-lytic switch activation in linkage with viral progeny production might partly account for this clinical observation. In the mouse model, a solitary stimulus that potently initiates murine CMV latent-lytic switch activation in vivo is not enough to effectively produce viral progeny (6).
Hematopoietic cells of monocytic and dendritic cell lineages are sites of HCMV latency. Naturally infected hematopoietic cells further subjected to differentiation and stimulation in vitro have subsequently produced infectious virus, but this outcome is inefficiently achieved using current methods. Hematopoietic cells are likely not the only cell types that support HCMV latency (7). In latently infected mice, periventricular primitive neuronal cell precursors are the source of latent virus that reactivates after culturing brain explants (8). The ventriculoencephalitis that results from HCMV reactivation in profoundly immunocompromised persons suggests a similar analogy. In these patients, the active HCMV infection is located in ependymal cells and subependymal neuronal cells (9, 10) that layer on a zone of neuronal stem cells.
HCMV infection of human embryonal NTera2-D1 cells (NT2) (11) maintained under drug-free stem cell growth conditions models HCMV quiescence in a primitive neuronal stem cell (12, 13). HCMV genomes effectively penetrate NT2 nuclei (14), and a subset of the nonreplicating viral genomes are configured as covalently closed circles with superhelical twists (12). As is the case in primary human neuronal stem cells, retinoic acid (R) exposure induces NT2 neuronal differentiation (15, 16) via R receptor-mediated signaling (16, 17). Differentiated NT2 permit an acute active HCMV infection that produces infectious viral progeny (13).
The HCMV major immediate early (MIE) gene products IE1-p72 and IE2-p86 are pivotal activators of the HCMV lytic cycle. In latently infected myeloid cells, expression of HCMV MIE genes is greatly restricted but is required for reactivating HCMV replication (18, 19). HCMV MIE gene expression is also tightly restricted in human NT2, embryonic stem cell lines, and primitive neuronal stem cells (12, 20, 21). Priming with R or adding R at the time of infection allows HCMV MIE gene expression in a subset of NT2 and primitive neuronal stem cells (12, 21). However, delaying R's application to NT2 after the infection fails to achieve this outcome (12). Stimulating quiescently infected NT2 with vasoactive intestinal peptide (V), an immunomodulatory neuropeptide, rapidly activates viral MIE gene expression in a cell subset via the cellular protein kinase A (PKA)-CREB-TORC2 signaling cascade and the repetition of MIE enhancer's cyclic AMP response elements (CRE) (22). Phorbol 12-myristate 13-acetate (P) also activates HCMV MIE gene expression in a subset of quiescently infected NT2, but through a different signaling pathway involving PKC-delta, CREB, and NF-κB (p65 and p50) and the MIE enhancer's CRE and NF-κB-binding sites (kB) (23). P also activates MIE gene expression in a small subset of human embryonic stem cells and Kasumi-3 myeloid progenitor cells that carry quiescent HCMV genomes (20, 24). Neither V nor P alone is able to productively activate HCMV in the broader population of quiescently infected cells. The global gene expression profile of NT2 closely resembles that of human embryonic stem cells (25–27). Cellular transcription factor OCT4, a pluripotency signature, maintains the NT2 undifferentiated state (28, 29). Neither V nor P disrupts the OCT4-maintained state of NT2 stemness during the peak time of active MIE gene expression (22, 23).
A specific combination of external cues and intrinsic cellular conditions is postulated to comprise the key for unlocking all restraints on reactivated HCMV replication. In this report, we describe a combination of stimuli that productively reverses HCMV quiescence in the NT2 primitive neuronal stem cell model. This combination launches multiple regulatory cascades and cellular differentiation. These concerted actions funnel through a triad of different MIE enhancer cis-acting element types that work in synergy to advance the active HCMV infection in restoring viral propagation.
MATERIALS AND METHODS
Cells and viruses.
Human NTera2/D1 cells (NT2) were kindly provided by E. Gonczol (13). NT2 were grown in Dulbecco's modified Eagle medium (DMEM) plus 3% of charcoal-treated HyClone fetal bovine serum (FBS) and 3% of Knockout Serum Replacement (Invitrogen) to minimize background levels of NT2 differentiation and HCMV MIE expression (12, 22). Mycoplasma-free NT2 were cultured in the presence of penicillin and streptomycin. The following inducers of HCMV activation were added to NT2 growth medium lacking Knockout Serum Replacement: retinoic acid (R; Sigma), 10 μM; phorbol 12-myristate 13-acetate (P; Sigma), 20 nM; and vasoactive intestinal peptide (V; EMD Millipore), 100 nM. For longer-term growth of NT2 after pulse induction with stimuli, the medium was changed to DMEM plus 10% HyClone FBS, with or without R (10 μM). Human foreskin fibroblasts (HFF) were isolated, propagated, and studied at passage number ≤6 (14). Differentiated NT2 cells (D-NT2) were generated by treatment of NT2 with R (10 μM) for ≥15 days in DMEM plus 10% FBS, and R was then removed 48 h prior to HCMV infection.
HCMV strains Towne and VR1814 were used. Strain VR1814 was maintained in human umbilical vein endothelial cells prior to one-step amplification in HFF. All viruses were partially purified by centrifugation of filtered infected-HFF cell supernatant through a 20% sorbitol cushion in phosphate-buffered saline (PBS) (12). NT2 were inoculated for 90 to 120 min with the indicated viruses at a multiplicity of infection (MOI) of 5 to 10 PFU per ml in DMEM plus 3% charcoal-treated FBS. Infected cells were then washed twice with Hanks' balanced salt solution without calcium or magnesium (HBSS). D-NT2 were infected in the same way, but at MOIs in high (1 to 3 PFU/ml) and low (0.03 to 0.05 PFU/ml) ranges.
HCMV-GFP has a green fluorescent protein (GFP) gene driven by the viral native UL127 promoter that is expressed with early/late kinetics (30). HCMV recombinants rCRE− (rC−), rkB− (rK−), rCRE−.kB− (rCK−), and rWTCK− have been reported previously (22, 23) and were derived from HCMV strain Towne in a bacterial artificial chromosome (BAC) (31). With the same procedures, we additionally constructed HCMV recombinants having site-directed base substitution mutations that functionally inactivate the retinoic acid response elements (RARE) (32), the serum response element (SRE) (33), and the ETS/ELK-1 (22, 33) binding site in the MIE enhancer. Placement of mutations in each of three copies of RARE created rRARE− (rR−). Combining CRE-kB mutations with RARE or SRE-ETS mutations created rCRE−.kB−.RARE− (rCKR−) or rCRE−.kB−.ETS−.SRE− (rCKES−), respectively. Replicates of recombinant viruses were produced from independent recombination procedures (22, 23). All recombinant virus genomes were analyzed by gel electrophoresis for size-mobility pattern of EcoRI restriction fragments and by DNA sequencing through the HCMV MIE locus. Only recombinant viruses at passage number ≤3 were used for phenotype analyses.
RNA analyses.
Whole-cell RNA was isolated according to the method of Chomczynski and Sacchi (34). Reverse transcription (RT) and quantitative real-time PCR (qPCR) were performed using methods described previously (22, 23). Spliced MIE RNA was quantified by using a primer set targeting MIE exons 1 and 2 and a fluorophore-conjugated probe spanning the mRNA splice junction (35). Spliced IE1 (36) and spliced IE2 (37) RNA levels were quantified by the TaqMan qPCR method, using analytes published previously.
Protein analyses.
Western blotting of whole-cell extracts supplemented with protease and phosphatase inhibitors and fractionated by SDS-PAGE were performed using methods described previously (22, 23). HCMV IE1-p72 and IE2-p86 were detected by monoclonal murine antibody MAB810 (EMD Millipore). GFP and beta-tubulin were detected by anti-GFP rabbit monoclonal antibody (Epitomics) and E7 murine monoclonal anti-beta-tubulin antibody (University of Iowa Hybridoma Bank, Iowa City, IA), respectively.
Immunofluorescence assay (IFA) was performed using published methods (22, 23). Primary antibodies MAB810 (Millipore), anti-pp28 (EastCoast Bio), and anti-gB (EastCoast Bio) were used for detection of HCMV MIE, pp28, and gB proteins, respectively. Secondary goat anti-mouse antibody conjugated to Alexa Fluor 555 or 488 (Molecular Probes Invitrogen) was applied. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; 1 mg/ml). An inverted Olympus IX 51 fluorescent microscope equipped with an X-Cite 120 fluorescence illumination system was used to capture images. The ratio of MIE+ cells to total DAPI+ cells was determined using NIH ImageJ 1.34s software.
Flow cytometric analysis was performed using a BD LSR II flow cytometer (BD Biosciences). Data were analyzed with BD FACS Diva software (where FACS is fluorescence-activated cell sorter) and FlowJo software. Cells were fixed in cold methanol, treated with 0.1% Tween 20 in PBS, and washed in PBS plus 2% FBS prior to the reaction with MAB810X conjugated to Alexa 488 (EMD Millipore) for detection of intracellular HCMV MIE proteins. Live/Dead Fixable Aqua staining was applied according to the company's instructions (Invitrogen).
Assay of infectious virus produced.
To quantify the amount of infectious virus in NT2, cells were washed, scraped, frozen, thawed, and gently sonicated. The cell extract, as well as the cell growth medium supernatant, was then passed through a 0.45-μm-pore-size filter. Filtered cell extract or supernatant was applied in a standard viral plaque assay on HFF to determine the infectious titer (in PFU per milliliter) (14). The spread of recombinant viruses from cell to cell was monitored in real time by live-cell fluorescence microscopy for detection of vGFP+ cells. All HCMV BAC recombinants express GFP from a simian virus 40 (SV40) early promoter-GFP gene located in the BAC DNA segment (31). HCMV-GFP is not BAC derived and expresses GFP from the viral UL127 promoter. Captured images were inverted in Adobe Photoshop to improve discrimination of GFP+ cells by eliminating the black background.
RESULTS
VP-joint synergism in HCMV infection activation fades over time.
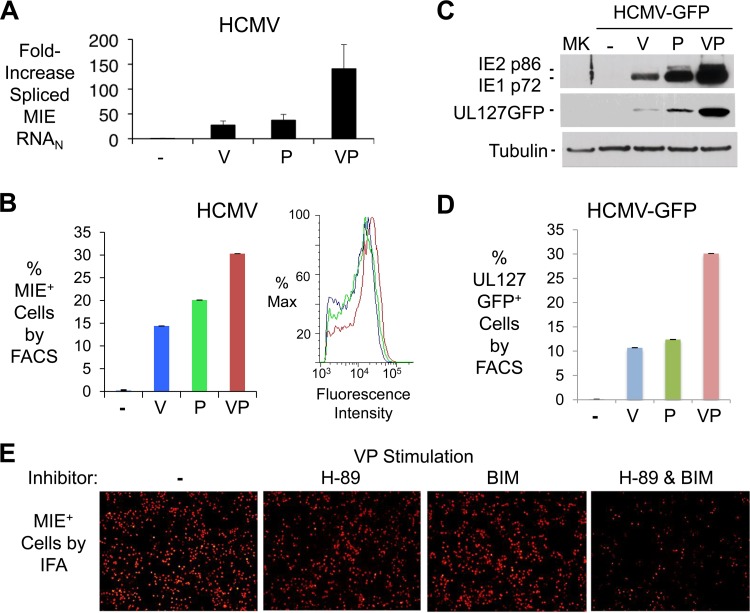
In quiescently infected NT2, either V-induced PKA/TORC-2/CREB activation (22) or P-induced PKC-delta/CREB/NF-κB activation (23) functions to partially relieve HCMV MIE gene silence. While PKA and PKC-delta separately drive signaling cascades that act through the same MIE enhancer cis-acting elements, the coactivation of these pathways may have unique downstream consequences. We show in Fig. 1 that costimulation with V and P (VP) generated a magnitude of HCMV immediate early and early gene expression that exceeded the sum of the individual responses produced by each component of the VP combination. The change in levels of HCMV-spliced MIE RNA (Fig. 1A) was matched with a commensurate change in levels of the resultant spliced IE1 and IE2 RNA subsets (data not shown). The VP combination also increased the proportion of NT2 expressing HCMV MIE proteins (MIE+ NT2), as well as the intracellular concentration of immunoreactive MIE protein in the MIE+ NT2 subpopulation (Fig. 1B). Analysis of crude extract from the entire NT2 population revealed that VP was superior to either stimulus alone in increasing the levels of expression of the dominant MIE proteins, IE1-p71 and IE2-p86 (Fig. 1C). A commensurate increase in the amount of the virally expressed GFP (vGFP) indicated that the MIE proteins were functioning in viral early kinetic-class gene activation (Fig. 1C). VP acted synergistically in expanding the vGFP+ NT2 population (Fig. 1D), which was evident over a wide range of V and P concentrations (data not shown). VP was also more effective at bringing about expression of other viral early kinetic-class gene products, such as pUL44 and gB (data not shown). As anticipated, inhibiting both PKA and PKC-delta signaling pathways greatly decreased VP's ability to activate MIE protein expression, whereas inhibiting just one of the pathways only modestly decreased this ability (Fig. 1E).
FIG 1.
The VP combination is superior to either stimulus alone in activating HCMV infection in quiescently infected NT2. (A) HCMV spliced MIE RNA level was quantified by RT-qPCR at 24 h poststimulation with the indicated agents. The bar graph depicts fold changes in RNA levels (means ± standard deviations [SD]), relative to that of mock treatment at 4 h, for triplicate biological samples after normalization to cellular 18S RNA amount (MIE RNAN). (B) Percentages of HCMV-infected NT2 containing IE1-p72/IE2-p86 (MIE+) were determined by a FACS-based method at 24 h poststimulation with the indicated agents (MOI of 5 PFU/cell). Fluorescence intensity profiles for MIE+ NT2 populations are shown in the panel insert; 20,000 gated cells were analyzed; error bars show SD for counting error. (C) Expression of IE1-p72, IE2-p86, and UL127GFP by HCMV-GFP 24 h after application of the indicated agents was assessed by Western blotting. (D) Percentages of live NT2 expressing UL127GFP were determined at 1 day after stimulation with indicated agents, based on FACS analysis of 30,000 propidium stain-negative cells. Unstimulated infected NT2 produced 0.1% GFP+ NT2. (E) Inhibition of VP-induced MIE+ NT2 (red fluorescence) by concentrations of H-89 (8 μM) and bisindolylmaleimide I (BIM, 150 nM) previously determined in NT2 to effectively inhibit PKA and PKC-delta activity, respectively (22, 23). IFA was performed at 24 h poststimulation. Cell density by DAPI counterstain did not differ significantly between groups (not shown).
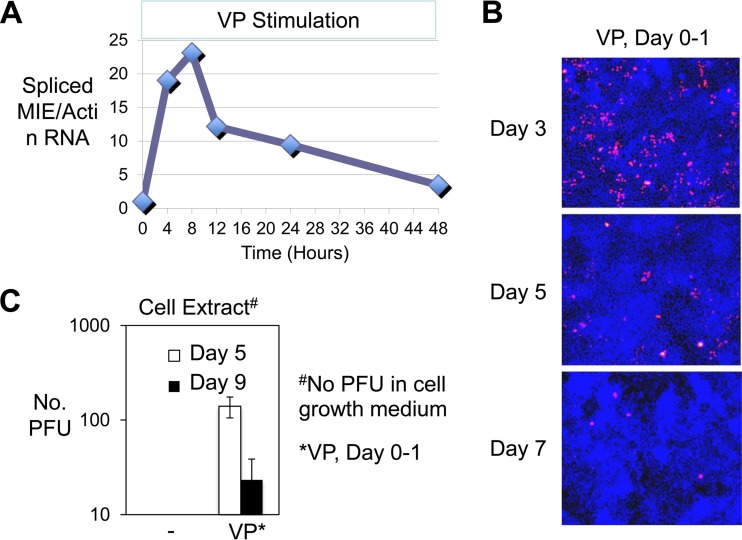
We next examined the durability of VP's effect on HCMV activation. While continuing the VP exposure, spliced MIE RNA amounts were found to peak at 8 h, decline steeply at 12 h, and then decline gradually over the course of 2 days, regardless of whether normalized to the level of cellular actin RNA (Fig. 2A) or 18S RNA (data not shown). The marked expansion in the MIE+ NT2 population size resulting from a 24-h pulse of VP treatment was followed as early as day 3 by a contraction in size of this population, which contracted further as the time span lengthened at days 5 and 7 after stimulation (Fig. 2B). Correspondingly, infectious HCMV progeny was found in greater amount inside NT2 at day 5 than at day 9 after VP stimulation (Fig. 2C). Infectious virus was not released into cell medium after VP stimulation, and virus was not recovered in the culture of the contents of the unstimulated infected NT2 population. Hence, the VP combination acts synergistically in launching the active HCMV infection, but this activity wanes over a fairly short time frame.
FIG 2.
VP-activated HCMV infection wanes over time. (A) HCMV spliced MIE RNA level was quantified by RT-qPCR at 4, 8, 12, 24, and 48 h after VP stimulation, which was continued for 48 h. RNAs from duplicate infections were pooled prior to quadruplicate measurements. The graph depicts fold changes in MIE RNA levels after normalization to actin mRNA (MIE/Actin RNA) relative to that of mock treatment at 4 h. (B) IFA of MIE+ cells (red) in HCMV-infected NT2 (MOI of 5 PFU/cell) at days 3, 5, and 7 after VP stimulation for 24 h (days 0 to 1). Cells were washed and cultured in DMEM containing 10% FBS plus R. Nuclear DNA was counterstained with DAPI (blue fluorescence). Original magnification, ×10. (C) Determination of HCMV PFU/ml in NT2 extract (PFU) at days 5 and 9 after VP stimulation for 24 h (day 0 to 1) or no stimulation, using standard plaque assay on HFF as described in Materials and Methods. Data represent results from 3 independent experiments; error bars show the SD. Cell growth medium supernatant was assayed in parallel.
R coupling to VP stimulation decreases loss of actively infected cells and promotes viral spread.
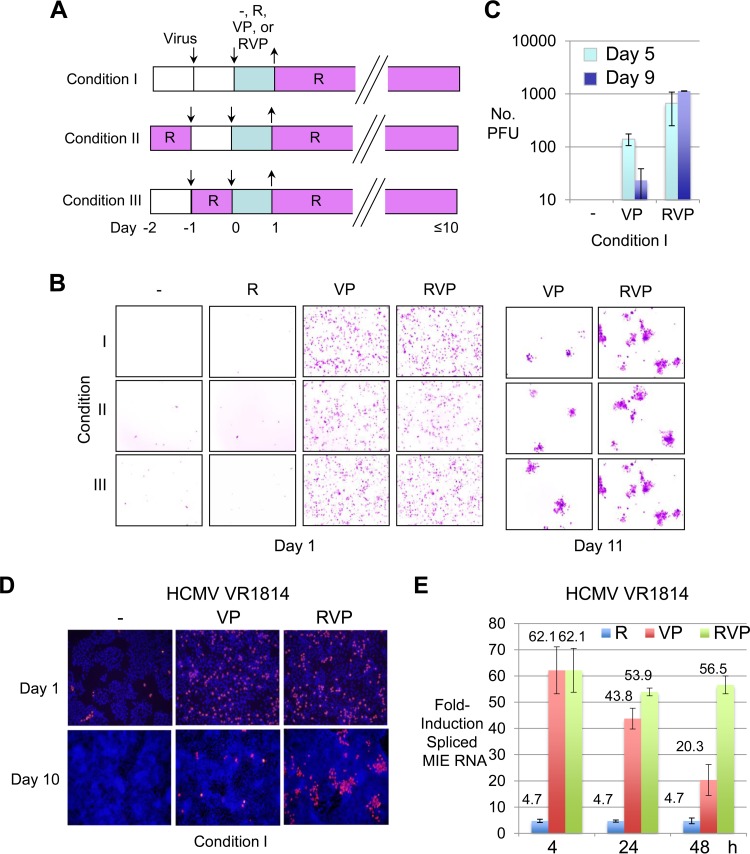
While R alone is unable to break HCMV quiescence in NT2 (12), it gradually yields NT2 differentiation along a neuronal lineage pathway (16). Subsequent infection of predifferentiated NT2 (D-NT2) produces infectious HCMV progeny (13). While exposing undifferentiated NT2 to R for a single day prior to infection is an insufficient length of time to render the cells differentiated, R quickly triggers changes in the cellular gene expression program that could conceivably influence the infection outcome. To determine whether the R-induced immediate signaling response or transformation in cellular differentiation might strengthen VP's ability to reverse HCMV quiescence in NT2, we tested three experimental conditions under which cells were first exposed to R at ≥1 day after infection (day 0 or 1 of stimulation), at the time of infection (day −1 before commencement of stimulation), or 1 day before infection (day −2 before commencement of stimulation) (Fig. 3A). All three experimental conditions included 4 treatment arms of a 24-h pulse stimulation with R, VP, RVP, or nothing, given at day 1 postinfection (p.i.) (day 0). R was applied the following day (day 1 after commencement of stimulation) to all treatment groups and was maintained thereafter, in order to promote cellular differentiation. As shown in Fig. 3B, pulse stimulation with R alone produced very few HCMV MIE+ NT2 at day 1 poststimulation. This outcome was not substantially changed by the priming of cells for 24 h with R at the commencement of infection (day −1) or 1 day before (day −2). Remarkably, VP and RVP initially produced similar-sized population expansions of HCMV MIE+ NT2 at day 1 poststimulation. The MIE+ NT2 population expansion gradually contracted over the course of days. A scattering of clusters of tightly packed vGFP+ NT2 (>6 vGFP+ cells) eventually emerged by day 7, and the number of vGFP+ cells per cluster increased with increasing days poststimulation. RVP outranked VP in both the number and the size of vGFP+ NT2 clusters that it produced (Fig. 3B). RVP also outperformed VP in the quantity of infectious HCMV progeny generated in NT2 at days 5 and 9 (Fig. 3C). RVP was unable to bring about the release of this virus into the culture medium (data not shown). Because priming with R had not substantially changed the result in the various treatment arms, all subsequent studies were performed without R priming.
FIG 3.
Adding R to VP later expands the active infection. (A) Schematic diagram of three different experimental conditions. Under conditions II and III, NT2 were primed with R for 24 h at days −2 and −1, respectively, relative to day 0 commencement of stimulation with nothing, R, VP, or RVP for 24 h. Cells were then washed and maintained in 10% FBS and R (10 μM). (B) HCMV-infected NT2 (MOI, 5) were subjected to conditions I, II, or III as described for panel A. Live-cell fluorescence microscopy was performed at days 1 and 11 poststimulation. The inverted image reveals HCMV-GFP+ NT2 (pink). Original magnification, ×10. (C) Determination via standard plaque assay of HCMV PFU/ml in NT2 lysate at days 5 and 9 after VP versus RVP stimulation for 24 h or no stimulation followed by continual R exposure (condition I). Data represent 3 independent experiments; error bars show the SD. (D) HCMV VR1814-infected NT2 (MOI, 10) were subjected to condition I in the manner described for panel A. MIE+ cells (pink) were analyzed by IFA at days 1 and 10 after stimulation for 24 h with nothing, VP, or RVP. Nuclear DNA was counterstained with DAPI (blue). Original magnification, ×10. (E) Comparison of R-, VP-, and RVP-induced expression of spliced MIE RNA by HCMV VR1814 at 4, 24, and 48 h poststimulation. Spliced MIE RNA levels were quantified from triplicate infections, using RT-qPCR and standard curve methods. The bar graph depicts fold increases in the spliced MIE RNA level (mean ± SD) relative to that produced in the absence of stimulation for each time point and normalized to the actin RNA level.
Testing of the endothelial-tissue-adapted HCMV strain VR1814, which is closer in form to an unmanipulated HCMV clinical isolate than is strain Towne, also revealed the superiority of RVP over VP in generating MIE+ NT2 clusters at day 10 poststimulation, but not in MIE+ cell numbers at day 1 poststimulation (Fig. 3D). Consistent with a previous report for HCMV Towne (12), R alone only minimally induced expression of spliced MIE RNA from HCMV VR1814, when assessed at 4, 24, and 48 h after commencement of R stimulation (Fig. 3E). In contrast, adding R to VP was remarkable in having lowered the rate of decline in the HCMV spliced MIE RNA level after the peak level of MIE RNA expression, despite continuing exposure to the stimuli.
Taken together, the results indicate that coupling of R to the VP pulse (i) limits the time-dependent decline in the amount of MIE RNA that occurs over the course of hours, (ii) limits the time-dependent decline in the number of actively infected NT2 that occurs over the course of several days, and (iii) enables the spread of the active infection to concentric layers of neighboring cells that occurs after a time lag of several days poststimulation.
The enhancer RARE contributes to R-induced differentiation-coupled HCMV activation and spread.
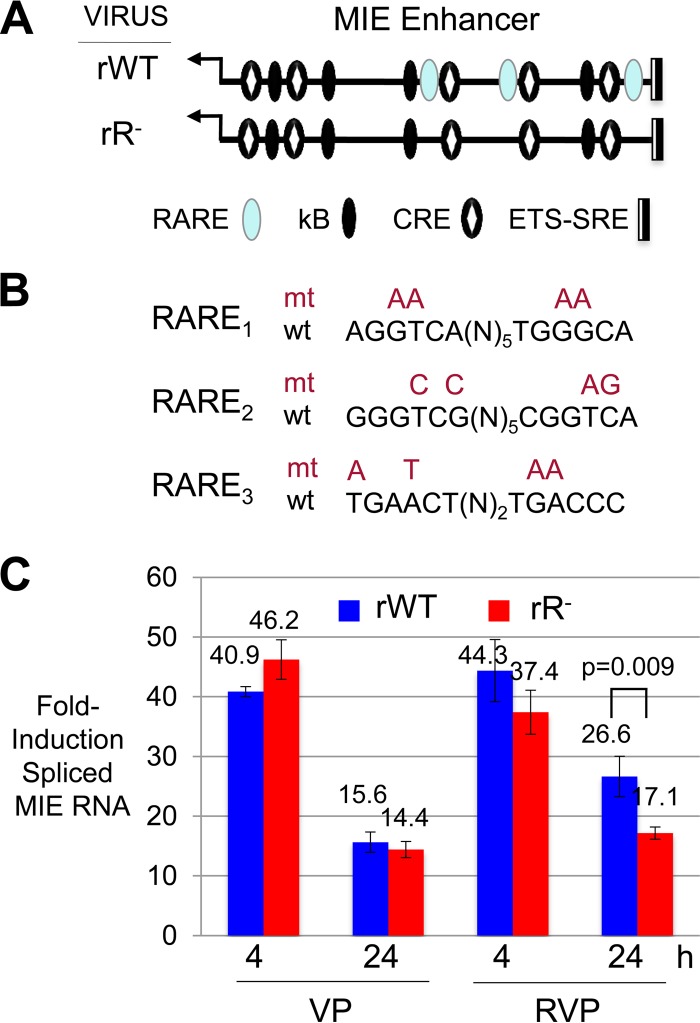
Previous reports have indicated that site-directed mutations placed in the three copies of retinoic acid response elements (RARE) in the MIE enhancer eliminates R-induced transcription from an HCMV MIE enhancer/promoter segment in a plasmid transiently transfected into NT2 (32, 38). To determine whether these observations translate to the virus and its response to RVP stimulation, we made the same base substitution mutations in the HCMV Towne genome (Fig. 4A and B). The application of equivalent infectious units of RARE-null (rR−) and parent wild-type (rWT) recombinant viruses to NT2 revealed that the two viruses produce similar amounts of spliced MIE RNA at 4 and 24 h of VP stimulation (Fig. 4C). The viruses differed minimally, but significantly (P < 0.001), in their response to RVP stimulation for 24 h. While enhancer RARE mutations only modestly lessened MIE gene expression in response to RVP stimulation in the short term, they could conceivably have additional effects on the virus that might not be manifest until later times after infection.
FIG 4.
Adding R to VP lowers the rate of decline in MIE RNA expression, which is partly dependent on the enhancer RARE. (A) Schematic diagram of location of RARE (3 copies), kB (4 copies), CRE (5 copies), and ETS-SRE (single set of sites) cis-acting elements in the HCMV MIE enhancer of the WT BAC recombinant (rWT) in comparison to the rR− BAC recombinant lacking functional RARE cis-acting elements. MIE enhancer cis-acting elements: RARE, blue oval; kB, black oval; CRE, black oval with white diamond; ETS-SRE, divided rectangle. (B) Diagram of specific base substitution mutations (mt) placed in each of three RARE (wt) in the HCMV rR− construct. (C) rWT versus rR− spliced MIE RNA levels at 4 and 24 h after VP or RVP stimulation were quantified from triplicate infections, using the methods described above.
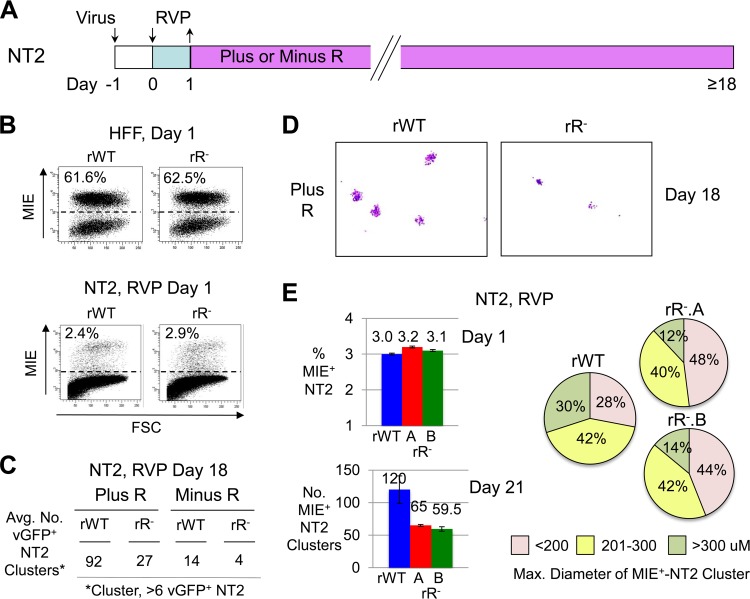
rR− and rWT were therefore compared in their abilities to develop vGFP+ NT2 clusters after RVP stimulation in relation to the presence or absence of continual R exposure (Fig. 5A). Balancing rWT and rR− by infectious units producing equivalent percentages of MIE+ HFF at day 1 p.i. (MOI, 0.6) revealed that the two viruses yielded equivalent percentages of MIE+ NT2 (MOI, 1) at day 1 of RVP stimulation (Fig. 5B). In the parallel group study of infected NT2 at day 18 after RVP pulse stimulation (MOI, 1), rR− was found to yield ∼70% fewer vGFP+ NT2 clusters than did rWT when cultured in either the presence or absence of R (Fig. 5C). Both rR− and rWT produced fewer vGFP+ NT2 clusters when R was omitted from the maintenance culture. rR− produced vGFP+ NT2 clusters that were visibly smaller in overall diameter than those produced by rWT (Fig. 5D). In a validation study, two independently constructed rR− recombinants, rR−.A and rR−.B, were concordant in the degree of disability in forming MIE+ NT2 clusters after RVP stimulation followed by R continuation, as gauged by the mildly decreased quantity and size of the MIE+ NT2 clusters in comparison to the rWT reference (Fig. 5E).
FIG 5.
RARE mutation-associated disability intensifies late after RVP induction. (A) Diagram of experimental design. Quiescently infected NT2 were pulsed with RVP for 1 day, followed by culturing in differentiation medium without or with R (Minus or Plus R, respectively). (B to D) NT2 were infected with equivalent infectious units of rWT and rR− (MOI, 1.0), as determined in parallel HFF infections at day 1 p.i. (MOI, 0.6 PFU/cell). MIE+ NT2 were analyzed by FACS at day 1 after RVP stimulation. Scatter plots show the distribution of MIE+ cells by fluorescence intensity versus forward side scatter. Percentages of MIE+ cells were determined from 20,000 and 100,000 gated HFF and NT2 cells, respectively (B). vGFP+ clusters (>6 cells per cluster) at day 18 after RVP stimulation were quantified for two separate infections, using live-cell fluorescence microscopy (C). Inverted images of representative micrographs show vGFP+ NT2 clusters (pink) at the original magnification of ×4 (D). (E) rWT and two independently constructed rR− recombinants, rR−.A and rR−.B, were subjected to the experimental conditions and analytical methods described above, with the exception of the use of IFA and image acquisition analysis to determine the spatial diameter of MIE+ NT2 clusters. The size distribution of 50 randomly selected MIE+ NT2 clusters is shown in a pie chart by category of range in diameter.
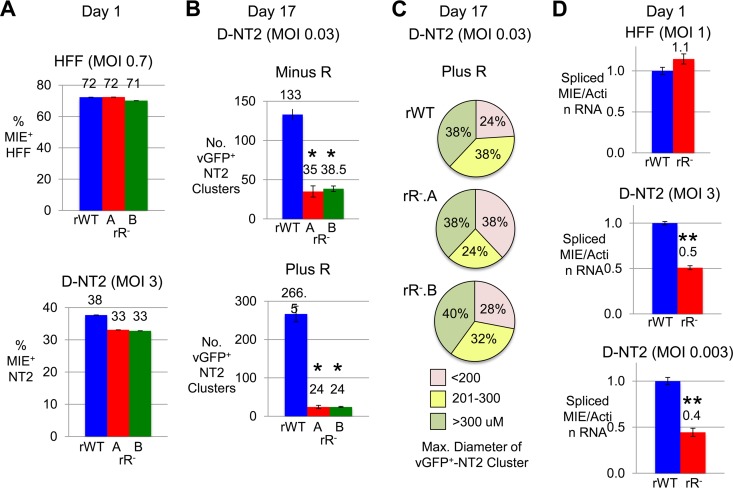
We further investigated whether the difference in rR− versus rWT phenotype was the result of the R stimulus or the indirect consequence of R-induced NT2 differentiation by study of the viruses in predifferentiated NT2 cells (D-NT2). D-NT2 were generated after 15 days of R exposure followed by washout of R for 2 days prior to infection. The balancing of rR−..A, rR−.B, and rWT for infectious units producing equivalent percentages of MIE+ HFF revealed that both rR−.A and rR−.B produce slightly fewer MIE+ D-NT2 (13%) at day 1 postinfection at an MOI of 3 in the absence of R exposure (Fig. 6A). In a parallel group, the MOI had been lowered to 0.03 to enable determination of whether the rR− viruses differed from rWT in the ability to form actively infected cell clusters in D-NT2. The infected cells were subsequently maintained in either the absence or the presence of R. As shown in Fig. 6B, both rR−.A and rR−.B produced significantly fewer vGFP+ D-NT2 clusters at day 17 postinfection (P < 0.005). Adding R after infection widened this difference (the differences between rR− and rWT vGFP+ D-NT2 cluster numbers were ∼3.5-fold and ∼11-fold in the absence and presence of R, respectively). The average diameter or size distribution of the clusters was not appreciably changed by the RARE mutations (Fig. 6C). Placing mutations in enhancer CRE (all five copies) instead of RARE had not produced an abnormality in MIE+/vGFP+ D-NT2 cluster formation (data not shown). In the absence of R stimulation, rR− also exhibited a modest but statistically significant ∼50 to 60% reduction in MIE RNA expression at 1 day p.i. at high and low MOIs (P < 0.005) (Fig. 6D).
FIG 6.
RARE mutation-associated disability also manifests in D-NT2. HFF and D-NT2 were infected with rWT and rR− at the indicated MOI. D-NT2 were derived by R-induced NT2 differentiation for 15 days, followed by washout of R for 2 days prior to infection. (A) At 1 day p.i., the percentage of MIE+ cells was determined by FACS analysis of 20,000 gated HFF (MOI, 0.7 PFU/ml) and 100,000 gated D-NT2 (MOI, 3.0) cells. (B) At 17 days p.i. at an MOI of 0.03, the average number of vGFP+ D-NT2 clusters was determined for separate biological replicates for each virus type. R (10 μM) was added to (Plus R) or omitted from (Minus R) the growth medium immediately after viral absorption. (C) Size distribution of 50 randomly selected vGFP+ D-NT2 clusters by category of diameter range. Results shown in panels A to C were produced from parallel infections. (D) At 1 day p.i., spliced MIE RNA levels normalized to actin RNA levels were quantified from triplicate infections of HFF (MOI, 1.0) and D-NT2 (MOI, 3.0 and 0.003) in the absence of R. *, P < 0.005; **, P < 0.001.
We surmise that the enhancer's RARE responds to both the intrinsic D-NT2 condition of cellular differentiation and R's stimulatory actions. The combined results also indicate that MIE enhancer-dependent viral productivity is driven by regulatory elements other than just the three RARE. Conversely, R is additionally acting via mechanistic pathways that are not disrupted by a string of RARE mutations in the MIE enhancer.
The enhancer CRE-kB combination sets the preinduction enhancer tone, initiates and amplifies MIE gene expression, and cooperates with RARE in driving differentiation-associated viral spread.
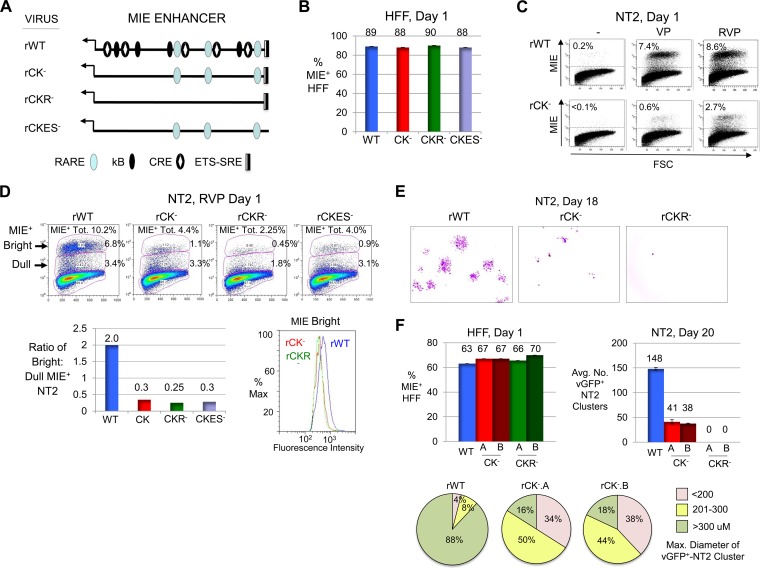
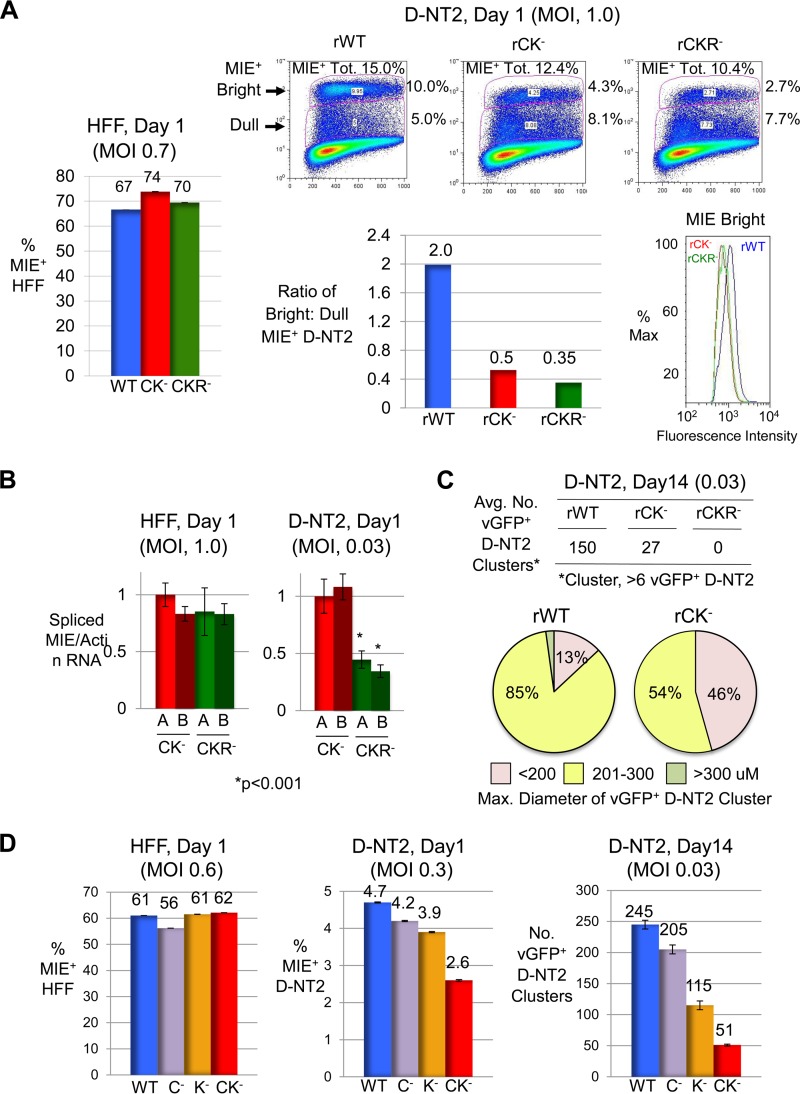
While the HCMV MIE gene regulatory elements that integrate RVP-stimulated signals are unknown, they are predicted to relay activities of PKA, PKC-delta, and R signaling cascades. To identify these elements, we assembled a panel of HCMV recombinants differing in specific transcription factor binding site candidates disabled by base substitution mutations. Because solitary targeting of the MIE enhancer's CRE, kB, ETS/ELK-1 binding site (ETS), or serum response element (SRE) had not disrupted RVP-activated MIE gene expression (data not shown), our focus turned to viruses carrying combinations of these mutations (Fig. 7A). The ETS-SRE mutations were fashioned after those that inactivated this unit's response to P- or V-induced signaling in other systems. Infectious titers of viruses without and with mutations in CRE-kB (rCK−), CRE-kB plus RARE (rCKR−), and CRE-kB plus ETS-SRE (rCKES−) were balanced in HFF with respect to producing equivalent levels of MIE+ cells (Fig. 7B) and IE1-p71 and IE2-p86 (data not shown) per FACS- and Western blot-based determinations, respectively. In the parallel group of quiescently infected NT2, we found that mutations in the CRE-kB combination nearly abolished VP's ability to produce MIE+ NT2 at day 1 of stimulation but were less effective at stopping RVP from initiating production of MIE+ NT2 (Fig. 7C). Adding RARE mutations to CRE-kB mutations further reduced by ∼50% the proportion of NT2 expressing any level of MIE protein at day 1 of RVP stimulation, whereas the addition of ETS-SRE mutations resulted in <10% difference (Fig. 7D). Strikingly, the CRE-kB mutations dropped the MIE protein amount expressed by individual members of the MIE+ NT2 population despite RVP stimulation. This drop is reflected in a marked lowering in the ratio of bright MIE+ (MIEBright) NT2 to dull MIE+ (MIEDull) NT2 subpopulations (ratios of 0.3 and 2.0 for rCK− and rWT, respectively) and a decrease in mean fluorescence intensity (MFI) of the MIEbright NT2 subpopulation. The add-on of RARE or ETS-SRE mutations does not substantially further reduce the MIE protein amount in the MIE+ NT2 population.
FIG 7.
CRE-kB mutations lower the initiation frequency and amplitude of RVP-induced MIE gene expression, whereas add-on RARE mutations nearly abolish viral activity later on. (A) Diagram of MIE enhancer cis-acting element types functionally neutralized by base substitution mutations in the context of recombinant HCMV genomes. Mutations were placed in CRE-kB (rCK−), CRE-kB plus RARE (rCKR−), and CRE-kB plus ETS-SRE (rCKES−). See Materials and Methods for details. (B to D) HFF (MOI, 0.8 PFU/cell) (B) and NT2 (MOI, 5.0) (C and D) were infected with rWT, rCK−, rCKR−, and/or rCKES−. FACS and FlowJo analyses were performed to characterize MIE+ HFF (20,000 gated cells) at day 1 p.i. and MIE+ NT2 (80,000 gated cells) at day 1 of stimulation with VP, RVP, or nothing. Input viral titers were adjusted to produce comparable amounts of MIE+ HFF (B). Percentages of MIE+ NT2 relative to all gated NT2 produced by VP- versus RVP-stimulated rWT and rCK− are depicted in scatter plots (C). Scatter plots of RVP-stimulated rWT, rCK−, rCKR−, and rCKES− depict the percentages of bright and dull MIE+ NT2 subsets, which make up the total MIE+ population (MIE+ Tot.) (D). The bar graph shows the ratio of bright to dull MIE+ NT2 for each of the viruses, whereas the histogram shows fluorescence intensity distributions of bright MIE D-NT2 subpopulations for rWT, rCK−, and rCKR− (D). (E) Inverted images of representative micrographs show vGFP+ NT2 clusters (pink color) for rWT, rCK−, and rCKR− at day 18 after commencement of RVP stimulation for 24 h followed by continual exposure to R; original magnification, ×4. rCKR− did not form NT2 clusters (>6 cells per cluster). The difference in input titers for the viruses was ≤3%, as determined in parallel analyses of MIE+ HFF at day 1 postinfection (not shown). (F) In a separate study, vGFP+ NT2 cluster formation (>6 cells per cluster) was analyzed for rWT, rCK−.A, rCK−.B, rCKR−.A, and rCKR−.B at day 20 after RVP stimulation followed by continual exposure to R, in relation to percentages of MIE+ HFF at 1 day p.i. The average number (Avg. No.) of clusters was determined from two separate infections. Size distributions of 50 randomly selected vGFP+ NT2 clusters are depicted by category of range in diameters.
Carrying forward the rWT, rCK−, and rCKR− infections for 18 days after RVP pulse stimulation followed by the continual presence of R revealed via inspection by inverted microscopy that rCK− produces smaller and ∼70% fewer vGFP+ NT2 clusters than does rWT (Fig. 7E). Moreover, rCKR− failed to produce clusters having >6 vGFP+ cells at day 18 and vGFP+ cells were rare. To confirm these results, two independently constructed viruses for rCK− (rCK−.A and rCK−.B) and rCKR− (rCKR−.A and rCKR−.B) were subjected to study in the same manner. As shown in Fig. 7F, rWT, rCK−.A, rCK−.B, rCKR−.A, and rCKR−.B were well balanced for infectious units used to establish quiescent NT2 infection. At day 20 after commencement of RVP pulse stimulation, rCK−.A and rCK−.B exhibited concordant behavior in yielding ∼73% fewer vGFP+ NT2 clusters and smaller clusters. Both rCKR−.A and rCKR−.B failed to produce vGFP+ NT2 clusters, whereas rWT produced 148 vGFP+ NT2 clusters on average.
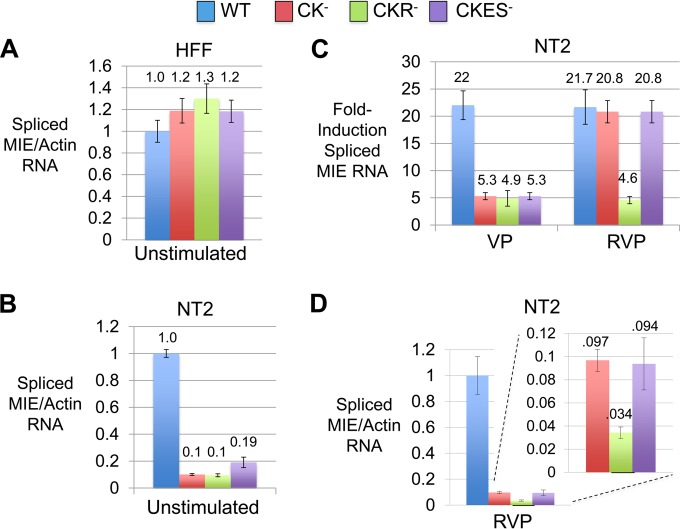
To better understand the mechanism behind the attenuated MIE protein expression by viruses containing CRE-kB mutations, we quantified spliced MIE RNA levels at 4 h after mock, VP, or RVP stimulation. Comparable inputs of infectious units of rWT, rCK−, rCKR−, and rCKES− were applied, as reflected in the spliced MIE RNA amount produced in a parallel infection of HFF (MOI, 1) (Fig. 8A). Remarkably, the infections of unstimulated NT2 (MOI, 5) revealed that CRE-kB mutations dropped basal levels of spliced MIE RNA production nearly 10-fold (Fig. 8B). The addition of RARE or ETS-SRE mutations did not further lower the basal MIE RNA level. Limiting mutations to either CRE or kB alone did not lower the basal production of spliced MIE RNA to the degree of lowering caused by mutations in both CRE and kB (data not shown). VP stimulation of rWT increased the spliced MIE RNA amount ∼22-fold over the levels produced in the unstimulated NT2 infection (Fig. 8C). Compared to this reference level of fold induction, mutations in CRE-kB lowered the fold induction level by ∼75%, and the add-on of RARE or ETS-SRE mutations had not further reduced the fold induction level. RVP stimulation of rWT also increased the spliced MIE RNA amount ∼22-fold. This magnitude of induction was not significantly lowered by mutations in CRE-kB or CRE-kB plus ETS-SRE. In contrast, combining CRE-kB and RARE mutations lowered the fold induction level by ∼75%. Separate studies showed that mutations in RARE alone had an insignificant effect on the MIE RNA level at 4 h of RVP stimulation (Fig. 4C). With RVP stimulation, CRE-kB mutations caused the total amount of spliced MIE RNA to drop 10-fold (Fig. 8D), commensurate with the drop of 10-fold in the basal level of MIE RNA expression in unstimulated cells. Mutations in CRE-kB plus RARE decreased the total amount of spliced MIE RNA 30-fold, which comprises the decreases in basal and inducible levels of MIE RNA expression. These results support the idea that the CRE-kB acts to determine the preinduction enhancer tone and, thereby, governs postinduction levels of MIE gene expression in undifferentiated NT2.
FIG 8.
CRE-kB mutations lower pre- and postinduction MIE RNA levels, whereas add-on RARE mutations blunt the inducible response. (A to D) HFF (MOI, 1.0 PFU/cell) (A) and NT2 (MOI, 5.0) (B to D) were infected in parallel with rWT, rCK−, rCKR−, and rCKES−. HCMV spliced MIE RNA level was quantified by RT-qPCR in triplicate biological samples at 4 h after HFF infection and at 4 h after stimulation of quiescently infected NT2 with VP, RVP, or nothing (Unstimulated). Bar graphs in panels A, B, and D depict spliced MIE RNA levels relative to that produced by rWT after normalization to cellular actin RNA amount (means ± SD). The panel C bar graph depicts fold changes in levels of spliced MIE RNA before and after stimulation for the respective virus type, normalized to actin RNA levels (means ± SD). RNA levels were quantified using TaqMan and standard curve methods.
Because rCK− and rCKR− differ the most in growth features that become apparent several days after commencement of R-coupled stimulation and induction of cellular differentiation, the activities of rWT, rCK−, and rCKR− were compared in predifferentiated D-NT2. At day 1 postinfection (MOI, 1), in the absence of R exposure, the mutations in CRE-kB and CRE-kB plus RARE were found to produce only ∼20% and ∼30% fewer MIE+ D-NT2, respectively (Fig. 9A). In contrast, the CRE-kB mutations greatly lowered MIE protein amounts in the vast majority of cells in the MIE+ D-NT2 population, as reflected in the lowering of the MIEbright/MIEdull NT2 ratio (0.5 and 2.0 for rCK− and rWT, respectively) and MFI of the MIEbright D-NT2 subpopulation. The addition of RARE mutations resulted in a further 30% reduction in MIEbright/MIEdull NT2 ratio beyond that caused by the CRE-kB mutations. This difference in MIE protein level accords with a 50 to 60% reduction in MIE RNA levels produced by rCKR−.A and rCKR−.B versus rCK−.A, and rCK−.B at day 1 postinfection in the absence of R (Fig. 9B). While the magnitude of difference between rCK− and rCKR− in the amplitude of MIE gene expression is similar for D-NT2 and NT2 (Fig. 7), the former situation is linked to cellular differentiation and the latter situation is dependent on RVP stimulation.
FIG 9.
Abnormalities caused by CRE-kB and add-on RARE mutations also manifest in D-NT2. (A) HFF (MOI, 0.7 PFU/cell) and D-NT2 (MOI, 1.0) were infected with rWT, rCK−, and rCKR− in the absence of R exposure. FACS was applied at day 1 p.i. to determine the percentages of MIE+ cells. Scatter plots show percentages of 250,000 gated cells in bright and dull MIE+ −D-NT2 subsets, which make up the total MIE+ population (MIE+ Tot.). The bar graph depicts the ratio of bright to dull MIE+ D-NT2. The histogram shows the fluorescence intensity profile for the MIEBright D-NT2 population. (B) At 1 day p.i. with rCK−.A, rCK−.B, rCKR−.A, and rCKR−.B, spliced MIE RNA levels were quantified from triplicate infections of HFF (MOI, 1.0) and D-NT2 (MOI, 0.03) in the absence of R and normalized to actin RNA levels. (C) D-NT2 (MOI, 0.03) infected with rWT, rCK−, and rCKR− in parallel with groups shown in panel A were maintained in the presence of R for 14 days p.i. The average number (Avg. No.) of vGFP+ D-NT2 clusters (>6 vGFP+ D-NT2 per cluster) was determined for two separate infections. Size distributions of 50 randomly selected vGFP+ NT2 clusters are depicted by category of diameter size. rCKR− did not form D-NT2 clusters at day 14. (D) HFF (MOI, 0.6 PFU/cell) and D-NT2 (MOI, 0.3 and 0.03) were infected with rWT, rC−, rK−, and rCK−. FACS determined the percentage of MIE+ D-NT2 at 1 day p.i. The average numbers of vGFP+ D-NT2 clusters were determined at day 14 p.i. for two separate infections.
A parallel group of D-NT2 infections (MOI, 0.03) carried forward for 14 days in the presence of R revealed that the CRE-kB mutations decrease both the quantity of vGFP+ NT2 clusters (by 83%) and the overall diameter of the clusters (Fig. 9C). The add-on of RARE mutations abolishes the virus's ability to form vGFP+ NT2 clusters. These findings were reproduced in separate studies of other independently constructed rCK− and rCKR− recombinants (data not shown). Thus, D-NT2 and RVP-stimulated NT2 undergoing differentiation yield matching patterns of growth differences for rWT, rCK−, and rCKR−. Lastly, the functional strength of the union between CRE and kB is also evident in D-NT2, as mutations in either CRE alone or kB alone are less effective at reducing MIE gene expression and formation of vGFP+ NT2 clusters than are mutations in both CRE and kB (Fig. 9D).
DISCUSSION
The regulatory mechanisms underlying differentiation-coupled activation of HCMV replication are not well understood. Here, we apply the differentiation-inducible NT2 model to characterize a set of stimuli whose combined actions effectively convert HCMV quiescence into productive infection. The pairing of VP stimuli increases both the likelihood and the amplitude of MIE gene expression, well exceeding that produced by stimulation with either individual component (Fig. 1). This outcome is dependent on a stretch of CRE and kB iterations in the MIE enhancer (Fig. 7) that respond to the mix of signals partly relayed through PKA and PKC delta (Fig. 1). Addition of R to the VP stimulation does not quantifiably increase MIE+ NT2 numbers or MIE protein amount in the short term (Fig. 3 and 7). However, it does lower the rate of MIE RNA level decline after 8 h poststimulation (Fig. 4) and off-loads work from CRE-kB to RARE in delivering the RVP-induced response (Fig. 7). In this HCMV quiescence system, fewer than 3 in 1,000 unstimulated NT2 contain MIE proteins detectable by FACS (Fig. 7), yet appreciable amounts of low-level expression of spliced MIE RNA are consistently detected in the unselected infected NT2 population (Fig. 1, 4, and 8). We surmise that spliced MIE RNA expression may occur below a threshold level whereby the FACS method of MIE protein detection is insensitive or the MIE protein is not produced. In the latter consideration, the threshold set point might be modulated in response to intrinsic cellular conditions or external stimuli via posttranscriptional mechanisms, such as microRNA-mediated silencing of MIE mRNA translation (39).
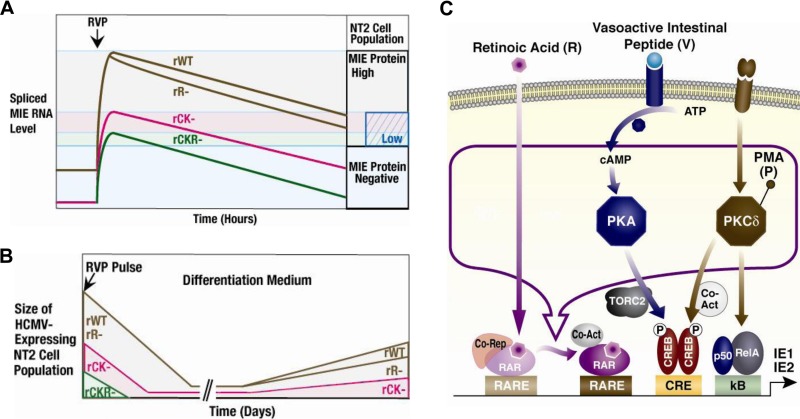
As illustrated in Fig. 10A, RVP stimulation increases spliced MIE RNA production in levels commensurate with the degree of gain in MIE+ NT2 population size in the first 24 h after onset of stimulation. The extent of attenuation of this response by CRE-kB mutations is proportional to the degree of lowering of preinduction levels of spliced MIE RNA expression (Fig. 7 and 8). Disrupting RARE in addition to CRE-kB via mutations does not further reduce spliced MIE RNA production at baseline (Fig. 8). Instead, the add-on of RARE mutations blunts the RVP-inducible fraction of output, which is reflected as further lowering in the maximum amount of spliced MIE RNA (Fig. 8) and MIE+ NT2 (Fig. 7). In contrast, the add-on of SRE-ETS mutations does not appreciably affect baseline or RVP-inducible outcome measurements (Fig. 7 and 8). Notably, CRE-kB mutations also lower the intracellular MIE protein amount in RVP-induced MIE+ NT2 (Fig. 7). Adding RARE mutations to CRE-kB mutations minimally changes this outcome (Fig. 7), whereas RARE mutations alone do not lower the number of MIE+ NT2 or the MIE protein amount in these cells (Fig. 5). RARE mutations alone increase somewhat the rate of MIE RNA level decline after RVP stimulation (Fig. 4), but whether this change is connected with RARE's role in boosting MIE RNA expression in D-NT2 counterparts (Fig. 6) remains to be determined. Thus, in the early stage following RVP induction, the enhancer CRE-kB unit's actions increase both the likelihood and the amplitude of HCMV MIE gene expression, whereas RARE functioning is largely superfluous unless the CRE-kB is absent.
FIG 10.
Model of differentiation-coupled induction of HCMV replication via enhancer RARE, CRE, and kB in quiescently infected NT2. (A) Synopsis of early time course results for RVP-induced MIE RNA and protein expression from WT virus (rWT) versus viruses without functional enhancer RARE (rR−), CRE-kB (rKB−), or CRE-kB-RARE (rCKR−). Our model proposes a threshold level of spliced MIE RNA expression, above which MIE protein is produced (MIE protein positive). The MIE RNA level is measured across the entire NT2 population. The MIE+ NT2 population size is proportional to this level. A substantial reduction in MIE RNA level also reduces the MIE protein level (MIE protein low). (B) Extended timeline of RVP's effects on the actively infected NT2 population in relation to locations of enhancer mutations. The size of the actively infected NT2 population represents the number of MIE+/vGFP+ NT2 clusters (>6 MIE+/vGFP+ cells) multiplied by the number of cells in these clusters. (C) V activates the PKA-CREB-TORC2-CRE signaling cascade (22). P activates the PKC-delta-CREB-NF-κB p65/p50-CRE-kB pathway (23). In transient assays, R-coupled retinoic acid receptor (RAR) binding to the retinoic acid receptor elements (RARE) activates viral MIE enhancer/promoter segments (32, 38). Corepressors (Co-Rep) and activators (Co-Act) regulate R-coupled RAR activity, and other signaling pathways regulate them (56, 60). We postulate that RVP produces an active R-liganded RAR coregulatory complex that strengthens MIE gene expression, which is not achieved with R alone.
The NT2 system models primitive neuronal stem cells that undergo differentiation upon exposure to R. V and P individually induce MIE gene expression without disrupting OCT4-mediated cellular pluripotency in the short term and only minimally produce viral progeny (22, 23). Combining V and P magnifies the level of HCMV activation, but this outcome is short-lived (Fig. 2) and also does not disrupt OCT4-mediated cellular pluripotency in the short term (data not shown). R-stimulated cellular differentiation, by itself, fails to reverse the silence of MIE gene expression (Fig. 3), and R plus V or P is less effective than RVP in bringing about the MIE gene expression (data not shown). Figure 10B schematically profiles the differential effects of mutation combinations in the CRE-kB-RARE core on the actively infected NT2 population over the course of many days following the RVP stimulation. The initial expansion and contraction are followed by reexpansion of the actively infected NT2 population. The reexpanded population's size represents the number of MIE+/vGFP+ NT2 clusters arising from individual MIE+/vGFP+ NT2 multiplied by the number of MIE+/vGFP+ NT2 in these clusters. CRE-kB mutations yield equivalent degrees of decreases in the number of MIE+ NT2 initially produced after RVP stimulation and the number of actively infected NT2 clusters that develop thereafter (Fig. 7). Mutations in RARE alone or RARE plus CRE-kB result in a greater degree of lowering of the actively infected NT2 cluster number than of the initial MIE+ NT2 number at day 1 of RVP stimulation (Fig. 5 and 7), suggesting that RARE may have a greater role in the pathway of cellular differentiation.
The presence of HCMV structural proteins gB and pp28 in cells at the outer limits of the actively infected NT2 cluster, as assessed by IFA (data not shown), suggests that the RVP-induced increase in infectious viral production over the course of many days (Fig. 3) is linked to cell-to-cell spread of the virus. The length of lag time to reexpansion of the actively infected NT2 population corresponds to the time needed to reach an advanced stage of R-induced NT2 differentiation. Four additional observations reflect an association between viral proliferation and cellular differentiation. First, RVP-generated actively infected NT2 clusters are fewer and smaller if R exposure is not continued to drive further differentiation (Fig. 5). Second, VP acquires the ability to generate actively infected NT2 clusters when R is added 1 day after VP stimulation, though cluster number and size are smaller than those produced by RVP stimulation (Fig. 3). Third, the direct infection of unstimulated D-NT2 produces actively infected cell clusters that expand at a similar rate (Fig. 6 and 9). Fourth, MIE gene expression is differentiation dependent. MIE gene expression in undifferentiated NT2 requires induction with specific stimuli (i.e., RVP), whereas such induction is unnecessary in differentiated cellular counterparts. As the RVP-induced NT2 progress toward cellular differentiation in the absence of continual R exposure, the introduction of RARE mutations impairs the development of MIE+/vGFP+ NT2 clusters (Fig. 5). In D-NT2 free of R exposure, MIE RNA levels are reduced by RARE mutations alone or when added to CRE-kB mutations (Fig. 6 and 9). The add-on of RARE mutations to CRE-kB mutations further lowers MIE protein levels in D-NT2, which correlates with the inability of this virus to subsequently form MIE+/vGFP+ NT2 clusters (Fig. 9). We infer from these findings that the intrinsic cellular condition of differentiation links MIE enhancer/promoter activation with viral replication via the actions of the enhancer CRE-kB-RARE. Notably, R also promotes viral replication through a pathway that does not involve the enhancer RARE but likely involves the cellular condition of differentiation (Fig. 5).
With this cell culture model, we bring into view the enhancer mechanics underlying the launching and maintenance phases of differentiation-coupled induction of MIE gene expression and viral propagation. Interplay in signaling responsive cis-activities of the MIE enhancer's CRE, kB, and RARE effectively overrides the forces behind MIE gene expression silence (Fig. 10C). The data indicate that the combination of CREB and NF-κB family member binding sites sets the tone of uninduced enhancer activity, rapidly induces MIE gene expression in response to stimuli, determines the likelihood and amplitude of this induced gene expression, and cooperates with retinoic acid receptor binding sites in strengthening MIE gene expression as cellular differentiation develops. This triple combination of stimuli is anticipated to change the activities of other cellular and possibly viral proteins that also contribute to the final outcome. Possible downstream cellular targets that could conceivably influence the result include, for example, other regulators or modulators of transcription, epigenetic function, signal transduction, and posttranscription processes. Whether the stimuli rectify the inability of viral pp71/UL82 to translocate to the undifferentiated NT2 nucleus for helping alleviate the cell-intrinsic repression of HCMV gene expression is not known (40). The MIE proteins themselves might even be affected in ways that change autoregulation of MIE gene expression (41, 42). A myriad of other possibilities abound and are beyond the scope of this discussion.
In fibroblasts, the same CRE mutations, whether or not combined with kB or ETS-SRE mutations, do not appreciably impair HCMV (strain Towne) MIE gene expression or viral replication (22, 23, 43). Under standard fibroblast growth conditions, the kB mutations do not disrupt MIE gene expression or viral replication for the multiple HCMV strains tested (strains Towne, VR1814/FIX, and Ad169), partly because of functional compensation provided by the ETS-SRE (23, 33, 44). In contrast, the kB mutations reduce HCMV VR1814/FIX's ability to produce MIE RNA and viral progeny in serum-deprived fibroblasts (45), suggesting that the kB have a greater relative role under conditions of cellular quiescence. Combining kB mutations with ETS-SRE or AP1-binding site mutations also decreases both MIE gene expression and viral replication (33, 44). The results described herein are the first to determine the functional role of enhancer RARE in HCMV infection. The murine CMV MIE enhancer also contains RARE repeats that respond to R-induced signaling in a plasmid-based reporter assay (46). Oral administration of R to mice worsens the outcome of acute murine CMV infection by increasing viral replication, disease severity, and death rate but does not worsen the outcome of acute vaccinia virus infection (46). A very recent report indicates that Toll-like receptor signaling activation cooperates with the R signaling pathway to boost MIE gene expression in acutely infected murine bone marrow-derived macrophages (47). This interactive response is also observed for a murine CMV having an HCMV MIE enhancer as replacement of the original MIE enhancer and is dependent on the RARE in the HCMV MIE enhancer (47).
R regulates diverse biological processes, including the functioning of human dendritic cells (DC) (48), which are a site of HCMV reactivation (49). Interleukin-4 (IL-4) and granulocyte-macrophage colony-stimulating factor (GM-CSF) trigger monocytic DC to make R (50) and IL-4 and R synergistically interact in inducing a regulatory phenotype of inflammatory DC (51). IL-4 and GM-CSF are commonly used to morph latently infected monocytes into an immature DC phenotype (52, 53). HCMV reactivation is triggered by IL-6 stimulation of the extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK-MAPK) signaling cascade resulting in CREB and histone phosphorylation (52–54). Phosphorylated CREB binds to enhancer CRE to activate MIE gene expression (54). CRE mutations abrogate the reactivation, whereas kB mutations do not (54). In neuronal cells, R induces a rapid and sustained increase in CREB phosphorylation and temporarily increases ERK1/2 phosphorylation (55). The ERK-MAPK signaling pathway phosphorylates nuclear receptors, including RAR and RXR, thereby increasing nuclear receptor-mediated transcriptional activity via the dissociation of corepressors and/or recruitment of coactivators (56). VP increases ERK1 and ERK2 phosphorylation in quiescently infected NT2 (J. Yuan and J. L. Meier, unpublished data). We postulate that VP-stimulated signaling cascades modify the activity of R-liganded RAR, the RXR binding partner, and/or associated corepressors/coactivators to enhance R-induced viral MIE gene expression. HCMV may exploit fundamentally similar mechanisms in diverse cellular systems to induce enduring levels of MIE gene expression, although system differences in the molecular details are anticipated.
Our NT2 results align with the longstanding recognition that HCMV reactivation from endogenously infected myeloid cells is tied to cellular differentiation (57, 58). They also accord with findings in human embryo-derived stem cells carrying quiescent HCMV genomes (lacking viral lytic gene expression), in which either P, R, or cellular differentiation generates signs of activated HCMV infection (20, 21). We acknowledge that the human embryonic NT2 model is imperfect. While the growth of NT2 in stem cell-like conditions has improved the model, NT2 do not fully phenocopy human embryonic stem cell lines. Use of human embryonic stem cell lines poses technical challenges in studying multiple viruses in parallel and applying multiple types of analyses to address the same issues in NT2. Human embryonic stem cell lines also have cell population heterogeneity that partly results from genomic and epigenetic instability, which may evolve with passage of cells in culture (59). Nevertheless, future studies are needed to determine whether the core principles operating in tipping the balance in favor of HCMV propagation in the NT2 model also apply to HCMV infection in cultured human primitive stem cells, as well as human myeloid precursors.
ACKNOWLEDGMENTS
This material is based upon work supported in part by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development 1BX001107-01 (J.L.M.), and the National Institutes of Health, National Institute of Infectious Diseases T32AI007533-12 (C.S.G.). The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the granting agencies.
We declare that we have no conflicts of interest.
We thank Mark Stinski for advice and insight and members of the Stinski and Meier laboratories for their comments and critiques.
Author contributions: J.Y. and J.L.M. conceived and designed the experiments; J.Y., M.L., and Y.R.T. performed the experiments; J.Y. and J.L.M. analyzed the data; J.Y., M.L., Y.R.T., and C.S.G. contributed reagents/materials/analysis tools; and J.Y. and J.L.M. wrote the paper.
REFERENCES
- 1.Cohen JI. 2010. Introduction to Herpesviridae, p 1937–1942. In Mandell GL, Bennett JE, Dolin R (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Churchill Livingston Elsevier, Philadelphia, PA. [Google Scholar]
- 2.Crumpacker CS, Zhang JL. 2010. Cytomegalovirus, p 1971–1987. In Mandell GL, Bennett JE, Dolin R (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Churchill Livingston Elsevier, Philadelphia, PA. [Google Scholar]
- 3.Lisco A, Munawwar A, Introini A, Vanpouille C, Saba E, Feng X, Grivel JC, Singh S, Margolis L. 2012. Semen of HIV-1-infected individuals: local shedding of herpesviruses and reprogrammed cytokine network. J Infect Dis 205:97–105. doi: 10.1093/infdis/jir700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Snydman DR, Allen U, Humar A, Transplantation Society International CMV Consensus Group. 2010. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation 89:779–795. doi: 10.1097/TP.0b013e3181cee42f. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson M, Lurain N, Hunt P. 2011. Cytomegalovirus viraemia in the modern antiretroviral era. HIV Med 12:387–388. doi: 10.1111/j.1468-1293.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 6.Simon CO, Seckert CK, Reddehase MJ, Grzimek NKA. 2006. Murine model of cytomegalovirus latency and reactivation: the silencing/desilencing and immune sensing hypothesis, p 483–500. In Reddehase MJ. (ed), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 7.Boeckh M, Geballe AP. 2011. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest 121:1673–1680. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsutsui Y, Kawasaki H, Kosugi I. 2002. Reactivation of latent cytomegalovirus infection in mouse brain cells detected after transfer to brain slice cultures. J Virol 76:7247–7254. doi: 10.1128/JVI.76.14.7247-7254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arribas JR, Storch GA, Clifford DB, Tselis AC. 1996. Cytomegalovirus encephalitis. Ann Intern Med 125:577–587. doi: 10.7326/0003-4819-125-7-199610010-00008. [DOI] [PubMed] [Google Scholar]
- 10.Wiley CA, Schrier RD, Denaro FJ, Nelson JA, Lampert PW, Oldstone MB. 1986. Localization of cytomegalovirus proteins and genome during fulminant central nervous system infection in an AIDS patient. J Neuropathol Exp Neurol 45:127–139. doi: 10.1097/00005072-198603000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Andrews PW, Damjanov I, Simon D, Banting GS, Carlin C, Dracopoli NC, Fogh J. 1984. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest 50:147–162. [PubMed] [Google Scholar]
- 12.Meier JL. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J Virol 75:1581–1593. doi: 10.1128/JVI.75.4.1581-1593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonczol E, Andrews PW, Plotkin SA. 1984. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science 224:159–161. doi: 10.1126/science.6322309. [DOI] [PubMed] [Google Scholar]
- 14.Meier JL, Stinski MF. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol 71:1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bani-Yaghoub M, Felker JM, Naus CG. 1999. Human NT2/D1 cells differentiate into functional astrocytes. Neuroreport 10:3843–3846. doi: 10.1097/00001756-199912160-00022. [DOI] [PubMed] [Google Scholar]
- 16.Andrews PW. 1984. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol 103:285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- 17.Angulo A, Suto C, Boehm MF, Heyman RA, Ghazal P. 1995. Retinoid activation of retinoic acid receptors but not of retinoid X receptors promotes cellular differentiation and replication of human cytomegalovirus in embryonal cells. J Virol 69:3831–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinclair J, Sissons P. 2006. Latency and reactivation of human cytomegalovirus. J Gen Virol 87:1763–1779. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 19.Reeves MB, Sinclair JH. 2010. Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J Gen Virol 91:599–604. doi: 10.1099/vir.0.015602-0. [DOI] [PubMed] [Google Scholar]
- 20.Penkert RR, Kaletja RF. 2013. Human embryonic stem cell lines model experimental human cytomegalovirus latency. mBio 4:e00298-13. doi: 10.1128/mBio.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belzile J-P, Stark TJ, Yeo GW, Spector DH. 2014. Human cytomegalovirus infection of human embryonic stem cell-derived primitive neural stem cells is restricted at several steps but leads to the persistence of viral DNA. J Virol 88:4021–4039. doi: 10.1128/JVI.03492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J, Liu X, Wu AW, McGonagill PW, Keller MJ, Galle CS, Meier JL. 2009. Breaking human cytomegalovirus major immediate-early gene silence by vasoactive intestinal peptide stimulation of the protein kinase A-CREB-TORC2 signaling cascade in human pluripotent embryonal NTera2 cells. J Virol 83:6391–6401. doi: 10.1128/JVI.00061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Yuan J, Wu AW, McGonagill PW, Galle CS, Meier JL. 2010. Phorbol ester-induced human cytomegalovirus MIE enhancer activation through PKC-delta, CREB, and NF-κB de-silences MIE gene expression in quiescently infected human pluripotent NTera2 cells. J Virol 84:8495–8508. doi: 10.1128/JVI.00416-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor CM, Murphy EA. 2012. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J Virol 86:9854–9865. doi: 10.1128/JVI.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. 2003. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A 100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Shin S, Zeng X, Zhan M, Gonzalez R, Mueller FJ, Schwartz CM, Xue H, Li H, Baker SC, Chudin E, Barker DL, McDaniel TK, Oeser S, Loring JF, Mattson MP, Rao MS. 2006. Genome wide profiling of human embryonic stem cells (hESCs), their derivatives and embryonal carcinoma cells to develop base profiles of U.S. Federal government approved hESC lines. BMC Dev Biol 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josephson R, Ording CJ, Liu Y, Shin S, Lakshmipathy U, Toumadje A, Love B, Chesnut JD, Andrews PW, Rao MS, Auerbach JM. 2007. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells 25:437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- 28.Matin MM, Walsh JR, Gokhale PJ, Draper JS, Morton ARBI, Moore HD, Andrews PW. 2004. Specific knockdown of Oct4 and B2-microglobulin expression by RNA interference in human embryonic stem cells. Stem Cells 22:659–668. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- 29.Greber B, Lehrach H, Adjaye J. 2007. Silencing of core transcription factors in human EC cells highlights the importance of autocrine FGF signaling for self-renewal. BMC Dev Biol 7:46. doi: 10.1186/1471-213X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundquist CA, Meier JL, Stinski MF. 1999. A strong transcriptional negative regulatory region between the human cytomegalovirus UL127 gene and the major immediate early enhancer. J Virol 73:9039–9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn W, Chou C, Li H, Hai R, Patterson D, Stoic V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angulo A, Suto C, Heymen RA, Ghazal P. 1996. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol Endocrinol 10:781–793. [DOI] [PubMed] [Google Scholar]
- 33.Caposio P, Luganini A, Bronzini M, Landolfo S, Gribaudo G. 2010. The Elk-1 and serum response factor binding sites in the major immediate-early promoter of the human cytomegalovirus are required for efficient viral replication in quiescent cells and compensate for inactivation of the NF-{kappa}B sites in proliferating cells. J Virol 84:4481–4493. doi: 10.1128/JVI.02141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocynate-phenol-chloroform extraction. Anal Biochem 162:156–159. [DOI] [PubMed] [Google Scholar]
- 35.Meier JL, Keller MJ, McCoy JJ. 2002. Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for activation of viral gene expression and replication. J Virol 76:313–320. doi: 10.1128/JVI.76.1.313-326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White EA, Clark CL, Sanchez V, Spector DH. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J Virol 78:1817–1830. doi: 10.1128/JVI.78.4.1817-1830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White EA, Del Rosario CJ, Sanders RL, Spector DH. 2007. The IE2 60-kilodalton and 40-kilodalton proteins are dispensable for human cytomegalovirus replication but are required for efficient delayed early and late gene expression and production of infectious virus. J Virol 81:2573–2583. doi: 10.1128/JVI.02454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghazal P, DeMattei C, Giulietti E, Kliewer SA, Umesono K, Evans RM. 1992. Retinoic acid receptors initiate induction of the cytomegalovirus enhancer in embryonal cells. Proc Natl Acad Sci U S A 89:7630–7634. doi: 10.1073/pnas.89.16.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connor CM, Vanicek J, Murphy EA. 2014. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol 88:5524–5532. doi: 10.1128/JVI.00481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol 80:3863–3873. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng MW, Bolovan-Fritts C, Dar RD, Womack A, Simpson ML, Shenk T, Weinberger LS. 2012. An endogenous accelerator for viral gene expression confers a fitness advantage. Cell 151:1569–1580. doi: 10.1016/j.cell.2012.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier JL, Stinski MF. 2013. Major immediate-early enhancer and its gene products, p 151–166. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, vol 1 Caister Academic Press/Horizon, Norfolk, United Kingdom. [Google Scholar]
- 43.Keller MJ, Wheeler DG, Cooper E, Meier JL. 2003. Role of the human cytomegalovirus major immediate-early promoter's 19-base-pair-repeat cAMP-response element in acutely infected cells. J Virol 77:6666–6675. doi: 10.1128/JVI.77.12.6666-6675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isern E, Gustems M, Messerle M, Borst E, Ghazal P, Angulo A. 2011. The activator protein 1 binding motifs within the human cytomegalovirus major immediate-early enhancer are functionally redundant and act in a cooperative manner with the NF-{kappa}B sites during acute infection. J Virol 85:1732–1746. doi: 10.1128/JVI.01713-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caposio P, Luganini A, Hahn G, Landolfo S, Gribaudo G. 2007. Activation of the virus-induced IKK/NF-kappaB signalling axis is critical for the replication of human cytomegalovirus in quiescent cells. Cell Microbiol 9:2040–2054. doi: 10.1111/j.1462-5822.2007.00936.x. [DOI] [PubMed] [Google Scholar]
- 46.Angulo A, Chandraratna RA, LeBlanc JF, Ghazal P. 1998. Ligand induction of retinoic acid receptors alters an acute infection by murine cytomegalovirus. J Virol 72:4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kropp KA, Hsieh WY, Isern E, Forster T, Krause E, Brune W, Angulo A, Ghazal P. 2015. A temporal gate for viral enhancers to co-opt Toll-like-receptor transcriptional activation pathways upon acute infection. PLoS Pathog 11:e1004737. doi: 10.1371/journal.ppat.1004737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manicassamy S, Pulendran B. 2009. Retinoic acid-dependent regulation of immune responses by dendritic cells and macrophages. Semin Immunol 21:22–27. doi: 10.1016/j.smim.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinclair J, Reeves M. 2014. The intimate relationship between human cytomegalovirus and the dendritic cell lineage. Front Microbiol 5:389. doi: 10.3389/fmicb.2014.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokota A, Takeuchi H, Maeda N, Ohoka Y, Kato C, Song S-Y, Iwata M. 2009. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int Immunol 21:361–377. doi: 10.1093/intimm/dxp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu B, Buttrick T, Bassil R, Zhu C, Olah M, Wu C, Xiao S, Orent W, Elyaman W, Khoury SJ. 2013. IL-4 and retinoic acid synergistically induce regulatory dendritic cells expressing Aldh1a2. J Immunol 191:3139–3151. doi: 10.4049/jimmunol.1300329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reeves MB, Compton T. 2011. Inhibition of inflammatory interleukin-6 activity via extracellular signal-regulated kinase-mitogen-activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells. J Virol 85:12750–12758. doi: 10.1128/JVI.05878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hargett D, Shenk TE. 2010. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci U S A 107:20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kew VG, Yuan J, Meier JL, Reeves M. 2014. Mitogen and stress activated kinases act cooperatively with CREB during induction of human cytomegalovirus immediate-early gene expression from latency. PLoS Pathog 10:e10004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canon E, Cosgaya JM, Scsucova S, Aranda A. 2004. Rapid effects of retinoic acid on CREB and ERK phosphorylation in neuronal cells. Mol Cell Biol 15:5583–5592. doi: 10.1091/mbc.E04-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zassadowski F, Rochette-Egly C, Chomienne C, Cassinat B. 2012. Regulation of the transcriptional activity of nuclear receptors by the MEK/ERK1/2 pathway. Cell Signal 24:2369–2377. doi: 10.1016/j.cellsig.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 57.Taylor-Wiedeman JA, Sissons JGP, Sinclair JH. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol 68:1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soderberg-Naucler C, Streblow DN, Fish KN, Allan-Yorke J, Smith PP, Nelson JA. 2001. Reactivation of latent human cytomegalovirus in CD14+ monocytes is differentiation dependent. J Virol 75:7543–7554. doi: 10.1128/JVI.75.16.7543-7554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, Noaksson K, Hyllner J, Schulz TC, Zeng X, Freed WJ, Crook J, Abraham S, Colman A, Sartipy P, Matsui S, Carpenter M, Gazdar AF, Rao M, Chakravarti A. 2005. Genomic alterations in cultured human embryonic stem cells. Nat Genet 37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 60.Gudas LJ. 2013. Retinoids induce stem cell differentiation via epigenetic changes. Semin Cell Dev Biol 24:701–705. doi: 10.1016/j.semcdb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]