ABSTRACT
Virus-specific interaction between the attachment protein (HN) and the fusion protein (F) is prerequisite for the induction of membrane fusion by parainfluenza viruses. This HN-F interaction presumably is mediated by particular amino acids in the HN stalk domain and those in the F head domain. We found in the present study, however, that a simian virus 41 (SV41) F-specific chimeric HPIV2 HN protein, SCA, whose cytoplasmic, transmembrane, and stalk domains were derived from the SV41 HN protein, could not induce cell-cell fusion of BHK-21 cells when coexpressed with an SV41 HN-specific chimeric PIV5 F protein, no. 36. Similarly, a headless form of the SV41 HN protein failed to induce fusion with chimera no. 36, whereas it was able to induce fusion with the SV41 F protein. Interestingly, replacement of 13 amino acids of the SCA head domain, which are located at or around the dimer interface of the head domain, with SV41 HN counterparts resulted in a chimeric HN protein, SCA-RII, which induced fusion with chimera no. 36 but not with the SV41 F protein. More interestingly, retroreplacement of 11 out of the 13 amino acids of SCA-RII with the SCA counterparts resulted in another chimeric HN protein, IM18, which induced fusion either with chimera no. 36 or with the SV41 F protein, similar to the SV41 HN protein. Thus, we conclude that the F protein specificity of the HN protein that is observed in the fusion event is not solely defined by the primary structure of the HN stalk domain.
IMPORTANCE It is appreciated that the HN head domain initially conceals the HN stalk domain but exposes it after the head domain has bound to the receptors, which allows particular amino acids in the stalk domain to interact with the F protein and trigger it to induce fusion. However, other regulatory roles of the HN head domain in the fusion event have been ill defined. We have shown in the current study that removal of the head domain or amino acid substitutions in a particular region of the head domain drastically change the F protein specificity of the HN protein, suggesting that the ability of a given HN protein to interact with an F protein is defined not only by the primary structure of the HN stalk domain but also by its conformation. This notion seems to account for the unidirectional substitutability among rubulavirus HN proteins in triggering noncognate F proteins.
INTRODUCTION
The parainfluenza viruses are classified into the genera Rubulavirus, Avulavirus, and Respirovirus in the family Paramyxoviridae (1, 2). Human parainfluenza virus 1 (HPIV1) and HPIV3 are members of the genus Respirovirus, while HPIV2, HPIV4A, HPIV4B, Simian virus 41 (SV41), and Parainfluenza virus 5 (PIV5) belong to the genus Rubulavirus; Mumps virus (MuV) is not a parainfluenza virus but is a member of the latter genus (1). Newcastle disease virus (NDV) is one of the 10 avian paramyxoviruses of the genus Avulavirus (1). These viruses have two kinds of glycoprotein spikes on the envelope: hemagglutinin-neuraminidase (HN) protein tetramers and fusion (F) protein trimers (2). The F protein mediates membrane fusion, such as cell-cell fusion or virus-cell fusion; cleavage of the F precursor (F0) by cellular proteases into F1 and F2 subunits is a prerequisite for its fusion activity (1, 2). The HN protein is responsible for binding to the sialoconjugate receptors on the cell surface and for enzymatic destruction of the receptors (1). In addition, the HN protein is required for the F protein to mediate membrane fusion (3, 4), although it is not precisely known how the HN protein promotes the F protein-mediated membrane fusion. It is appreciated, at least, that fusion is induced through a series of conformational changes of the F protein that has been triggered by specific interaction with the homologous HN protein (5–7).
The stalk domain of the HN protein contains the site that determines F protein specificity in promoting cell-cell fusion; thus, it would be involved in the functional interaction with the F protein (8–10); in the case of the PIV5 HN protein, the putative F-activating region (FAR) has been identified in the stalk domain (11). Indeed, it has been demonstrated by coimmunoprecipitation that the NDV HN stalk domain is responsible for the physical interaction with the cognate F protein (12). On the other hand, the HN head region carries both the receptor-binding and -destroying activities (13, 14). Importantly, the headless HN proteins of PIV5, NDV, and MuV have been found to efficiently trigger their cognate F proteins and induce extensive cell-cell fusion (11, 15), indicating that the HN stalk domain harbors sufficient elements for interacting with and triggering the F protein. It should be noted in this context, however, that both the head and stalk domains are required for the HPIV2 HN protein to exhibit its triggering activity toward noncognate HPIV4A F protein (10).
According to the model based on the structural studies on PIV5 and NDV HN proteins (16), the HN protein tetramer converts from the “4-heads-down conformation” to the “4-heads-up conformation” after interacting with the receptors, by which an otherwise hidden FAR in the stalk domain is exposed and becomes accessible to the F protein. Attempts to detect the physical HN-F interaction by coimmunoprecipitation so far have been unsuccessful for most of the parainfluenza viruses, presumably due to the nature of the interaction that may take place transiently and/or with very low avidity. As a substitute for coimmunoprecipitation, bimolecular fluorescence complementation has been successfully employed for detecting the HN-F interaction of HPIV3 (17), whereas it was not available for PIV5 (18), preventing the molecular mechanism of physical HN-F interaction from reaching a consensus (16, 19).
Recently, we and Bose et al. have independently identified two separate regions of the PIV5 F head domain that would be involved in the interaction with the HN stalk domain (20, 21). During this process, we created a chimeric PIV5 F protein by replacing 21 amino acids of the head domain with the SV41 F counterparts. This chimeric PIV5 F protein, designated no. 36, was efficiently triggered by the SV41 HN protein and induced extensive cell-cell fusion, but not by the PIV5 HN protein. On the other hand, we previously obtained a chimeric HPIV2 HN protein, CH95-571, by replacing the cytoplasmic/transmembrane domains and the stalk domain with those of the SV41 HN protein; CH95-571 triggered the SV41 F protein nearly 10 times as efficiently as it triggered the HPIV2 F protein (10). Therefore, we anticipated that the SV41 F-specific chimera CH95-571 would efficiently trigger the SV41 HN-specific chimera no. 36 and induce fusion. Surprisingly, we have found in the current study that CH95-571 is unable to trigger chimera no. 36, suggesting that the SV41 HN-derived stalk domain of CH95-571 does not have enough elements to mediate functional HN-F interaction between these SV41-like chimeric proteins. We have investigated this issue and eventually revealed that some factor, as well as the primary structure, is critical for the HN stalk domain to functionally interact with the F protein.
MATERIALS AND METHODS
Cells and antibodies.
BHK-21 (BHK) cells were maintained in Eagle's minimum essential medium supplemented with 5% fetal calf serum. The monoclonal antibodies (MAb) 173-1A and M1-1A, specific for the HPIV2 HN protein, were reported previously (22). MAb 127A-1, specific for the SV41 HN protein, was reported elsewhere (23). MAb M1-1A recognizes at least two residues, N83 and K91, of the HPIV2 HN stalk region (10, 24), while the epitope for MAb 127A-1 resides between residues 291 and 443 of the SV41 HN head domain (10). The epitope for MAb 173-1A has been identified in this study to reside between residues 295 and 447 of the HPIV2 HN head domain, as analyzed by indirect immunofluorescence assay using the previously reported chimeric HN proteins of HPIV2 and SV41 (10). The culture supernatant of the hybridoma cells secreting either of these antibodies was used without dilution. Monoclonal anti-FLAG M2 (clone M2; Sigma-Aldrich) was used at a concentration of 5 ng/ml. Biotinylated horse immunoglobulin specific for mouse IgG was purchased from Vector Laboratories.
Plasmid vectors.
The pcDL-SRα expression vectors encoding the HN and F proteins of HPIV2, SV41, and PIV5 were described previously (10, 25). The expression vectors encoding the chimeric HN proteins CH1-94 and CH95-571 were also reported previously (10). The expression vectors encoding the HN and F proteins of MuV strain Enders were prepared in the present study by the method described for those of MuV strain Miyahara (25). The expression vector, p3XFLAG-myc-CMV-26, was bought from Sigma-Aldrich and used as the template for preparing cDNA for the FLAG epitope tag. The cDNA for the modified terminal tetramerization module, which was based on a derivative of yeast GCN4 (26), was synthesized and cloned in the pMD20T plasmid by TaKaRa Custom Services (TaKaRa Bio, Shiga, Japan).
Quantification of cell-cell fusion.
Subconfluent BHK cell monolayers in six-well culture plates were cotransfected with 0.2 μg/well of HN-encoding expression vector and 2 μg/well of F-encoding expression vector by using an X-tremeGENE 9 DNA transfection kit (Roche Diagnostics). After 24 h of incubation at 37°C, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), washed three times with PBS, and stained with Giemsa's solution. A photomicrograph which involved approximately 2.5 × 104 cells was taken, and the areas (number of pixels) occupied by fused cells (syncytia) were measured with the aid of a graphics software, NIH ImageJ ver.1.45s. The extent of cell fusion then was estimated as a percentage of the syncytial areas out of the total area of the photograph. Ten randomly taken photographs were measured for each sample, and the average fusion index (in percent) and standard deviations were determined. Our preliminary experiment indicated that the transfection of more than 0.2 μg/well of HN-encoding expression vector usually resulted in the reduction of the fusion index.
Western blotting.
In order to detect truncated and headless HN proteins by Western blotting, FLAG epitope tags were attached to their cytoplasmic tails. Subconfluent BHK cell monolayers were transfected with 2 μg/well of HN-encoding expression vector as described above. After 24 h of incubation at 37°C, the cells were lysed on ice for 30 min with 400 μl/well of lysis buffer (50 mM HEPES [pH 7.3], 10 mM lauryl maltoside, 1 mM phenylmethylsulfonyl fluoride, 100 mM NaCl). Aliquots (15 μl) of each cell lysate were subjected to SDS-PAGE under reducing conditions, and the separated proteins were electroblotted to a Protran BA 85 nitrocellulose membrane (GE Healthcare Life Sciences). The membrane then was successively treated with anti-FLAG M2 monoclonal antibody, biotinylated anti-mouse IgG horse immunoglobulin, and streptavidin-biotin-peroxidase complex (Vectastain ABC kit; Vector Laboratories). The HN protein bands then were visualized by enhanced chemiluminescence (ECL) using the Western blotting luminol reagent (Santa Cruz Biotechnology), followed by exposure to X-ray film (Konica Minolta, Tokyo, Japan).
Cell surface biotinylation and immunoprecipitation.
Subconfluent BHK cell monolayers in six-well culture plates were transfected with 0.2 to 4.0 μg/well of HN-encoding expression vector. When the amount of transfectant was below 2 μg, the appropriate amount of pcDL-SRα vector was added to it so that the total amount became 2 μg. After 24 h of incubation at 37°C, the cells were treated with 0.3 mg/ml of sulfo-NHS-LC-biotin (Thermo Scientific) solution in PBS supplemented with 0.1 mM CaCl2 and 1 mM MgCl2 at 23°C for 30 min and lysed on ice with 400 μl/well of lysis buffer. The biotinylated proteins in 150 μl of the cell lysates then were immunoprecipitated with monoclonal antibodies specific for either the HPIV2 HN protein (MAb M1-1A and 173-1A), the SV41HN protein (MAb 127A-1), or the FLAG epitope tag (anti-FLAG M2). The biotinylated and immunoprecipitated HN proteins then were electroblotted to either the nitrocellulose membrane or Hybond-P polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences) and detected by ECL after treatment with streptavidin-biotin-peroxidase complex. The intensity of the HN protein band was quantified with the aid of the graphics software NIH ImageJ ver.1.45s, and the relative surface expression level was estimated.
Native gel electrophoresis.
In order to detect native HN proteins by Western blotting after native gel electrophoresis, FLAG epitope tags were attached to the cytoplasmic tails of representative HN proteins. Subconfluent BHK cell monolayers in 12-well culture plates were transfected with 1 μg/well of HN-encoding expression vector. After 24 h of incubation at 37°C, the cells were lysed on ice for 30 min with 160 μl/well of 1× NativePAGE sample buffer (Life Technologies) containing 1% lauryl maltoside and 1 mM phenylmethylsulfonyl fluoride. After centrifugation at 15,000 × g for 30 min, the supernatant was treated with 1 U/μl of endonuclease (Benzonase; Sigma-Aldrich) at room temperature for 30 min. The aliquot (12 μl) was subjected to native gel electrophoresis under nonreducing conditions by using a NativePAGE Novex Bis-Tris gel system (Life Technologies), and the migrated proteins were blotted to a Hybond-P PVDF membrane according to the manufacturer's instructions. The HN proteins on the membrane were detected with anti-FLAG M2 monoclonal antibody as described above for Western blotting.
Molecular modeling.
Molecular modeling of the HN proteins was performed on the automated comparative protein modeling server SWISS-MODEL (http://swissmodel.expasy.org) by using the crystal structure of the PIV5 (W3A) HN protein in the “2-heads-up/2-heads-down” conformation (PDB entry 4JF7) as the template. This conformation is considered to represent a hybrid between the 4-heads-down and 4-heads-up conformations (25). The three-dimensional structures of the HN proteins were analyzed with the aid of a graphics software, Waals (Altif Laboratories, Tokyo, Japan).
RESULTS
The HN proteins of HPIV2 and MuV can trigger noncognate F proteins.
It has been revealed previously that the HN protein of MuV (Miyahara strain) can efficiently trigger either the PIV5 F protein or the HPIV2 F protein, whereas neither the PIV5 HN protein nor the HPIV2 HN protein is capable of triggering the MuV F protein (11, 25, 27). On the other hand, the HPIV2 HN protein is able to trigger the SV41 F protein, whereas the SV41 HN protein cannot trigger the HPIV2 F protein (10). In order to investigate this strange issue (that is, unidirectional substitutability among the rubulavirus HN proteins), we retested the F-triggering activities of these HN proteins.
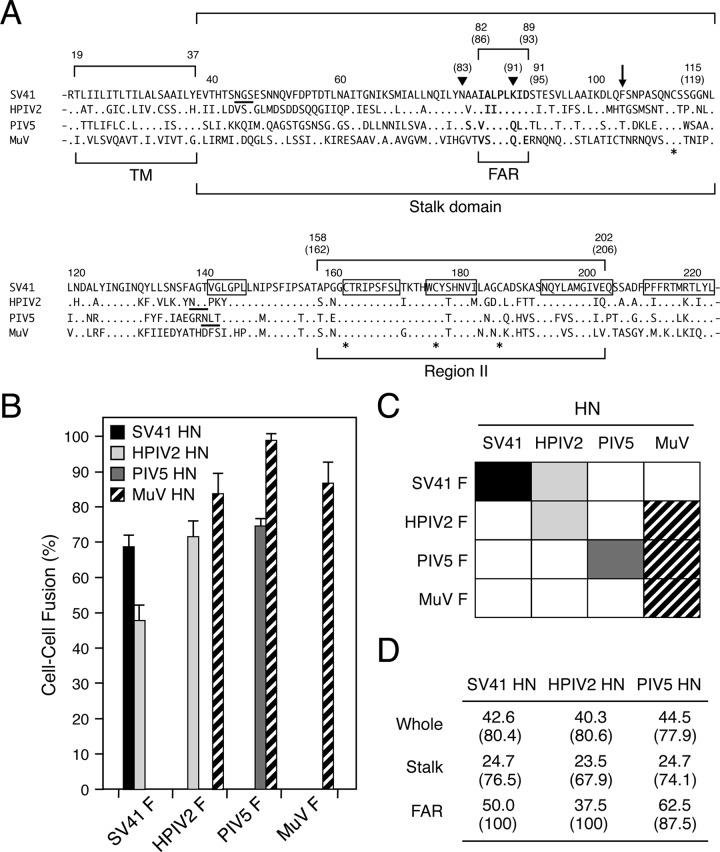
As shown in Fig. 1B and C, we confirmed our previous observation that the HPIV2 HN protein induced cell-cell fusion when coexpressed with the noncognate SV41 F protein but not vice versa (10). Similarly, the MuV (Enders strain) HN protein induced prominent fusion either with the HPIV2 F protein or with the PIV5 F protein but not vice versa (Fig. 1B and C), as reported for the MuV (Miyahara strain) HN protein (11, 27). It is worth noting in this context that the SV41 HN protein showed higher amino acid sequence identity to the MuV HN protein than that of the HPIV2 HN protein did, even when the primary structures of the stalk domains including the FAR were compared (Fig. 1A and D). Furthermore, although a threonine residue (the counterpart of F104 in the SV41 HN stalk domain) was conserved only among the HN proteins of MuV, PIV5, and HPIV2 (Fig. 1A) and might be critical for the MuV HN protein to trigger the F proteins of PIV5, HPIV2, and MuV, this residue seemed insufficient for the PIV5 HN protein or the HPIV2 HN protein to trigger the MuV F protein. These findings indicate that the primary structure of the HN stalk domain (including the FAR) cannot simply explain the ability or inability of a given rubulavirus HN protein to substitute for other rubulavirus HN proteins.
FIG 1.
HN proteins of HPIV2 and MuV can trigger noncognate F proteins. (A) Amino acid sequence alignment of the rubulavirus HN proteins. The dots in the sequences indicate the amino acids that are identical to the counterparts of the SV41 HN protein, and the asterisks below the sequences denote the positions of cysteine residues that are conserved among all HN proteins. The positions of the PIV5 HN stalk domain (28) and HPIV2 HN region II (10) are indicated. The amino acid numbers of the SV41 HN protein are shown above the sequences; those of the HPIV2 HN protein are shown in parentheses. Filled triangles indicate the positions of N83 and K91 of the HPIV2 HN protein, which are the constituents of the MAb M1-1A epitope (10, 24). The downward arrow indicates the position of F104 of the SV41 HN protein; its counterparts of the other four HN proteins are conserved threonine residues. The amino acid regions that comprise the five β strands (β1) in the first blade of the 6-bladed propeller (32) are boxed. Consensus sequences for N-linked glycosylation are underlined. TM, transmembrane domain; FAR, F-activating region. (B) F-triggering activity of the rubulavirus HN proteins. Subconfluent BHK cells in six-well culture plates were transfected with 0.2 μg/well of HN expression vector together with 2 μg/well of cognate or noncognate F expression vector, except that 1 μg/well of expression vector was used for the PIV5 HN protein. After 24 h of incubation at 37°C, the average fusion index was determined as described in Materials and Methods; error bars indicate standard deviations. (C) Substitutability of the rubulavirus HN proteins. Events of functional HN-F interaction are indicated as filled or hatched boxes on the basis of the results presented in panel B. (D) Amino acid sequence identity and similarity among the rubulavirus HN proteins. Percent identity (or percent similarity) of the amino acid sequences between the MuV HN protein and the other HN proteins was calculated individually.
Amino acid substitutions in region II variously affect F protein specificity.
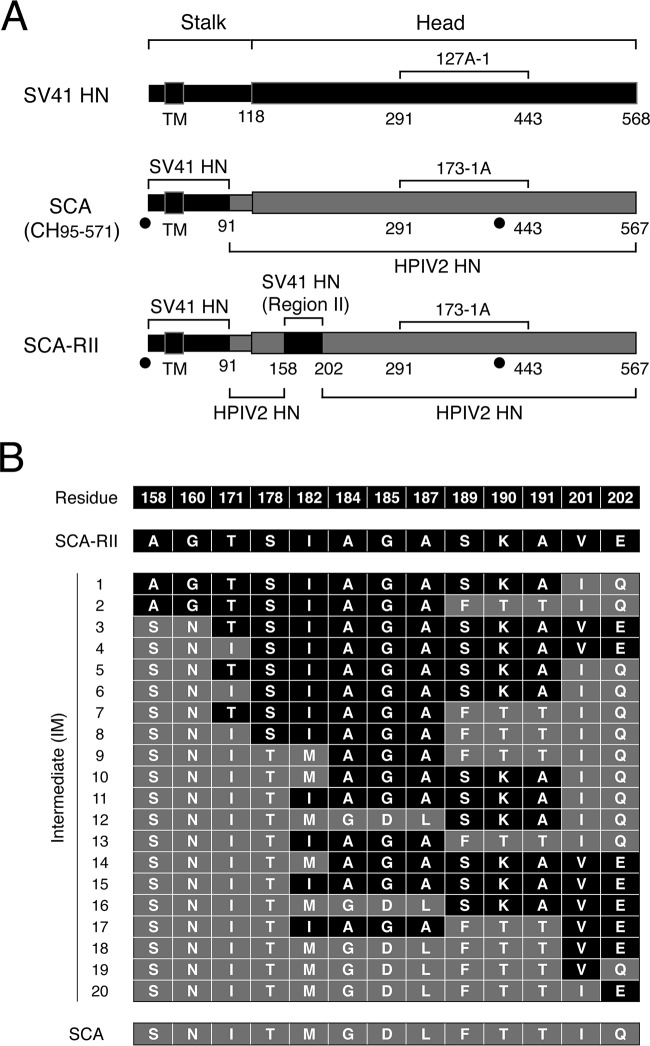
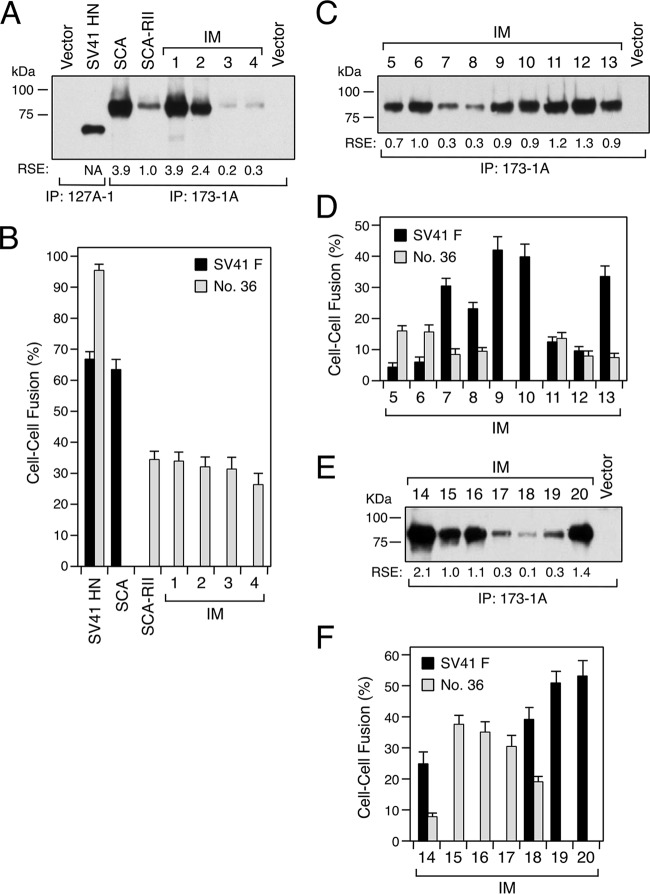
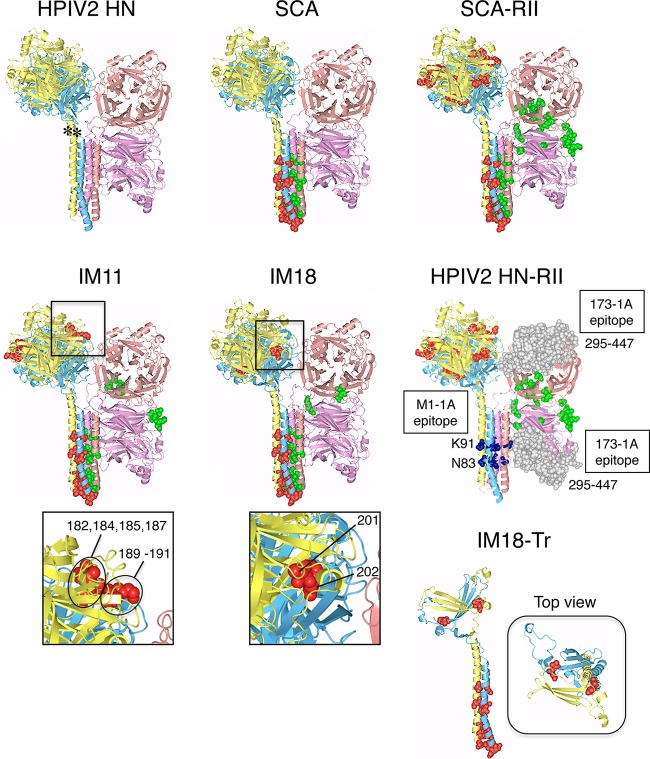
We have recently found that a chimeric PIV5 F protein, no. 36, whose 21 amino acids in the head domain are replaced with the SV41 F counterparts, is triggered by the SV41 HN protein but not by the PIV5 HN protein (21). On the other hand, we previously created an SV41 F-specific chimeric HPIV2 HN protein, CH95-571, whose N-terminal 94 residues including the FAR were replaced with the SV41 HN counterparts (10); CH95-571 was renamed SCA for convenience sake in the current study (Fig. 2A). Thus, we anticipated that the SV41 F-specific SCA would be able to trigger the SV41 HN-specific chimera no. 36. Unexpectedly, however, coexpression of SCA with chimera no. 36 in BHK cells resulted in no detectable cell-cell fusion (Fig. 3B), suggesting that the C-terminal part (residues 95 to 571) of the SV41 HN protein contains some elements that are involved in the interaction with chimera no. 36. This phenomenon reminded us of our previous finding concerning the two regions of the HPIV2 HN protein that are required to substitute for the HPIV4A HN protein; that is, the stalk domain and region II (residues 162 to 206) in the head domain (10). To examine whether region II of the SV41 HN protein would be responsible for triggering chimera no. 36, we created a chimeric HN protein, SCA-RII, on the SCA background in which the HPIV2 HN-derived region II is replaced with that of the SV41 HN protein, which harbored 13 amino acids derived from SV41 HN that were not shared with SCA (or the HPIV2 HN protein) (Fig. 1A and 2). To create this chimeric HN protein, desired portions of the SV41 HN- or SCA-encoding expression vector were amplified and joined by PCR. The resulting PCR product was inserted into the SCA-encoding expression vector (between the two EcoRI sites shown in Fig. 2A) by using an In-Fusion HD cloning kit (Clontech Laboratories). Region II is located in the amino acid region, which comprises the first blade of the 6-bladed β propeller, and most of the unconserved 13 amino acids are located above or below the β strands (β1) of the first blade (Fig. 1A and 4). Although the amount of cell surface-expressed SCA-RII in the transfected BHK cells was about one-fourth that of SCA (Fig. 3A), it was able to trigger chimera no. 36 as intended, leading to moderate cell-cell fusion, but failed to trigger the SV41 F protein (Fig. 3B). This surprising conversion in F protein specificity led us to expect that an intermediate between SCA and SCA-RII is able to trigger both the SV41 F protein and chimera no. 36, similar to the SV41 HN protein. Since region II of SCA-RII harbored 13 amino acids that were not shared with SCA as mentioned above, we replaced them stepwise with the counterparts of SCA, resulting in 20 intermediates between SCA-RII and SCA (Fig. 2B). When these intermediates were individually expressed in BHK cells, they were successfully expressed on the cell surface yet at different levels (Fig. 3A, C, and E). In order to examine the F protein specificity of these intermediates, they then were expressed in BHK cells together with the SV41 F protein or with chimera no. 36. As revealed by the intermediate IM2, replacement of five unconserved residues (from 189 to 202) with the SCA counterparts did not affect the F protein specificity of SCA-RII (Fig. 3B). Similarly, replacement of three unconserved residues (from 158 to 171) did not change the F protein specificity, as represented by the intermediate IM4. It is worth pointing out that these intermediates triggered chimera no. 36 as efficiently as SCA-RII did, although their expression levels were much greater or much less than that of SCA-RII, respectively (Fig. 3A and B). These observations suggested that five unconserved residues (from 178 to 187) of SCA-RII would harbor indispensable residues for triggering chimera no. 36. On the other hand, the intermediates IM15, IM16, and IM17 displayed F protein specificity similar to that of SCA-RII: they triggered chimera no. 36 as efficiently as SCA-RII did irrespective of their surface expression levels (Fig. 3E and F). Interestingly, as revealed by IM16, replacement of eight unconserved residues (from 158 to 187) proved to have no impact on the F protein specificity of SCA-RII despite the fact that all five of the unconserved residues (from 178 to 187) that had been anticipated to be critical for SCA-RII to trigger chimera no. 36 as described above were replaced. In contrast, the replacement of seven unconserved residues (from 158 to 182) and those at positions 201 and 202 resulted in the intermediate IM10, which specifically triggered the SV41 F protein (Fig. 3D). Surprisingly, when M182 of intermediate IM10 had been retroreplaced with I182, the resulting intermediate, IM11, equally triggered the SV41 F protein and chimera no. 36, but it did so with low efficiency. Region II of IM11 harbored seven amino acids derived from SCA-RII (Fig. 2B), which were located in the vicinity of the dimer interface as two clusters (Fig. 5). Retroreplacement of one of the clusters (composed of residues 182, 184, 185, and 187) with the SCA counterpart did not significantly change the F protein specificity (Fig. 3D, IM12), while retroreplacement of the other cluster (residues 189 to 191) resulted in the intermediate IM13 that exhibited much higher triggering activity toward the SV41 F protein (Fig. 3D). Finally, nine intermediates (from IM5 to IM8, from IM11 to IM14, and IM18) proved to exhibit triggering activity toward both F proteins, with that of M18 being the most efficient (Fig. 3D and F). It was noteworthy that region II of IM18 harbored only two SCA-RII-derived residues, V201 and E202, that were located at the dimer interface (Fig. 2B, 4, and 5). Importantly, retroreplacement of either of these residues resulted in selective loss of triggering activity toward chimera no. 36, as represented by IM19 and IM20 (Fig. 2B and 3F). Intriguingly, the intermediate IM15 harbored these two SCA-RII-derived residues as well as the seven SCA-RII-derived residues that were shared with IM11, but it specifically triggered the SV41 HN protein (Fig. 2B and 3F). Taking these findings together, we concluded that no amino acid in HN region II was directly involved in the interaction with the F proteins, since every amino acid in region II of a given intermediate behaved differently from that in another intermediate. It should be noted in this context that none of the unconserved 13 amino acids are in direct contact with the stalk domain of the HN protomers in the 2-heads-up conformation (Fig. 5, SCA-RII). Given that the HN stalk domain can get access to the F protein only when the HN protein has adopted the 4-heads-up conformation (28), these results suggest that subtle differences in the amino acid sequence of region II results in a conformational change of the HN head domain, which then exerts indirect yet drastic effects on the HN stalk conformation that might be critical for determining the F protein specificity.
FIG 2.
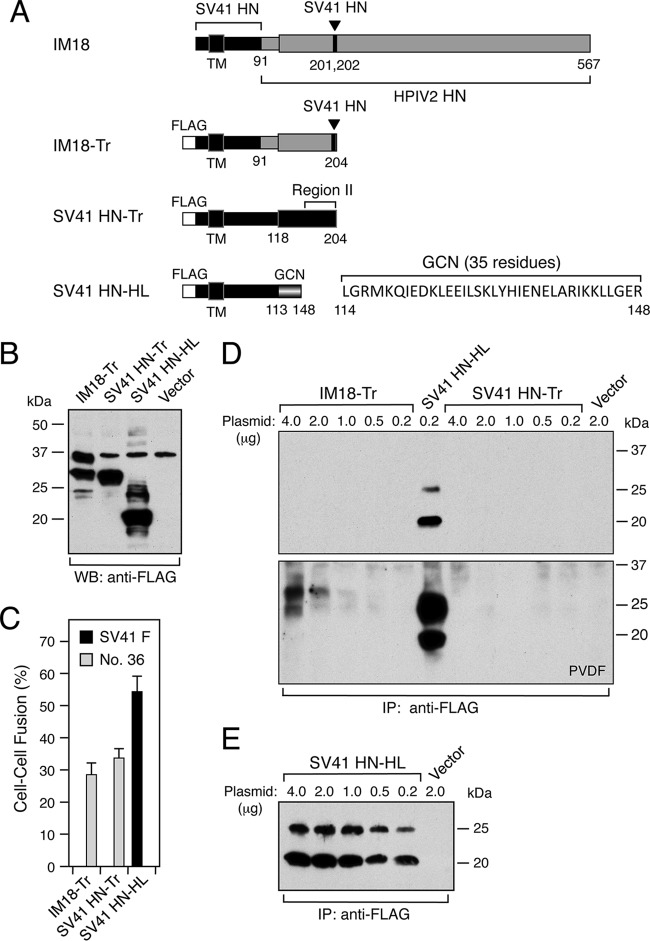
Creation of chimeric SV41 HN proteins. (A) Schematic diagram. Positions of the epitopes for the SV41 HN-specific MAb (127A-1) and HPIV2 HN-specific MAb (173-1A) are indicated. The previously reported chimeric HN protein CH95-571 was renamed SCA after the restriction enzyme ScaI, which had been utilized for the chimeric recombination (10). Filled circles below the diagrams of SCA and SCA-RII indicate the positions of EcoRI sites in the cDNA encoding these chimeric proteins. (B) Primary structures of 20 intermediates between SCA-RII and SCA. The residue numbers of 13 amino acids of region II that are not conserved between SCA-RII and SCA (that is, between the SV41 HN protein and the HPIV2 HN protein) are denoted. The primary structure of each intermediate is represented by a combination of these 13 amino acids: black boxes indicate amino acids characteristic of SCA-RII, while gray boxes indicate those characteristic of SCA.
FIG 3.
Amino acid substitutions in region II variously affect the F protein specificity of SCA. (A, C, and E) Detection of the HN proteins expressed on the cell surface. Subconfluent BHK monolayers in six-well culture plates were transfected with 2 μg/well of HN expression vector. This amount (2 μg/well) was chosen instead of 0.2 μg/well, because our preliminary experiment indicated that the latter amount was not enough to detect the intermediates IM3 and IM4. At 24 h posttransfection, the transfected cells were biotinylated, the cell lysates subjected to immunoprecipitation (IP) with respective anti-HN MAbs, and the precipitates analyzed by SDS-PAGE under reducing conditions. Relative surface expression (RSE) levels of the HN proteins are presented below each lane. The RSE value given by each HN protein was normalized to that given by SCA-RII, IM6, or IM15, which showed a moderate expression level among the respective HN proteins run in the same gel. NA, not applicable. (B, D, and F) F-triggering activity of the HN proteins. The average fusion index was determined as described in the legend to Fig. 1B; error bars indicate standard deviations.
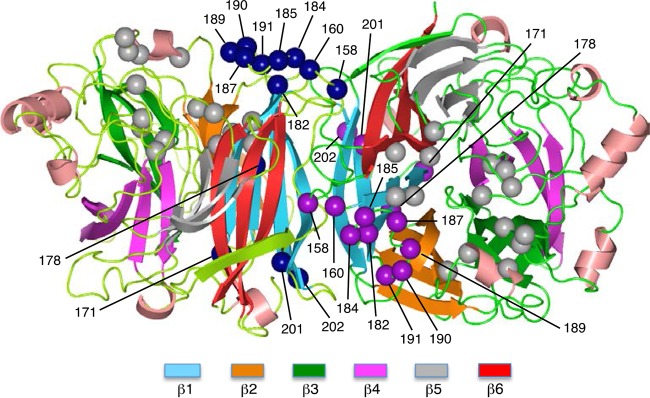
FIG 4.
Side view of the SV41 HN head domain. Two HN protomers in the 2-heads-up conformation are shown as ribbon models on the basis of the crystal structure of the PIV5 (W3A) HN protein as described in Materials and Methods. The positions of α-carbons of the unconserved 13 amino acids in region II are shown as purple or dark blue balls. The positions of amino acids that are considered involved in binding to sialyllactose, a neuraminidase substrate (32), are shown as dark gray balls; residue 187 in SV41 HN region II is a constituent of these amino acids. The β strands in each blade of the six-bladed propeller are colored as indicated below the diagram.
FIG 5.
Predicted three-dimensional structure of the HN ectodomains. The side view of each HN tetramer in the 2-heads-up/2-heads-down conformation are drawn on the basis of the crystal structure of the PIV5 (W3A) HN protein as described in Materials and Methods. Two protomers in the 2-heads-up conformation are shown in yellow and blue as ribbon models, while those in the 2-heads-down conformation are shown in magenta and salmon, respectively. The SV41 HN-derived amino acids in the former two protomers are depicted in red as space-filling models, while those in the latter two protomers are depicted in green. In the figure for the HPIV2 HN protein, the positions of the conserved threonine residues (shown in Fig. 1A) are indicated in the two protomers in the 2-heads-up conformation as asterisks. In the figure for HPIV2 HN-RII, residues N83 and K91 (the constituents of the MAb M1-1A epitope) are shown in dark blue as space-filling models, while residues from 295 to 447 (which harbor the MAb 173-1A epitope) are shown in gray; the MAb 173-1A epitopes in the protomers in the 2-heads-up conformation are not shown for clarity. The figure for IM18-Tr is created on the basis of the IM18 structure; only two protomers in the 2-heads-up conformation are depicted. SV41 HN-derived residues in the IM18-Tr stalk domains are not indicated in the top view for clarity.
Oligomeric stability of the HN protein does not correlate with F protein specificity.
The paramyxovirus HN tetramer is composed of two dimers (1). In the case of the SV41 HN protein, each dimer is linked by a disulfide bond, most likely via a cysteine residue at position 112 in the stalk domain that is conserved among rubulaviruses (Fig. 1A) (23); thus, Cys-112 is preserved in all of the chimeric and intermediate HN proteins used in this study. Therefore, it seemed interesting to examine whether the amino acid substitutions affect the oligomeric stability, which might be important for determining the F protein specificity. To this end, we performed native gel electrophoresis of the representative HN proteins with various F protein specificities. In order to detect these HN proteins in the Western blot after native gel electrophoresis, FLAG epitope tags were attached to their cytoplasmic tails. It was noteworthy that chimeras SCA and SCA-RII and all of the representative intermediates were detected predominantly as dimers, whereas the SV41 HN protein was detected mainly as tetramers (Fig. 6A), indicating that the replacement of the head domain in itself lowered the oligomeric stability of the SV41 HN protein. As shown in Fig. 6C, the F protein specificities of the tagged HN proteins were similar to those of their untagged parent HN proteins (Fig. 3B, D, and F), except for tagged IM12, which triggered the SV41 F protein 3-fold more efficiently than no. 36 while its untagged parent equally triggered these F proteins (Fig. 3D and 6C). The altered F protein specificity of IM12 with FLAG epitope tag suggests that this epitope transduces some inside-out signal to the ectodomain, which would result in modification of the conformation. This assumption may be supported by the observation that cell surface-localized IM12 was more efficiently precipitated by MAb 173-1 than by anti-FLAG monoclonal antibody, whereas these antibodies precipitated the other HN proteins to similar extents (Fig. 6B). Most importantly, these results indicate that there is no correlation between the oligomeric stability and F protein specificity of the HN proteins.
FIG 6.
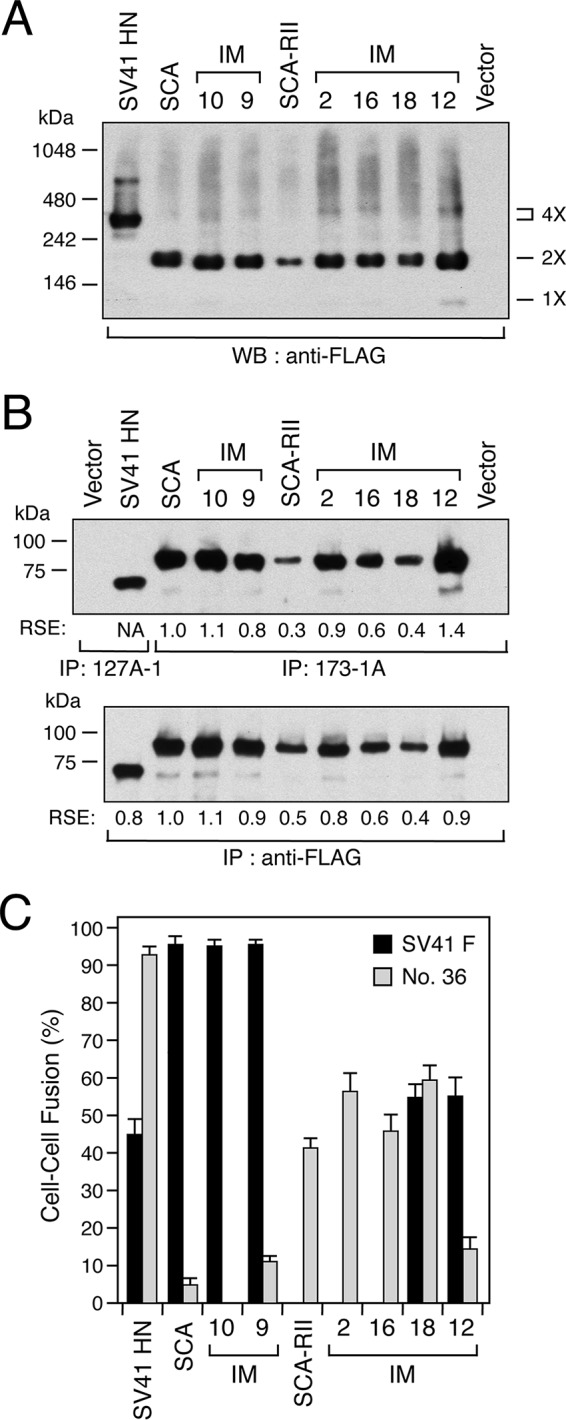
Oligomeric stability of the HN proteins. In order to detect native HN proteins by Western blotting, FLAG epitope tags were attached to the cytoplasmic tails of representative HN proteins used in this figure. (A) Native gel electrophoresis. Subconfluent BHK monolayers in 12-well culture plates were transfected with 1 μg/well of HN expression vector. At 24 h posttransfection, the cell lysate was subjected to native gel (4 to 16% gradient gel) electrophoresis under nonreducing conditions, followed by Western blotting (WB) with anti-FLAG monoclonal antibody as described in Materials and Methods. (B) Detection of cell surface-localized HN proteins. Cell surface biotinylation, immunoprecipitation, and SDS-PAGE analyses were performed as described in the legend for Fig. 3, except that anti-FLAG monoclonal antibody was used in addition to the anti-HN MAbs. The RSE value given by each HN protein was normalized to that given by SCA. (C) F-triggering activity of the HN proteins. The average fusion index was determined as described in the legend for Fig. 1B; error bars indicate standard deviations.
Headless and truncated forms of the SV41 HN protein exhibit altered F protein specificity.
To investigate further the possible role of region II in determining the F protein specificity, we created truncated forms of IM18 and the SV41 HN protein, designated IM18-Tr and SV41 HN-Tr, respectively, in which most of the head domain downstream of region II had been deleted (Fig. 5 and 7A). In addition, a headless form of the SV41 HN protein, SV41 HN-HL, was created; a modified terminal tetramerization module (GCN) was attached to its C terminus for stabilization (Fig. 7A), since it could not trigger the SV41 F protein without this module (our unpublished observation). In order to detect these HN proteins, FLAG epitope tags were attached to their cytoplasmic tails (Fig. 7A). These proteins showed comparable expression levels, as analyzed by Western blotting, when BHK cells were transfected with 2 μg/well of individual expression vector (Fig. 7B). As we had expected from the results obtained by using SCA (Fig. 3B), SV41 HN-HL efficiently triggered the SV41 F protein and did not trigger chimera no. 36 (Fig. 7C). In contrast, IM18-Tr and SV41 HN-Tr were able to trigger chimera no. 36 but not the SV41 F protein. Intriguingly, in this context, cell surface-localized SV41 HN-Tr was not detected even when the cells were transfected with 4 μg/well of expression vector and the biotinylated proteins were blotted to PVDF membrane, which gave much greater signals than those of nitrocellulose membrane, while IM18-Tr was detectable by using PVDF membrane only when the cells were transfected with more than 2 μg/well of the recombinant vector (Fig. 7D, lower). In contrast, SV41 HN-HL was easily detectable using nitrocellulose membrane even when the cells were transfected with 0.2 μg/well of the recombinant vector (Fig. 7E). Nonetheless, these results indicate that removal of all or most of the head domain from the HN protein significantly alters the F protein specificity.
FIG 7.
Headless and truncated forms of the SV41 HN protein exhibit altered F protein specificity. (A) Schematic diagram of the HN proteins. FLAG, FLAG epitope tag; GCN, a modified terminal tetramerization module. (B) Detection of the HN proteins in the transfected cells. Subconfluent BHK cell monolayers in six-well culture plates were transfected with 2 μg/well of HN expression vector and lysed at 24 h posttransfection, and the cell lysate was subjected to SDS-PAGE under reducing conditions, followed by Western blotting with anti-FLAG monoclonal antibody. (C) F-triggering activity of the HN proteins. The average fusion index was determined as described in the legend for Fig. 1B; error bars indicate standard deviations. (D and E) Detection of cell surface-localized HN proteins. Cell surface biotinylation, immunoprecipitation with anti-FLAG monoclonal antibody, and SDS-PAGE analyses were performed as described in the legend to Fig. 3, except that 0.2 to 4.0 μg/well of expression vector was used for transfection and that PVDF membrane was employed for electroblotting in addition to the nitrocellulose membrane.
F-triggering activities of the truncated HN proteins are independent of their cell surface expression levels.
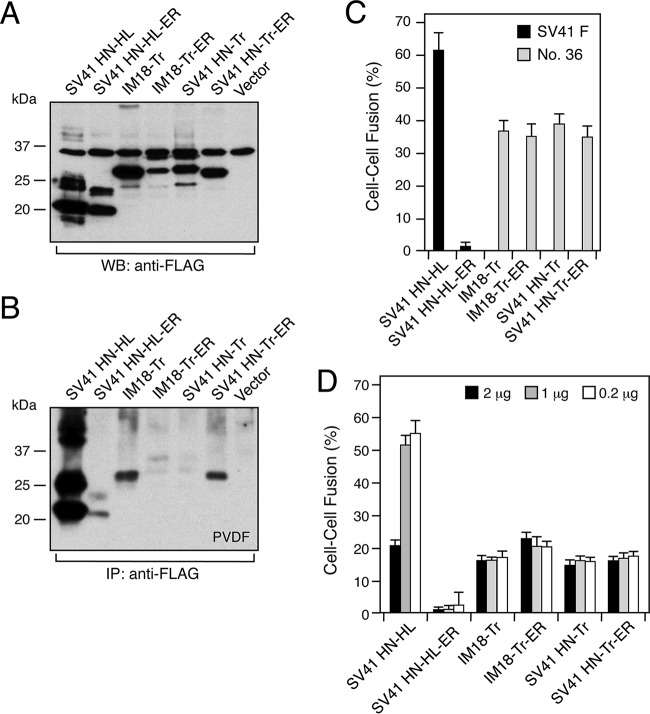
To evaluate the possibility that the truncated HN proteins are able to trigger chimera no. 36 before reaching the cell surface, we replaced the first methionine of the FLAG epitope tag of the headless or truncated HN protein with a short peptide, MRKR, which harbored an endoplasmic reticulum (ER) retention/retrieval signal motif (29). As shown in Fig. 8B, cell surface-localized IM18-Tr became undetectable due to the attached ER retention/retrieval signal, as represented by IM18-Tr-ER. Surprisingly, on the other hand, SV41 HN-Tr became detectable on the cell surface by the addition of the ER retention/retrieval signal as represented by SV41 HN-Tr-ER (Fig. 8B). Most notably, however, the triggering activities of these truncated HN proteins were similar to one another regardless of their cell surface expression levels (Fig. 8C), suggesting that they are able to trigger chimera no. 36 before reaching the cell surface. On the other hand, the F-triggering activity of SV41 HN-HL was extremely reduced by addition of the ER retention/retrieval signal (Fig. 8C), seemingly reflecting its significantly reduced cell surface expression level (Fig. 8B). Interestingly, increasing the amount of HN-encoding vector from 0.2 μg to 2 μg resulted in an unchanged or even lower fusion index (Fig. 8D), indicating that 0.2 μg of vector encoding the truncated or headless HN protein was more than enough to maximally induce fusion with 2 μg of F-encoding vector under our experimental conditions, similar to the vectors encoding full-length HN proteins described in Materials and Methods.
FIG 8.
F-triggering activities of the truncated HN proteins are independent of their cell surface expression levels. (A) Detection of the headless and truncated HN proteins in the transfected cells. Western blotting was carried out as described in the legend to Fig. 7B. (B) Detection of cell surface-localized HN proteins. Cell surface biotinylation, immunoprecipitation, and SDS-PAGE analyses were carried out as described in the legend to Fig. 3, except that PVDF membrane was employed for electroblotting. (C) F-triggering activity of the HN proteins. The average fusion index was determined as described in the legend to Fig. 1B; error bars indicate standard deviations. (D) F-triggering activity of various amounts of HN proteins. The average fusion index was determined as described in the legend to Fig. 1B, except that various amounts of expression vector encoding SV41HN-HL or SV41 HN-HL-ER were used for transfection together with 2 μg/well of SV41 F-encoding vector, while various amounts of expression vector encoding either of the four truncated HN proteins were used together with 2 μg/well of chimera no. 36-encoding vector; the total amount of transfectant was adjusted to 4 μg/well by adding the appropriate amount of pcDL-SRα vector. Error bars indicate standard deviations.
Replacement of region II or removal of the head domain converts the F protein specificity of the HPIV2 HN protein.
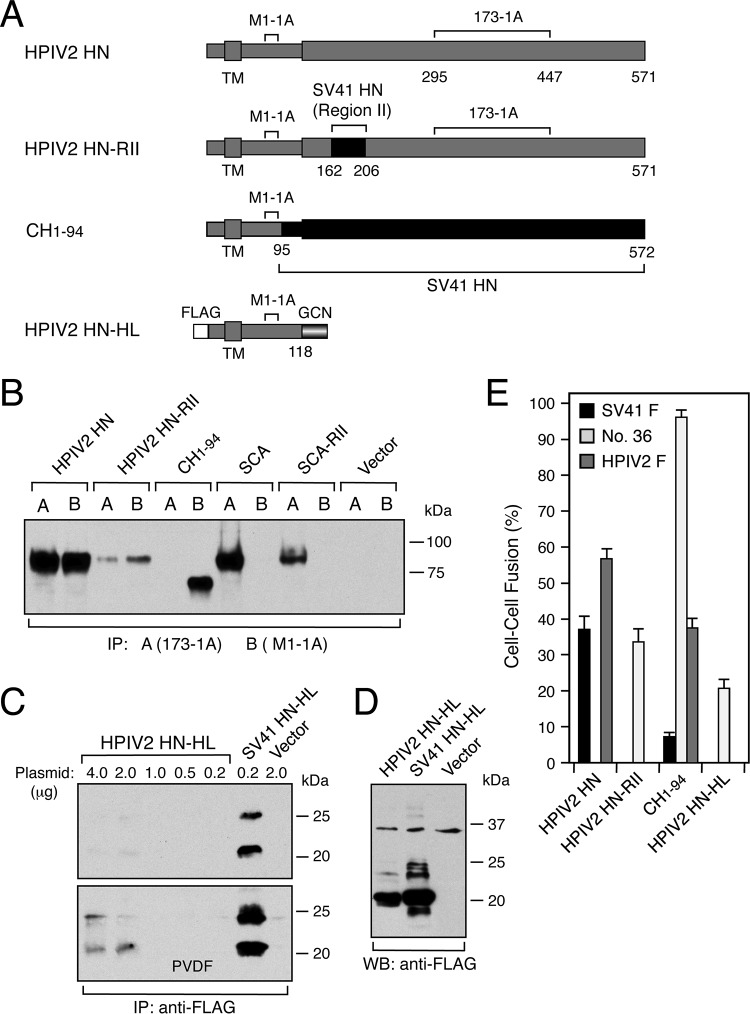
The results obtained so far have indicated that, in addition to the stalk domain, particular amino acids in region II are required for the SV41 HN protein to trigger the SV41 HN-specific chimera no. 36. To examine the possibility that region II would be directly involved in the interaction with the F protein, we replaced region II of the HPIV2 HN protein, which was unable to trigger chimera no. 36 (Fig. 9E), with that of the SV41 HN protein. Interestingly, the resulting chimera, HPIV2 HN-RII, failed to trigger the F proteins of HPIV2 and SV41 but was able to trigger chimera no. 36 (Fig. 9A and E) despite its remarkably low cell surface expression level compared to that of the HPIV2 HN protein (Fig. 9B). It was worth noting in this context that the stalk domain-specific antibody, MAb M1-1A, exhibited seemingly stronger reactivity to HPIV2 HN-RII than the head domain-specific MAb 173-1A, while these antibodies reacted equally to the HPIV2 HN protein (Fig. 9B), suggesting that these HN proteins differ in their conformation or stability in the presence of detergent. Interestingly, on the other hand, a previously reported chimeric SV41 HN protein, CH1-94, whose N-terminal 90 residues were derived from the HPIV2 HN protein (10), exhibited prominent triggering activity toward chimera no. 36 while showing moderate triggering activity toward the HPIV2 F protein (Fig. 9A and E). Intriguingly, although the headless form of the HPIV2 HN protein, HPIV2 HN-HL, was not efficiently transported to the cell surface (Fig. 9C), it was able to trigger chimera no. 36 but not the HPIV2 F protein, similar to HPIV2 HN-RII (Fig. 9A and E). Provided that the apparent inability of HPIV2 HN-RII and HPIV2 HN-HL to trigger the HPIV2 HN protein is due to their comparatively weak F-triggering activity compared to that of CH1-94, their F protein specificity could be regarded as being similar to that of CH1-94. These results indicate that SV41 HN region II can convert the F protein specificity of the HPIV2 HN protein by substitution, but that this region is not directly involved in the interaction with the F protein.
FIG 9.
Replacement of region II or removal of the head domain converts the F protein specificity of the HPIV2 HN protein. (A) Schematic diagram of the HN proteins. Positions of the epitopes for the HPIV2 HN-specific MAbs M1-1A and 173-1A are indicated. FLAG, FLAG epitope tag; GCN, a modified terminal tetramerization module; TM, transmembrane domain. (B) Detection of cell surface-localized chimeric HPIV2 HN proteins. Cell surface biotinylation, immunoprecipitation, and SDS-PAGE analyses were carried out as described in the legend to Fig. 3A. (C) Detection of the headless HPIV2 HN protein expressed on the cell surface. Cell surface biotinylation, immunoprecipitation, and SDS-PAGE analyses were carried out as described in the legend to Fig. 7D. (D) Detection of the headless HPIV2 HN protein in the transfected cells. Western blotting was carried out as described in the legend to Fig. 7B. (E) F-triggering activity of the HN proteins. The average fusion index was determined as described in the legend to Fig. 1B; error bars indicate standard deviations.
DISCUSSION
We previously reported that replacement of the stalk domain of the SV41 HN protein with that of the HPIV2 HN protein was enough to convert the SV41 HN protein to an HPIV2 F-specific protein (10). Among the unconserved amino acids of the HPIV2 HN stalk domain, two isoleucine residues in the FAR and a valine residue immediately upstream of the FAR (Fig. 1A) proved to be critical, because these amino acids bestowed the ability to trigger the HPIV2 F protein on the SV41 HN protein by substitution (10). However, they were not enough to convert the SV41 HN protein to an HPIV2 F-specific protein, indicating that the amino acids located between the transmembrane domain and the FAR also were indispensable for determining the F protein specificity (10). In support of this notion, it has recently been reported that a chimeric NDV HN protein whose FAR had been replaced with that of the PIV5 HN protein failed to trigger the noncognate PIV5 F protein and retained the ability to trigger the cognate NDV F protein (11).
We have shown in the present study that amino acid substitutions in region II of the HN head domain drastically change the F protein specificity of the HN protein. For instance, as represented by chimera SCA-RII, the replacement of the HPIV2 HN-derived region II of chimera SCA with the SV41 HN counterpart results in the loss of triggering activity toward the SV41 F protein and instead results in the acquisition of the triggering activity toward chimera no. 36 (Fig. 3B). We also have shown that the removal of a great part of the head domain from the SV41 HN protein results in the loss of triggering activity toward the SV41 F protein while preserving the activity toward chimera no. 36 (Fig. 7A and C). In contrast, the complete removal of the head domain of the SV41 HN protein results in the loss of triggering activity toward chimera no. 36 while preserving activity toward the SV41 F protein (Fig. 7A and C). Interestingly, moreover, removal of the head domain of the HPIV2 HN protein results in a loss of triggering activity toward the HPIV2 F protein but in an unexpected acquisition of the triggering activity toward chimera no. 36 (Fig. 9A and E), suggesting that the stalk domain of the HPIV2 HN protein is intrinsically capable of triggering chimera no. 36 but that this activity can be displayed only when it has adopted a given conformation which is no longer fittable to the cognate HPIV2 F protein. It also should be pointed out that the chimeric HPIV2 HN protein, HPIV2 HN-RII, which harbors the SV41 HN-derived region II, fails to trigger the cognate HPIV2 F protein but triggers chimera no. 36 (Fig. 7A and C). Thus, these findings let us hypothesize that either replacement of the HN region II or removal of all (or a great part of) the HN head domain somehow alters the conformation of the HN stalk domain, resulting in the conversion of the F protein specificity. Thus, we propose that the HN head domain plays a dual role in the fusion event: one is its transition from the 4-heads-down conformation to the 4-heads-up conformation after receptor binding that allows the HN stalk domain to interact with the F protein (16, 28), and the other is an uncharacterized function of the head domain in the 4-heads-up conformation that modulates the conformation of the HN stalk domain. It is worth noting in this context that the headless MuV HN protein triggers the noncognate PIV5 F protein far less efficiently than the full-length MuV HN protein (11).
Interestingly, chimeras SCA and SCA-RII exhibit significantly lower oligomeric stability than the SV41 HN protein (Fig. 6A). Since the stalk and head domains of these chimeric HN proteins are heterologous (Fig. 2A and 5), the noncovalent interactions between these domains in the 4-heads-down conformation (16, 28) may be weakened, which somehow leads to the destabilization of the dimer and then that of the tetramer. This notion also would be the case with the intermediates shown in Fig. 6. Importantly, nonetheless, no correlation was found between the oligomeric stability and the F protein specificity of the HN proteins.
In the present study, we have confirmed previously reported substitutability among rubulavirus HN proteins, simultaneously providing novel evidence that the HN protein of MuV strain Enders has a broad substitutability that is the same as that of MuV strain Miyahara (11, 25, 27): they can substitute for the HN proteins of HPIV2 and PIV5, while the latter proteins are unable to substitute for the MuV HN proteins. Since none of the rubulavirus HN proteins can substitute for the avulavirus NDV HN protein (11, 27), the observed substitutability seems to take place on the basis of the similarity in their primary structures. However, such similarity by itself clearly is not sufficient to explain the observed unidirectional nature of the substitutability. Interestingly, in this context, the truncated HPIV2 HN protein exhibits triggering activity toward chimera no. 36, while the full-length HPIV2 HN protein does not (Fig. 9E). Therefore, it is conceivable that all four rubulavirus HN proteins listed in Fig. 1C are intrinsically substitutable for one another and that a subtle difference in the HN stalk conformation defines whether a given HN protein can trigger a noncognate F protein, resulting in the observed unidirectional substitutabilities. Notably, it has been proven that the HN stalk domain is highly flexible (30), which would be beneficial to conform different shapes at least transiently.
It has been reported recently that various headless HN proteins can efficiently trigger their cognate F proteins without interacting with receptors (11, 15). This observation suggests that the headless HN proteins prematurely trigger the F protein in the trans-Golgi network (TGN), where the F protein undergoes proteolytic cleavage (2). However, since the suppression of cell surface expression of the headless SV41 HN protein by the ER retention/retrieval signal significantly reduces the extent of cell-cell fusion (Fig. 8B and C), the headless SV41 HN molecules seem to function predominantly on the cell surface. One possibility is that in acidic pH environments, such as the Golgi apparatus, TGN, and secretory vesicles, the headless HN molecules undergo a reversible conformational change, thereby becoming unable to interact with the F protein until reaching the cell surface. Intriguingly, on the other hand, the truncated HN protein IM18-Tr-ER, which has the ER retention/retrieval signal, can trigger chimera no. 36 as efficiently as IM18-Tr, whereas its cell surface expression is not detectable (Fig. 8B and C). It should be noted in this context that even the surface expression of IM18-Tr is undetectable when the cells are transfected with 0.2 μg of the vector encoding it (Fig. 7D), which is more than enough to maximally induce cell-cell fusion with 2 μg of chimera no. 36-encoding vector (Fig. 8D). Therefore, it seems that extremely small amounts of the truncated HN proteins on the cell surface, far below the detection level of cell surface biotinylation and immunoprecipitation, is able to trigger chimera no. 36 efficiently. Alternatively, chimera no. 36 may be destabilized in the TGN by the truncated HN proteins before the HN molecules undergo retrieval to the ER, resulting in the induction of HN-independent cell-cell fusion on the cell surface that is the same as that of the PIV5 F mutant L22P (25).
It is very curious that the addition of the ER retention/retrieval signal to SV41 HN-Tr results in an increase in its cell surface expression level, as demonstrated by SV41 HN-Tr-ER (Fig. 8B). Since SV41 HN-Tr cannot be detected on the cell surface despite our efforts while IM-18-Tr was barely detectable (Fig. 7D), it may tend to undergo misfolding more than IM-18-Tr does. It is worth noting in this context that SV41 HN-Tr-ER migrates faster than SV41 HN-Tr, whereas IM18-Tr-ER and IM18-Tr migrate similarly (Fig. 8A), suggesting that the majority of the SV41 HN-Tr molecule does not fold correctly and would be arrested by ER-folding chaperones. In all likelihood, the addition of the ER signal to SV41 HN-Tr may somehow facilitate correct folding, which would overcome the signal-mediated ER retention.
Finally, we conclude that the F protein specificity of the HN protein is not defined solely by the primary structure of the stalk domain. It is important in this context to note that the HN protein specificity of the F protein also is not solely defined by the primary structure of the F head domain, as we have reported previously (21, 31).
ACKNOWLEDGMENT
This work was supported by a Grant-in-Aid for Scientific Research (grant 26460552) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Lamb RA, Parks GD. 2013. Paramyxoviridae: the viruses and their replication, p 957–995. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Karron RA, Collins PL. 2013. Parainfluenza viruses, p 996–1023. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology 6th ed vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Heminway BR, Yu Y, Galinski MS. 1994. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res 32:1–16. doi: 10.1016/0168-1702(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 4.Hu X, Ray R, Compans RW. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol 66:1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iorio RM, Melanson VR, Mahon PJ. 2009. Glycoprotein interactions in paramyxovirus fusion. Future Virol 4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb RA, Jardetzky TS. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol 17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison TG. 2003. Structure and function of a paramyxovirus fusion protein. Biochim Biophys Acta 1614:73–84. doi: 10.1016/S0005-2736(03)00164-0. [DOI] [PubMed] [Google Scholar]
- 8.Deng R, Mirza AM, Mahon PJ, Iorio RM. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch Virol 13Suppl:115–130. [DOI] [PubMed] [Google Scholar]
- 9.Tanabayashi K, Compans RW. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J Virol 70:6112–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsurudome M, Kawano M, Yuasa T, Tabata N, Nishio M, Komada H, Ito Y. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190–203. doi: 10.1006/viro.1995.1559. [DOI] [PubMed] [Google Scholar]
- 11.Bose S, Song AS, Jardetzky TS, Lamb RA. 2014. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol 88:3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melanson VR, Iorio RM. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol 80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirza AM, Sheehan JP, Hardy LW, Glickman RL, Iorio RM. 1993. Structure and function of a membrane anchor-less form of the hemagglutinin neuraminidase glycoprotein of Newcastle disease virus. J Biol Chem 268:21425–21432. [PubMed] [Google Scholar]
- 14.Thompson SD, Portner A. 1987. Localization of functional sites on the hemagglutinin-neuraminidase glycoprotein of Sendai virus by sequence analysis of antigenic and temperature-sensitive mutants. Virology 160:1–8. doi: 10.1016/0042-6822(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 15.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A 109:E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose S, Jardetzky TS, Lamb RA. 2015. Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479-480C:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porotto M, Palmer SG, Palermo LM, Moscona A. 2012. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J Biol Chem 287:778–793. doi: 10.1074/jbc.M111.298059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. 2009. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol 83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gui L, Jurgens EM, Ebner JL, Porotto M, Moscona A, Lee KK. 2015. Electron tomography imaging of surface glycoproteins on human parainfluenza virus 3: association of receptor binding and fusion proteins before receptor engagement. mBio 6:e02393–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bose S, Heath CM, Shah PA, Alayyoubi M, Jardetzky TS, Lamb RA. 2013. Mutations in the parainfluenza virus 5 fusion protein reveal domains important for fusion triggering and metastability. J Virol 87:13520–31351. doi: 10.1128/JVI.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsurudome M, Nakahashi M, Matsushima Y, Ito M, Nishio M, Kawano M, Komada H, Nosaka T. 2013. Full conversion of the hemagglutinin-neuraminidase specificity of the parainfluenza virus 5 fusion protein by replacement of 21 amino acids in its head region with those of the simian virus 41 fusion protein. J Virol 87:8342–8350. doi: 10.1128/JVI.03549-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsurudome M, Nishio M, Komada H, Bando H, Ito Y. 1989. Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 171:38–48. doi: 10.1016/0042-6822(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 23.Tsurudome M, Bando H, Nishio M, Iwamoto Y, Kawano M, Kondo K, Komada H, Ito Y. 1990. Antigenic and structural properties of a paramyxovirus simian virus 41 (SV41) reveal a close relationship with human parainfluenza type 2 virus. Virology 179:738–748. doi: 10.1016/0042-6822(90)90141-D. [DOI] [PubMed] [Google Scholar]
- 24.Yuasa T, Kawano M, Tabata N, Nishio M, Kusagawa S, Komada H, Matsumura H, Ito Y, Tsurudome M. 1995. A cell fusion-inhibiting monoclonal antibody binds to the presumed stalk domain of the human parainfluenza type 2 virus hemagglutinin-neuraminidase protein. Virology 206:1117–1125. doi: 10.1006/viro.1995.1035. [DOI] [PubMed] [Google Scholar]
- 25.Ito M, Nishio M, Kawano M, Kusagawa S, Komada H, Ito Y, Tsurudome M. 1997. Role of a single amino acid at the amino terminus of the simian virus 5 F2 subunit in syncytium formation. J Virol 71:9855–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol 87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsurudome M, Ito M, Nishio M, Kawano M, Okamoto K, Kusagawa S, Komada H, Ito Y. 1998. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for haemagglutinin-neuraminidase proteins to promote cell fusion. J Gen Virol 79:279–289. doi: 10.1099/0022-1317-79-2-279. [DOI] [PubMed] [Google Scholar]
- 28.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. 2013. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog 9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerangue N, Schwappach B, Jan YN, Jan LY. 1999. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K (ATP) channels. Neuron 22:537–548. doi: 10.1016/S0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 30.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky T, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol 85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsurudome M, Ito M, Nishio M, Nakahashi M, Kawano M, Komada H, Nosaka T, Ito Y. 2011. Identification of domains on the fusion (F) protein trimer that influence the hemagglutinin-neuraminidase specificity of the F protein in mediating cell-cell fusion. J Virol 85:3253–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]