ABSTRACT
The phylogeographic history of the Brazilian HIV-1 subtype C (HIV-1C) epidemic is still unclear. Previous studies have mainly focused on the capital cities of Brazilian federal states, and the fact that HIV-1C infections increase at a higher rate than subtype B infections in Brazil calls for a better understanding of the process of spatial spread. A comprehensive sequence data set sampled across 22 Brazilian locations was assembled and analyzed. A Bayesian phylogeographic generalized linear model approach was used to reconstruct the spatiotemporal history of HIV-1C in Brazil, considering several potential explanatory predictors of the viral diffusion process. Analyses were performed on several subsampled data sets in order to mitigate potential sample biases. We reveal a central role for the city of Porto Alegre, the capital of the southernmost state, in the Brazilian HIV-1C epidemic (HIV-1C_BR), and the northward expansion of HIV-1C_BR could be linked to source populations with higher HIV-1 burdens and larger proportions of HIV-1C infections. The results presented here bring new insights to the continuing discussion about the HIV-1C epidemic in Brazil and raise an alternative hypothesis for its spatiotemporal history. The current work also highlights how sampling bias can confound phylogeographic analyses and demonstrates the importance of incorporating external information to protect against this.
IMPORTANCE Subtype C is responsible for the largest HIV infection burden worldwide, but our understanding of its transmission dynamics remains incomplete. Brazil witnessed a relatively recent introduction of HIV-1C compared to HIV-1B, but it swiftly spread throughout the south, where it now circulates as the dominant variant. The northward spread has been comparatively slow, and HIV-1B still prevails in that region. While epidemiological data and viral genetic analyses have both independently shed light on the dynamics of spread in isolation, their combination has not yet been explored. Here, we complement publically available sequences and new genetic data from 13 cities with epidemiological data to reconstruct the history of HIV-1C spread in Brazil. The combined approach results in more robust reconstructions and can protect against sampling bias. We found evidence for an alternative view of the HIV-1C spatiotemporal history in Brazil that, contrary to previous explanations, integrates seamlessly with other observational data.
INTRODUCTION
Since its emergence in the human population in central Africa around 1920 (1), HIV-1 group M has diversified into nine subtypes and numerous circulating recombinant forms (CRFs) through a series of founder effects and recombination events (2, 3). Although HIV-1 subtype B (HIV-1B) dominates in many countries in Europe and the Americas (2), more than 50% of the infections worldwide are caused by HIV-1 subtype C (HIV-1C), which is the most prevalent subtype in southern African countries and India and is increasing in prevalence in China and South America (2, 4).
The epidemic in Brazil is mainly driven by HIV-1B, followed by lower frequencies of HIV-1C, -F1, and -BF1 recombinants (5). In the southern regions of Brazil, however, the spread of HIV-1B is matched by that of HIV-1C, which cocirculates in similar proportions and can even be responsible for up to 80% of infections (4). In addition, the two southernmost Brazilian states, Rio Grande do Sul (RS) and Santa Catarina (SC), have the highest AIDS incidence in the country (6). These patterns have motivated several investigations into the history and dynamics of the Brazilian HIV-1C epidemic (HIV-1C_BR), which is estimated to have originated in the 1960s and 1970s in the state of Paraná (PR) (7, 8, 9). The fact that HIV-1C incidence in more northern states has only recently begun to increase (4, 10–14) suggests viral diffusion is driven by unknown factors that promote fast dissemination to the south while constraining spread to the north.
HIV infections are characterized by a dynamic viral population of closely related variants that can quickly adapt to changing selective pressures, which manifests in a formidable speed at which genetic diversity accumulates within hosts (15). This rapid accumulation of genetic diversity makes HIV-1 a prime example of “measurably evolving populations” (16). As a consequence, there has been an important role for phylogenetic tools to shed light on the epidemiological history of HIV. In fact, this has stimulated many developments in the field of statistical phylodynamics, such as molecular clock models to incorporate sampling time as calibration information (17, 18) and coalescent models to infer the changes in viral population size over time (19–21). More recently, such genealogy-based population genetic inferences have also been complemented by state-of-the-art phylogeographic tools (22–24). Phylodynamic analyses of HIV-1 have culminated in a relatively rich statistical account of its evolutionary and epidemiological history (1).
While these statistical models and inference tools have proven invaluable for testing hypotheses using virus genetic data (25, 26), they are limited in their ability to link epidemic processes to pathogen evolution because nongenetic data are usually not directly incorporated into the models. For phylogeography, this has recently been addressed by extending a Bayesian discrete phylogenetic diffusion approach in order to incorporate covariates in the process of spread (27). This approach estimates phylogeographic history while identifying the contributions of several potential explanatory variables (predictors) of spatial spread and allows cross talk between the spatial genetic distribution and the relevant predictors. The predictors are selected for the ability to explain the location transition history, but by helping to inform the process parameters, they can also assist in shaping ancestral reconstructions. This approach has already proven useful in elucidating the drivers of both human and swine influenza virus dispersal (27, 28).
In the present study, we reconstruct the phylogeographic history of HIV-1C in Brazil, incorporating newly obtained sequence data. While previous studies mostly included sequences from state capital cities, here, we expanded the spatial sampling by including HIV-1C sequences from 10 rural locations in the southernmost states, RS and SC. Our study demonstrates for the first time that augmenting the molecular sequence data with relevant epidemiological information can contribute to the robustness of phylogeographic reconstructions.
MATERIALS AND METHODS
Patients, samples, and new sequences.
A total of 360 HIV-1-seropositive patients from 13 cities in the states of SC and RS (Fig. 1) were enrolled in this study, which was approved by the ethics committees of the Federal University of Santa Catarina and the Foundation of Research and Production in Health of the state of Rio Grande do Sul. Between October 2009 and February 2014, blood samples were collected, and HIV-1 envelope (HXB2 bp 6846 to 7360) and polymerase (HXB2 bp 2274 to 3545) fragments were amplified from whole cellular DNA by nested PCR and sequenced as described previously (29). The sequences were subtyped using the REGA, RIP, and SCUEAL online subtyping tools (30–32) and by performing maximum-likelihood phylogenetic inference incorporating HIV-1 subtype reference sequences available from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov). Recombinant sequences were identified through bootscanning analysis using Simplot 3.5.1 (33) (see Text S1 in the supplemental material for methodological details).
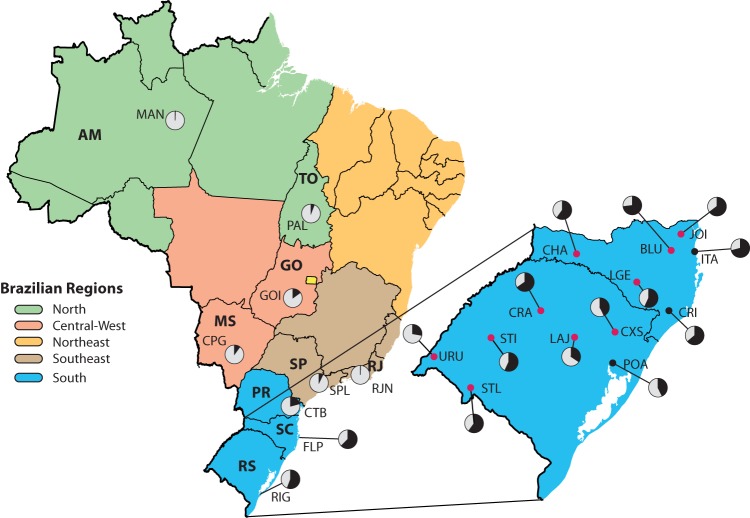
FIG 1.
Administrative map of Brazil indicating the locations from which HIV-1C sequences were obtained. The pie charts show the HIV-1C (black) percentages of infections relative to other HIV-1 strains (gray) in all cities with pol or env sequences included in this study. State name abbreviations are shown in boldface. The inset shows an enlarged map with the sampling locations (black and red dots) in Santa Catarina and Rio Grande do Sul from which new sequence data were generated. Red dots, cities sampled for the first time; black dots, sampling locations from which sequence data from other studies were also available. The Brazilian regions are colored according to the legend. States: AM, Amazonas; GO, Goiás; MS, Mato Grosso do Sul; PR, Paraná; RJ, Rio de Janeiro; SC, Santa Catarina; SP, São Paulo; TO, Tocantins; RS, Rio Grande do Sul. Cities: BLU, Blumenau; CHA, Chapecó; CPG, Campo Grande; CRA, Cruz Alta; CRI, Criciúma; CTB, Curitiba; CXS, Caxias do Sul; FLP, Florianópolis; GOI, Goiania; ITA, Itajaí; JOI, Joinville; LAJ, Lajeado; LGE, Lages; MAN, Manaus; PAL, Palmas; POA, Porto Alegre; RIG, Rio Grande; RJN, Rio de Janeiro; SPL, São Paulo; STI, Santiago; STL,Santana do Livramento; URU, Uruguaiana. (The maps were drawn by T. Graf using QGIS software and source maps from Natural Earth.)
Sequence data set compilation.
Briefly, we complemented our new sequence data with all publically available Brazilian HIV-1C sequences (HIV-1C_BR) from pol and env genes (n = 385). Non-Brazilian HIV-1C sequences were selected using BLAST+ (34). For this purpose, we created a local BLAST database that contained all HIV1-C sequences minus those from Brazil. We performed a similarity search on this database using every HIV-1C_BR sequence as a query, and the 50 best hits for each search were logged. Duplicate entries were removed from these hits and compiled as the international database. After extensive data cleaning, this resulted in data sets with 1,522 pol and 621 env sequences that were downsampled to around 500 sequences each to reduce the computational burden in subsequent Bayesian statistical analyses (see Text S1 in the supplemental material for full details of the procedure followed). Six additional subsamplings containing only Brazilian sequences were made for pol and env to allow assessment of the robustness of the phylogeographic reconstructions (see below). For this, we aimed at reducing sampling bias by creating three random downsamples in two groups: (i) Rand10, with a maximum of 10 sequences by location, and (ii) Rand20, with a maximum of 20 sequences by location (see Table S1 in the supplemental material for the HIV-1C_BR sequences in each data set).
Phylogenetic divergence time estimation and population dynamics inference.
Time-scaled phylogenetic trees were reconstructed using a Bayesian inference method implemented in the BEAST/BEAGLE software (35, 36). All analyses were performed using the GTR + I + Γ4 nucleotide substitution model and an uncorrelated lognormal relaxed molecular clock under the Bayesian Skyride coalescent model (18, 37). Due to the low temporal signals of the data sets, the use of an informative prior on the time to the most recent common ancestor (tMRCA) of the Brazilian subtype C clade was required. For this purpose, we specified a normal distribution with a mean (1976) and a standard deviation (5.1 years) based on previous estimates of the time of introduction of subtype C in Brazil (8). When exact sampling dates were unknown, the dates were integrated over a known sampling time interval (38). Monte Carlo Markov chains (MCMC) were run for sufficiently long to ensure stationarity and an adequate effective sample size (ESS) of >200, as diagnosed by Tracer (http://beast.bio.ed.ac.uk/tracer). Maximum clade credibility (MCC) trees were summarized using the TreeAnnotator tool and visualized in Figtree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). A representative sample of 1,000 trees was collected and used as an empirical tree distribution for estimating the virus migration processes (see below). To ensure that subsequent phylogeographic analyses were based on histories specific to Brazil, we pruned sequences clustering outside the HIV-1C_BR cluster and non-Brazilian sequences clustering inside the HIV-1C_BR cluster from these trees using PAUP (http://paup.csit.fsu.edu) (see Text S1 in the supplemental material for methodological details).
Phylogeny-trait association.
We tested for significant phylogenetic clustering by location in two different ways. First, we calculated the association index (AI), parsimony score (PS), and monophyletic clade (MC) measures using BaTS (39). For our second approach, we introduced the use of the path-sampling and stepping stone (SS) marginal likelihood estimators as implemented in BEAST (40, 41) to evaluate the extent to which the topology is required as a correlation structure to explain traits. For this, we specified a discrete symmetric (reversible) model of location transitioning and incorporated a Bayesian stochastic search variable selection (BSSVS) procedure (22), fitting the trait diffusion process on (i) a fixed MCC tree summarized from the Bayesian phylogenetic analysis of the complete data set and (ii) a star-like tree with the same trait annotations as the MCC tree.
Phylogeographic inference with epidemiological predictors.
To assess the impacts of potential explanatory variables (predictors) of the viral diffusion process on phylogeographic reconstructions, we made use of the recent generalized linear model (GLM) extension of Bayesian discrete phylogeographic models (27). This allows reconstruction of the spatial diffusion history throughout the tree while simultaneously evaluating the contributions of various potential predictors. Support for predictors is estimated using a BSSVS procedure, and the contribution of each predictor is quantified as a GLM coefficient that has an impact (effect size) on the transition rate among the locations.
Using this approach, we tested the following predictors (see Text S1 in the supplemental material for methodological details): (i) geographic distance (the great-circle distance between each pair of cities), (ii) passenger air traffic (the number of passengers traveling between each pair of airports), (iii) HIV population size (the total number of AIDS notifications in a period of 10 years reported in each city), (iv) HIV prevalence [(HIV population size/city population size) × 100,000 inhabitants], (v) HIV-1C population size (HIV population size times the proportion of HIV-1C as reported in the literature [4, 10–14]), (vi) HIV-1C prevalence [(HIV-1C population size/city population size) × 100,000 inhabitants], and (vii) sample size (the number of sequences by location).
Because not all sampling locations have an airport, we specified a different geographic partitioning for evaluating predictor 2 (passenger air traffic). This partitioning is not well suited to the epidemiological predictors, which led us to test predictor 2 in separate analyses including only sample size as an additional potential predictor.
GLM analyses were run in BEAST using previously recommended prior specifications on the set of empirical trees obtained by the Bayesian phylogenetic analysis (27). Bayes factors (BFs) were calculated to determine the support for the inclusion of each predictor in the model, and predictor contributions are reported as effect sizes conditional on the effect being included in the model.
A phylogeographic analysis with BSSVS was performed, with asymmetric transition rates informed by the predictors supported by the GLM analysis.
In other words, for each subsampled data set, we used the rate estimates for prior specification based on the corresponding GLM analysis. SPREAD software was used to identify the well-supported transition rates based on BFs of >3 (42). We complemented this analysis with Markov jump estimation of the number of location transitions throughout the evolutionary history (43). RStudio (http://www.rstudio.org/) was used to calculate the Bayes factors and effect sizes and to summarize the posterior densities of the highly supported transitions from the BEAST log files.
Nucleotide sequence accession numbers.
The sequences generated in the present study were deposited in GenBank under accession numbers KR065788 to KR066336 and KP224476 to KP224501.
RESULTS
Sequence data set compilation.
We sequenced 140 pol and 202 env HIV-1C isolates in 13 locations in the states SC and RS, 10 of which had not been sampled before (Fig. 1). By combining the generated sequence data with publicly available Brazilian and international HIV-1C sequences, we were able to compile comprehensive pol- and env-based data sets for reconstructing the spatiotemporal history of HIV-1C in Brazil. In summary, the complete pol data set contained 380 Brazilian and 120 international sequences, while the env data set totaled 293 Brazilian and 170 international sequences (see Data Set S1 in the supplemental material for complementary information about sequences retrieved from public data banks). The Brazilian pol sequences are distributed over 21 locations, and the env sequences represent 17 locations, totaling 22 locations represented by pol or env sequences, most of them in SC and RS (15/21 for pol and 14/17 for env). Considering the complete Brazilian data set, the sequences represent the period between 2002 and 2014 (see Table S1 in the supplemental material).
Phylogeny-trait association.
Because our preliminary analyses suggested a considerable degree of phylogenetic mixing by location, we formally tested whether the data sets containing only Brazilian sequences still supported spatial population structure. The hypothesis of a panmictic population could be rejected for the pol and env data sets based on the AI and PS statistics (P < 0.05), but the MC scores revealed that for 12/21 (57%) pol locations and 12/17 (71%) env locations, random clustering could not be rejected (see Table S2 in the supplemental material for the MC scores). The results of the approach based on model testing also provided strong support against the absence of phylogenetic association by sampling location in the pol and env data sets (Bayes factors of 74 and 39, respectively).
Inconsistencies in root state estimates.
The results of the phylogeographic reconstruction showed, with strong agreement between most data sets and models applied (50/56 analyses), that the epidemic originated in SC or RS. Its exact location of introduction, however, could not be unambiguously determined using only virus genetic data. Whereas in the complete pol and env data sets Florianópolis (FLP) was consistently estimated to be the most likely location at the root, other cities—most notably Porto Alegre (POA) (7/48) and Criciúma (CRI) (7/48)—were implicated in 60% (29/48) of the analyses based on the Rand10 and Rand20 subsampled data sets (Table 1).
TABLE 1.
Modal root state and posterior probability estimates resulting from different discrete Bayesian phylogeographic analyses applied to different data sets
| Sequencet | Method | Estimate for data seta: |
||||||
|---|---|---|---|---|---|---|---|---|
| Complete | Rand10A | Rand10B | Rand10C | Rand20A | Rand20B | Rand20C | ||
| pol | Symmetric-BSSVS | FLP (1.00) | SPL (0.99) | CTB (0.99) | RJN (0.99) | POA (0.99) | RIG (0.99) | RIG (0.99) |
| Symmetric | FLP (0.99) | CRI (0.99) | FLP (0.97) | CRI (0.97) | FLP (0.99) | FLP (0.98) | ITA (0.99) | |
| Asymmetric-BSSVS | FLP (1.00) | CRI (1.00) | FLP (0.99) | CTB (0.99) | ITA (1.00) | FLP (0.99) | POA (0.99) | |
| Asymmetric | FLP (0.99) | RJN (0.99) | FLP (0.99) | CTB (0.99) | ITA (1.00) | FLP (0.99) | FLP (0.99) | |
| env | Symmetric-BSSVS | FLP (0.97) | FLP (0.99) | POA (0.99) | POA (0.99) | FLP (0.99) | FLP (0.99) | CXS (0.99) |
| Symmetric | FLP (0.97) | FLP (0.99) | POA (0.99) | FLP (0.96) | FLP (0.99) | FLP (0.99) | CRI (0.99) | |
| Asymmetric-BSSVS | FLP (0.99) | CRI (0.99) | POA (0.99) | LAJ (0.99) | CRI (1.00) | POA (0.99) | FLP (1.00) | |
| Asymmetric | FLP (0.99) | BLU (0.99) | FLP (0.99) | CRI (0.69) | FLP (0.99) | CRA (0.99) | FLP (1.00) | |
Cities: BLU, Blumenau; CRI, Criciúma; CTB, Curitiba; CXS, Caxias do Sul; FLP, Florianópolis; ITA, Itajaí; LAJ, Lajeado; POA, Porto Alegre; RIG, Rio Grande; RJN, Rio de Janeiro.
Predictors of viral spread.
Using a phylogeographic GLM approach, we evaluated which measures predicted the rates of location exchange in the complete and subsampled data sets (Table 2). In the pol and env complete data sets, only the origin and destination sample sizes yielded strong BF support, reflecting the fact that only sample sizes and their heterogeneity are needed to explain the number of location transitions. We also found strong support for destination sample size in the model as a predictor in all pol and env subsampled data sets (Rand10 and Rand20). This indicates that, despite the more homogeneous distribution of sequences by sampling locations in these subsampled data sets, the remaining heterogeneity still has an impact on the phylogeographic reconstructions.
TABLE 2.
Bayes factor support for an explanatory role in the HIV-1C_BR diffusion process for all tested predictors in all data sets
| Sequence | Predictora | BFb for data set: |
||||||
|---|---|---|---|---|---|---|---|---|
| Complete | Rand10A | Rand10B | Rand10C | Rand20A | Rand20B | Rand20C | ||
| pol | Geographical distance | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 |
| Origin sample size | Inf | 0.5 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | |
| Destination sample size | Inf | 6,583.3 | 5,758.4 | 1,674.5 | Inf | Inf | Inf | |
| Origin HIV population size | 0.6 | 1.4 | 1.1 | 1.6 | 1.2 | 1.1 | 1.2 | |
| Destination HIV population size | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Origin HIV prevalence | 0.2 | 6.1 | 10.8 | 16.1 | 17.3 | 13.1 | 7.9 | |
| Destination HIV prevalence | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Origin HIV-1C population size | 0.3 | 14.4 | 22.4 | 9.6 | 18.1 | 23.4 | 38.8 | |
| Destination HIV-1C population size | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | |
| Origin HIV-1C prevalence | 0.3 | 7.2 | 1.3 | 4.1 | 1.1 | 1.4 | 1.0 | |
| Destination HIV-1C prevalence | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| env | Geographical distance | 0.0 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 |
| Origin sample size | Inf | 0.3 | 0.4 | 0.9 | 0.3 | 0.3 | 0.4 | |
| Destination sample size | Inf | 20.6 | 32.5 | 29.4 | Inf | Inf | Inf | |
| Origin HIV population size | 0.3 | 0.8 | 1.1 | 2.6 | 0.8 | 0.9 | 0.9 | |
| Destination HIV population size | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Origin HIV prevalence | 0.3 | 19.6 | 18.9 | 15.3 | 22.3 | 26.0 | 23.4 | |
| Destination HIV prevalence | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Origin HIV-1C population size | 0.3 | 13.8 | 14.0 | 15.5 | 13.0 | 11.3 | 12.9 | |
| Destination HIV-1C population size | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Origin HIV-1C prevalence | 0.4 | 0.4 | 0.6 | 1.0 | 0.7 | 0.6 | 0.5 | |
| Destination HIV-1C prevalence | 0.0 | 0.6 | 0.6 | 0.6 | 0.0 | 0.0 | 0.0 | |
Epidemiological predictors included in all Rand10 and Rand20 data sets are in boldface.
BFs of ≥3 are in boldface. Inf, infinite.
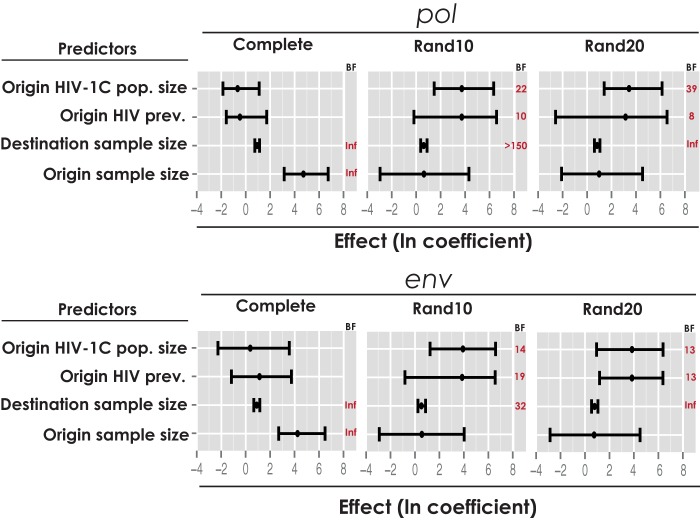
Two predictors, “origin HIV prevalence” and “origin subtype C population size,” were included in all pol and env Rand10 and Rand20 data sets with Bayes factor estimates ranging from moderate (BF = 6) to strong (BF = 39) support and with positive mean conditional effect sizes (Fig. 2). Hence, locations with higher HIV prevalence and larger HIV-1C populations tend to act as sources for onward spread.
FIG 2.
Significant predictors of the Brazilian HIV-1C epidemic spread among subsampling data sets of pol and env genes. Inclusion probabilities are represented as BFs, and a BF threshold of 3 was used as a positive indication of predictor inclusion. The effect of each predictor, conditional on its inclusion, is represented by the posterior mean (black dot) and the 95% HPD of the GLM coefficients in log scale. Inf, infinite.
In addition to epidemiological predictors, we also tested geographical distance or air transportation data (in a separate analysis [data not shown]) as a predictors of HIV-1C diffusion, but they did not result in noticeable support by any of the analyzed data sets.
Interestingly, incorporating relevant epidemiological information into the phylogeographic reconstructions resulted in consistent root state estimates: using the GLM model, we found POA to be the modal root state in all (12/12) pol and env Rand10 and Rand20 data sets. Only in the complete data sets, where the sampling bias is more severe, was FLP still estimated to be the modal root state.
To assess the robustness of the phylogeographic reconstructions with respect to the root height prior (see Materials and Methods), we also performed ancestral reconstruction using genealogies estimated under priors that specified a mean tMRCA that was 10 years longer or shorter. We found that differences in tree depths did not impact the outcome: POA was consistently the modal root state, and the same predictors found substantial Bayes factor support of the extended and shortened histories in all pol and env Rand10 and Rand20 data sets.
Porto Alegre as a central hub of the HIV-1C epidemic.
We subsequently estimated the most likely migration patterns, using an asymmetrical phylogeographic analysis with BSSVS and priors on the location exchange rate that are based on the GLM rate estimates. The robustness of the ancestral reconstructions was somewhat lower, because in this analysis, the predictors can only influence the analysis through the prior specification. POA was found to be the root state location in 10/12 pol and env Rand10 and Rand20 data sets (data not shown). Nonetheless, POA was strongly linked to all other locations (Bayes factors ≥ 3), while only a few additional well-supported transitions were found. Because this suggests a central role for POA in the Brazilian HIV-1C dissemination, we address its role in more detail.
The time of arrival of HIV-1C in POA was estimated as 1973 (95% highest posterior density [HPD], 1966 to 1980) for pol and 1971 (95% HPD, 1963 to 1978) for env, and the spread to other cities started around 1980. The timing of these events reveals a consistent pattern. Nearby locations within RS (Rio Grande and Uruguaiana cities) were initially affected, followed by export to southern and southeastern state capitals (e.g., to Florianópolis, Curitiba, Rio de Janeiro, and São Paulo) in the early 1980s. More distant locations were affected at a later stage, first in the central-western region (Campo Grande) in the mid-1980s and later in the northern region (Palmas and Manaus) in the late 1980s and early 1990s. Only two exceptions to this pattern were found (one in the pol and one in the env data sets): the capital city Goiânia, where HIV-1C appears to have been introduced from POA in 1981 (pol), and Rio de Janeiro, where the introduction of HIV-1C was more recent, according to the env data sets (see Table S3 in the supplemental material for the time of first transition from Porto Alegre).
More insights into the temporal pattern of spread were obtained by mapping the densities of location transitions from POA to the other state capital cities through time. This revealed a period of higher density of viral influx to the southern region capital cities, Florianópolis and Curitiba, 25 to 30 years ago. Among the sampled capitals in the southeastern region, a similar pattern emerged for Rio de Janeiro, but there is a more evenly distributed transition density through time to São Paulo. Such a shift of transition density toward more recent times is slightly noticeable for the capital cities of the central-western and northern regions (see Fig. S1 in the supplemental material for transitions by time from Porto Alegre).
DISCUSSION
We reconstructed the phylogeographic history of HIV-1C in Brazil using a comprehensive set of pol and env subtype C sequences from 22 different cities, 10 of which were sampled for the first time. Using a new model-testing-based approach and by calculating several phylogeny-trait association measures using BaTS (39), we could reject random mixing in both data sets. However, as seen in MC scores, not all locations contributed equally to the phylogenetic signal, resulting in a considerable degree of uncertainty in the phylogeographic inferences. Nevertheless, after balancing the number of samples per location to mitigate the confounding effects of sampling biases, we were able to identify support for two epidemiological predictors of the viral spatial diffusion process.
Specifically, we found higher migration intensities from cities with larger numbers of HIV-1C-infected patients and higher HIV prevalence. Interestingly, this is in agreement with a pattern of HIV-1C spread toward the north of Brazil, where the prevalence of HIV is lower and only a few cases of HIV-1C infection have been found (4, 6). An intriguing result illustrating the complexity of modeling human mobility is that neither “geographical distance” nor “passenger air traffic” predicted viral spread. The sample sizes of source and/or recipient locations, on the other hand, were always included in the model (in isolation or together) (Table 2). Samples sizes are expected to predict the number of transitions to some extent, and it was not our intention to formally demonstrate this. Rather, we wanted to avoid the possibility that other predictors would be supported simply because of correlation with sample size. In other words, we do not expect that the support for HIV prevalence and subtype C population size in the origin locations is an artifact of the potential correlation with sample size, as it is already accommodated explicitly in the GLM analysis.
To explore how sampling heterogeneity also impacts ancestral reconstructions, we analyzed six random downsampled data sets in parallel with the complete pol and env data sets. This highlighted substantial variability in the root state estimates (Table 1) and confirmed that the sampling scheme can indeed have a profound effect on the inferred location state probabilities at the internal nodes of the tree. The impact of sampling biases was most likely aggravated by the relatively high degree of mixing observed in the pol and env data sets (see Table S2 in the supplemental material for MC scores). The geographical partitioning is also an important factor in discrete phylogeographic analyses, because it determines the level of spatial detail that can be recovered. Whereas previous studies investigating the spread of HIV-1C in Brazil categorized locations according to federal states or geopolitical regions (7–9), we opted for a higher-resolution scheme and defined cities as the locations of interest. This allowed us to include more precise predictors in the GLM.
We were able to largely resolve sampling-bias-related inconsistencies by informing the phylogeographical reconstructions with relevant epidemiological information. Our results consistently identified POA, and not the state of Paraná (7–9), as the point of introduction. Several lines of evidence support this hypothesis. The population in the metropolitan area of POA is about 4 million, the largest in the southern region, and the AIDS incidence rate in POA and its metropolitan area is the highest in Brazil (6). This suggests that the virus found ideal circumstances for transmission and explains why the HIV-1C prevalence in Paraná's capital, Curitiba, is much lower (∼22%) than that in POA (∼40% and up to ∼60% if the proportion of CRF31_BC, a local circulating form with a small subtype B insertion in a subtype C backbone, is considered) (4, 44).
Differences in risk group associations between the subtype B and C epidemics in Brazil also seem to support our findings. Whereas in POA, the association between men having sex with men (MSM) and HIV-1B disappeared in more recent sampling because of an expansion of HIV-1C in heterosexual (HET) and MSM groups (45), compartmentalized epidemics are still observed in other cities in the southern region, including Paraná, which could be explained by later introduction of HIV-1C (4, 46–48).
Finally, a central role for POA is also reflected in the high support for transitions from POA to all other locations and the reconstructed temporal pattern of dissemination. After its introduction in the early 1970s, HIV-1C started spreading to other cities in the early 1980s, first to nearby locations and then to locations progressively further away. It is interesting that we could recover a noticeably larger fraction of recent jumps from POA to São Paulo compared to transitions from POA to other southern or southeastern region capital cities, which points to a strong, longstanding epidemiological link between the cities.
Although our analysis provides support for POA as the central dissemination point of HIV-1C in Brazil, some caution is required when analyzing the number of transitions in star-like trees, such as those typically found for HIV-1. The absence of clear phylogenetic structure deeper in the trees also offers little opportunity to capture clear spatial structuring and transitions beyond those out of the location state at the root. In the current work, our sampling strategy focused on broad geographic coverage rather than on dense sampling, and a small sample from a large and diverse population that has grown exponentially through time generally results in star-like topologies. Thus, despite the support for a central role of POA, we can recover little detail on viral spread beyond transitions out of this location.
In conclusion, we present a comprehensive reconstruction of the spatial and temporal dynamics of HIV-1C in Brazil based on pol and, for the first time, env sequence data and included data from 10 newly sampled cities. By augmenting the viral genetic information with epidemiological data, we revealed a central role for the city of POA in the spread of HIV-1C in Brazil. In addition, we also identified locations with high HIV prevalence and large subtype C population sizes as key in the epidemic expansion toward the north of Brazil.
Supplementary Material
ACKNOWLEDGMENTS
We thank all collaborating municipal health centers from Santa Catarina and Rio Grande do Sul.
This study was supported by the Fundação de Amparo à Pesquisa e Inovação do Estado de Santa Catarina (FAPESC), the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 278433-PREDEMICS and ERC grant agreement no. 260864, the National Institutes of Health under grant R01 AI107034, and the National Science Foundation under grant DMS 1264153.
We have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01681-15.
REFERENCES
- 1.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. The early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 3.Ariën KK, Vanham G, Arts EJ. 2007. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol 5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gräf T, Pinto AR. 2013. The increasing prevalence of HIV-1 subtype C in Southern Brazil and its dispersion through the continent. Virology 435:170–178. doi: 10.1016/j.virol.2012.08.048. [DOI] [PubMed] [Google Scholar]
- 5.Inocencio LA, Pereira AA, Sucupira M, Fernandez J, Jorge CP, Souza DF, Fink HT, Diaz RS, Becker IM, Suffert TA, Arruda MB, Macedo O, Simão MB, Tanuri A. 2009. Brazilian Network for HIV Drug Resistance Surveillance: a survey of individuals recently diagnosed with HIV. J Int AIDS Soc 12:20. doi: 10.1186/1758-2652-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazilian Ministry of Health. 2014. AIDS epidemiological bulletin July 2013-June 2014. Brazilian Ministry of Health, Brasília, Brazil. [Google Scholar]
- 7.Veras NMC, Gray RR, de Macedo Brigido LF, Rodrigues R, Salemi M. 2011. High-resolution phylogenetics and phylogeography of human immunodeficiency virus type 1 subtype C epidemic in South America. J Gen Virol 92:1698–1709. doi: 10.1099/vir.0.028951-0. [DOI] [PubMed] [Google Scholar]
- 8.Delatorre E, Couto-Fernandez JC, Guimarães ML, Vaz Cardoso LP, de Alcantara KC, Martins de Araújo SM, Romero H, Freire CCM, Iamarino A, de Zanotto PMA, Morgado MG, Bello G. 2013. Tracing the origin and northward dissemination dynamics of HIV-1 subtype C in Brazil. PLoS One 8:e74072. doi: 10.1371/journal.pone.0074072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bello G, Zanotto PMA, Iamarino A, Gräf T, Pinto AR, Couto-Fernandez JC, Morgado MG. 2012. Phylogeographic analysis of HIV-1 subtype C dissemination in Southern Brazil. PLoS One 7:e35649. doi: 10.1371/journal.pone.0035649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brígido LFM, Ferreira JLP, Almeida VC, Rocha SQ, Ragazzo TG, Estevam DL, Rodrigues R. 2011. Southern Brazil HIV type 1 C expansion into the state of São Paulo, Brazil. AIDS Res Hum Retroviruses 27:339–344. doi: 10.1089/aid.2010.0157. [DOI] [PubMed] [Google Scholar]
- 11.Cardoso LPV, Pereira GAS, Viegas AA, Schmaltz LEPR, Martins de Araújo SM. 2010. HIV-1 primary and secondary antiretroviral drug resistance and genetic diversity among pregnant women from central Brazil. J Med Virol 82:351–357. doi: 10.1002/jmv.21722. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho BC, Cardoso LPV, Damasceno S, Martins de Araújo SM. 2011. Moderate prevalence of transmitted drug resistance and interiorization of HIV type 1 subtype C in the inland north state of Tocantins, Brazil. AIDS Res Hum Retroviruses 27:1081–1087. doi: 10.1089/aid.2010.0334. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira AS, Cardoso LPV, Martins de Araújo SM. 2011. Moderate prevalence of transmitted drug resistance and high HIV-1 genetic diversity in patients from Mato Grosso state, Central Western Brazil. J Med Virol 83:1301–1307. doi: 10.1002/jmv.22128. [DOI] [PubMed] [Google Scholar]
- 14.da Silveira AA, Cardoso LPV, Francisco RBL, Martins de Araújo SM. 2012. HIV type 1 molecular epidemiology in pol and gp41 genes among naive patients from Mato Grosso do Sul State, Central Western Brazil. AIDS Res Hum Retroviruses 28:304–307. doi: 10.1089/aid.2011.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambaut A, Posada D, Crandall KA, Holmes EC. 2004. The causes and consequences of HIV evolution. Nat Rev Genet 5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 16.Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. 2003. Measurably evolving populations. Trends Ecol Evol 18:481–488. doi: 10.1016/S0169-5347(03)00216-7. [DOI] [Google Scholar]
- 17.Rambaut A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- 18.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pybus OG, Rambaut A, Harvey PH. 2000. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strimmer K, Pybus OG. 2001. Exploring the demographic history of DNA sequences using the generalized skyline plot. Mol Biol Evol 18:2298–2305. doi: 10.1093/oxfordjournals.molbev.a003776. [DOI] [PubMed] [Google Scholar]
- 21.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemey P, Rambaut A, Welch JJ, Suchard MA. 2010. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol 27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan TG, Kuhnert D, Popinga A, Welch D, Drummond AJ. 2014. Efficient Bayesian inference under the structured coalescent. Bioinformatics 30:2272–2279. doi: 10.1093/bioinformatics/btu201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pybus OG, Drummond AJ, Nakano T, Robertson BH, Rambaut A. 2003. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol Biol Evol 20:381–387. doi: 10.1093/molbev/msg043. [DOI] [PubMed] [Google Scholar]
- 26.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, Suchard MA. 2014. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog 10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, Rambaut A, Suchard MA, Holmes EC, Lemey P. 2015. Global migration of influenza A viruses in swine. Nat Commun 6:6696. doi: 10.1038/ncomms7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Librelotto CS, Gräf T, Simon D, Almeida SEM, Lunge VR. 2015. HIV-1 epidemiology and circulating subtypes in the countryside of South Brazil. Rev Soc Bras Med Trop 48:249–257. doi: 10.1590/0037-8682-0083-2015. [DOI] [PubMed] [Google Scholar]
- 30.de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, Snoeck J, van Rensburg EJ, Wensing AMJ, van de Vijver DA, Boucher CA, Camacho R, Vandamme A-M. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 31.KosakovskyPond SL, Posada D, Stawiski E, Chappey C, Poon AFY, Hughes G, Fearnhill E, Gravenor MB, Brown AJL, Frost SDW. 2009. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siepel AC, Halpern AL, Macken C, Korber BTM. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses 11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 33.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, Huelsenbeck JP, Ronquist F, Swofford DL, Cummings MP, Rambaut A, Suchard MA. 2012. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol 61:170–173. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro B, Ho SYW, Drummond AJ, Suchard MA, Pybus OG, Rambaut A. 2011. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol Biol Evol 28:879–887. doi: 10.1093/molbev/msq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol 8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielejec F, Rambaut A, Suchard MA, Lemey P. 2011. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brien JD, Minin VN, Suchard MA. 2009. Learning to count: robust estimates for labeled distances between molecular sequences. Mol Biol Evol 26:801–814. doi: 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Passaes CPB, Bello G, Lorete RS, Matos Almeida SE, Junqueira DM, Veloso VG, Morgado MG, Guimarães ML. 2009. Genetic characterization of HIV-1 BC recombinants and evolutionary history of the CRF31_BC in Southern Brazil. Infect Genet Evol 9:474–482. doi: 10.1016/j.meegid.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Almeida SEM, de Medeiros RM, Junqueira DM, Gräf T, Passaes CPB, Bello G, Morgado MG, Guimarães LM. 2012. Temporal dynamics of HIV-1 circulating subtypes in distinct exposure categories in southern Brazil. Virol J 9:306. doi: 10.1186/1743-422X-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raboni SM, de Matos Almeida SM, Rotta I, Elisa C, Ribeiro L, Rosario D, Vidal LR, Nogueira MB, Riedel M, Winhescki G, Ferreira KA, Ellis R. 2010. Molecular epidemiology of HIV-1 clades in Southern Brazil. Mem Inst Oswaldo Cruz 105:1044–1049. doi: 10.1590/S0074-02762010000800015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gräf T, Passaes CPB, Ferreira LGE, Grisard EC, Morgado MG, Bello G, Pinto AR. 2011. HIV-1 genetic diversity and drug resistance among treatment naïve patients from Southern Brazil: an association of HIV-1 subtypes with exposure categories. J Clin Virol 51:186–191. doi: 10.1016/j.jcv.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 48.Silveira J, Santos AF, Martínez AMB, Góes LR, Mendoza-Sassi R, Muniz CP, Tupinambás U, Soares MA, Greco DB. 2012. Heterosexual transmission of human immunodeficiency virus type 1 subtype C in southern Brazil. J Clin Virol 54:36–41. doi: 10.1016/j.jcv.2012.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.